Phenotypic and Genotypic Characterization of Colistin, ESBL, and Multidrug Resistance in Escherichia coli Across the Broiler Production Chain in Karnataka, India
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Location and Sampling Sources
2.2. Isolation and Identification of E. coli
2.3. Phenotypic Characterization of Antimicrobial Resistance in E. coli Isolates
2.4. Determination of Minimum Inhibitory Concentration (MIC) of Colistin in E. coli Isolates
2.5. Phenotypic Confirmation of Extended-Spectrum Beta-Lactamase (ESBL) in E. coli Isolates Using Double-Disk Diffusion Test
2.6. Extraction of DNA from E. coli Isolates
2.7. Determination of AMR Genes in E. coli Isolates
2.8. Statistical Analysis
3. Results
3.1. Phenotypic Antimicrobial Resistance
3.1.1. Colistin Resistance
3.1.2. Extended-Spectrum Beta-Lactamase (ESBL)-Producing E. coli
3.1.3. Clustering Dendrogram of E. coli Isolates
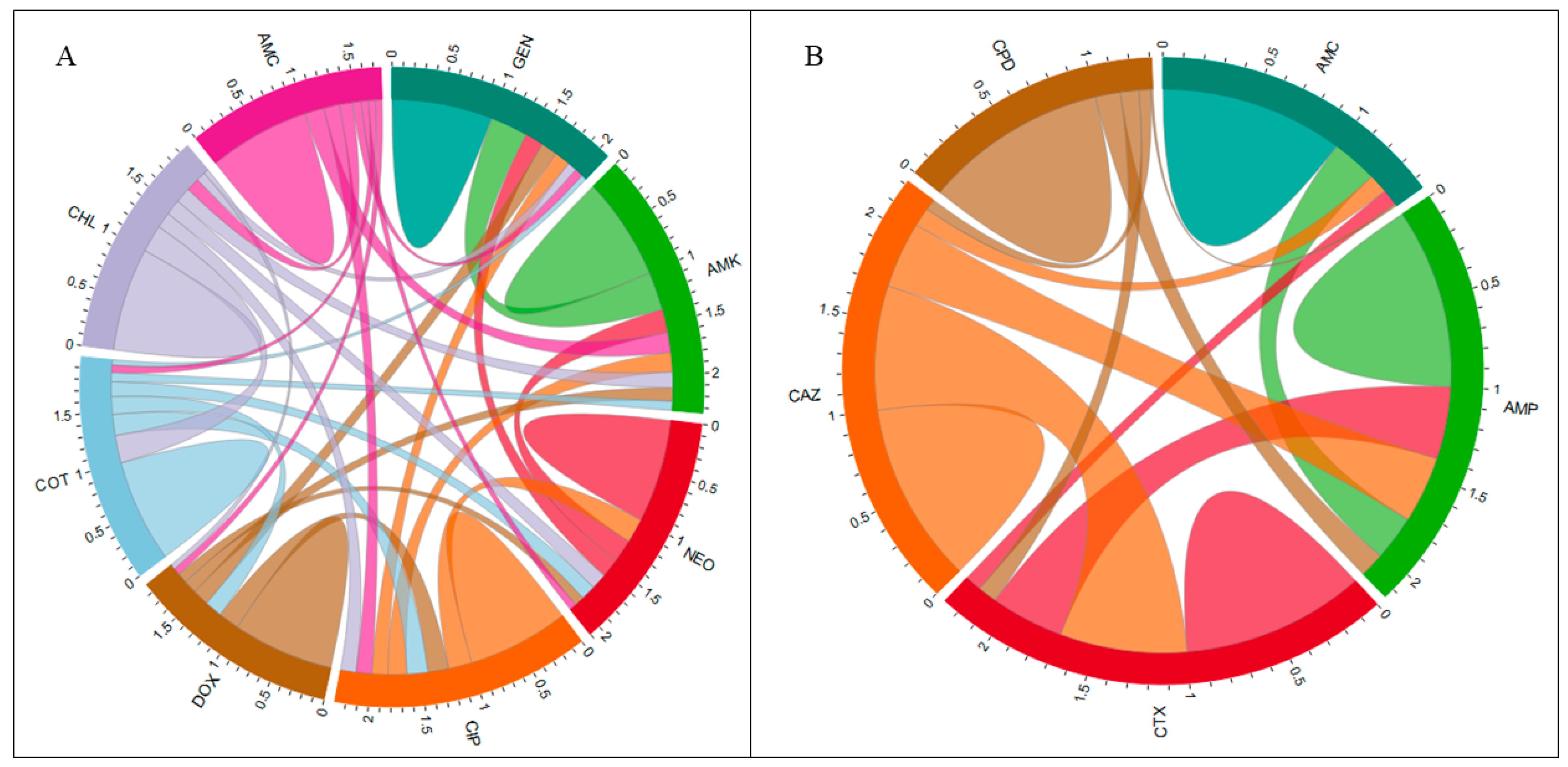
3.1.4. Assessing Pairwise and Total Correlations Among AMR of E. coli Isolates
3.1.5. Regression Analysis
3.2. Prevalence of ARGs in E. coli Isolates in the Broiler Production Chain
3.3. Prevalence of ARGs in E. coli Isolates in the Broiler Crop Cycle
Antimicrobial Resistance Genes (ARGs) in E. coli Isolated from Different Sample Types
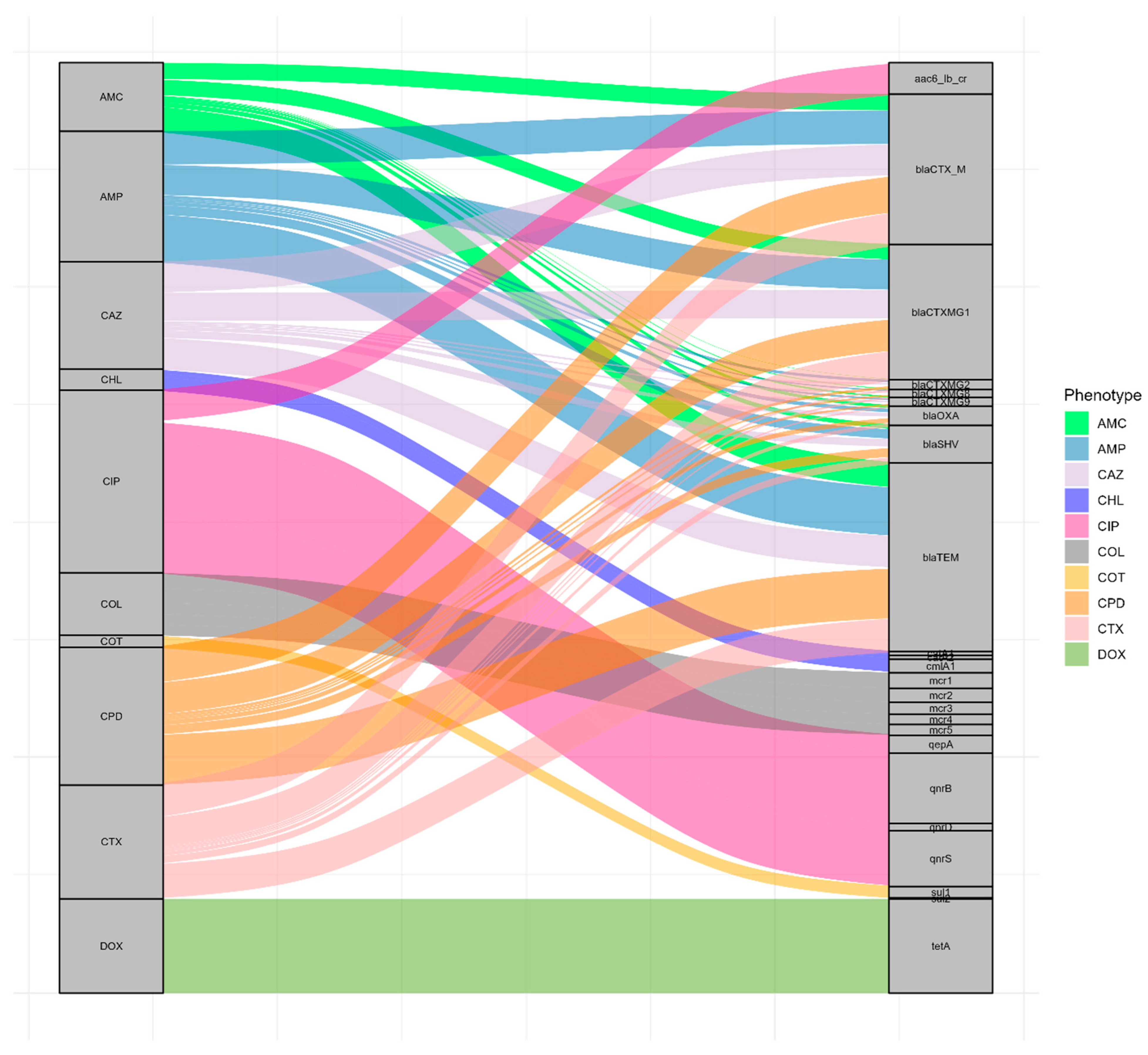
3.4. Correlation Between AMR Phenotype and Genotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gulati, A.; Juneja, R. Poultry Revolution in India: Lessons for Smallholder Production Systems; Center for Development Research: Bonn, Germany, 2023. [Google Scholar]
- Solano-Blanco, A.L.; González, J.E.; Medaglia, A.L. Production Planning Decisions in the Broiler Chicken Supply Chain with Growth Uncertainty. Oper. Res. Perspect. 2023, 10, 100273. [Google Scholar] [CrossRef]
- Femin, S.; Akash, R.; Sachin, C.; Sumayya, S.R.; Aarathi, S.; Deepa, P.M. Evaluation of Factors Associated with Biosecurity, Antimicrobial Use and Mortality in Broiler Farms in Northern Kerala. J. Vet. Anim. Sci. 2023, 54, 583–588. [Google Scholar] [CrossRef]
- Sharma, G.; Dey, T.K.; Hazarika, R.A.; Shome, B.R.; Shome, R.; Singh, V.P.; Deka, R.P.; Grace, D.; Lindahl, J.F. Knowledge and Practices Related to Antibiotics among Poultry Producers and Veterinarians in Two Indian States. One Health 2024, 18, 100700. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; de Lurdes Enes Dapkevicius, M.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.M.; Barbieri, N.L.; Oliveira, A.L.; de Willis, D.; Nolan, L.K.; Logue, C.M. Characterizing Avian Pathogenic Escherichia coli (APEC) from Colibacillosis Cases, 2018. PeerJ 2021, 9, e11025. [Google Scholar] [CrossRef]
- Joseph, J.; Zhang, L.; Adhikari, P.; Evans, J.D.; Ramachandran, R. Avian Pathogenic Escherichia coli (APEC) in Broiler Breeders: An Overview. Pathogens 2023, 12, 1280. [Google Scholar] [CrossRef] [PubMed]
- Varga, C.; Brash, M.L.; Slavic, D.; Boerlin, P.; Ouckama, R.; Weis, A.; Petrik, M.; Philippe, C.; Barham, M.; Guerin, M.T. Evaluating Virulence-Associated Genes and Antimicrobial Resistance of Avian Pathogenic Escherichia coli Isolates from Broiler and Broiler Breeder Chickens in Ontario, Canada. Avian Dis. 2018, 62, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.D.; Agunos, A.; Gow, S.P.; Deckert, A.E.; Varga, C. Associations between Antimicrobial Resistance in Fecal Escherichia coli Isolates and Antimicrobial Use in Canadian Turkey Flocks. Front. Microbiol. 2022, 13, 954123. [Google Scholar] [CrossRef]
- Anjum, M.F.; Schmitt, H.; Börjesson, S.; Berendonk, T.U.; Donner, E.; Stehling, E.G.; Boerlin, P.; Topp, E.; Jardine, C.; Li, X.; et al. The Potential of Using E. coli as an Indicator for the Surveillance of Antimicrobial Resistance (AMR) in the Environment. Curr. Opin. Microbiol. 2021, 64, 152–158. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Agrawal, I.; Yudhanto, S.; Varga, C. Longitudinal Analysis of Differences and Similarities in Antimicrobial Resistance among Commensal Escherichia coli Isolated from Market Swine and Sows at Slaughter in the United States of America, 2013–2019. Int. J. Food Microbiol. 2023, 407, 110388. [Google Scholar] [CrossRef]
- WHO List of Medically Important Antimicrobials (2024). Available online: https://cdn.who.int/media/docs/default-source/gcp/who-mia-list-2024-lv.pdf?utm_source=chatgpt.Com (accessed on 6 October 2025).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- World Health Organization (WHO) Critically Important Antimicrobials for Human Medicine: 6th Revision. 2019. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 22 July 2025).
- WHO Global Report on Surveillance 2014; WHO 2014 AMR Report; World Health Organization: Geneva, Switzerland, 2014; pp. 1–8.
- Bhargavi, D.; Sahu, R.; Nishanth, M.A.D.; Doijad, S.P.; Niveditha, P.; Kumar, O.R.V.; Sunanda, C.; Girish, P.S.; Naveena, B.M.; Vergis, J.; et al. Genetic Diversity and Risk Factor Analysis of Drug-Resistant Escherichia coli Recovered from Broiler Chicken Farms. Comp. Immunol. Microbiol. Infect. Dis. 2023, 93, 101929. [Google Scholar] [CrossRef] [PubMed]
- Mikhayel, M.; Leclercq, S.O.; Sarkis, D.K.; Doublet, B. Occurrence of the Colistin Resistance Gene Mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey. Microbiol. Spectr. 2021, 9, e00025-21. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Antimicrob. Resist. Bact. Livest. Companion Anim. 2018, 6, 289–316. [Google Scholar] [CrossRef]
- Dessie, H.K.; Bae, D.H.; Lee, Y.J. Characterization of Integrons and Their Cassettes in Escherichia coli and Salmonella Isolates from Poultry in Korea. Poult. Sci. 2013, 92, 3036–3043. [Google Scholar] [CrossRef]
- Wadaskar, B. Detection of Antimicrobial Resistance in Escherichia coli and Salmonella Isolated from Flies Trapped at Animal and Poultry Farm Premises; Maharashtra Animal and Fishery Sciences University: Nagpur, India, 2021; Volume 11. [Google Scholar]
- Kumar, H.; Chen, B.H.; Kuca, K.; Nepovimova, E.; Kaushal, A.; Nagraik, R.; Bhatia, S.K.; Dhanjal, D.S.; Kumar, V.; Kumar, V.; et al. Understanding of Colistin Usage in Food Animals and Available Detection Techniques: A Review. Animals 2020, 10, 1892. [Google Scholar] [CrossRef]
- Sharma, E.; Chen, Y.; Kelso, C.; Sivakumar, M.; Jiang, G. Navigating the Environmental Impacts and Analytical Methods of Last-Resort Antibiotics: Colistin and Carbapenems. Soil Environ. Health 2024, 2, 100058. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Wang, J.; Yassin, A.K.; Butaye, P.; Kelly, P.; Gong, J.; Guo, W.; Li, J.; Li, M.; et al. Molecular Detection of Colistin Resistance Genes (Mcr-1, Mcr-2 and Mcr-3) in Nasal/Oropharyngeal and Anal/Cloacal Swabs from Pigs and Poultry. Sci. Rep. 2018, 8, 3705. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.S.; Chen, Y.T.; Wang, Y.W.; Liu, Y.Y.; Kuo, H.C.; Tu, Y.H.; Lin, A.C.; Liao, Y.S.; Hong, Y.P. Dissemination of Mcr-1-Carrying Plasmids among Colistin-Resistant Salmonella Strains from Humans and Food-Producing Animals in Taiwan. Antimicrob. Agents Chemother. 2017, 61, e00338-17. [Google Scholar] [CrossRef]
- Ghafur, A.; Shankar, C.; GnanaSoundari, P.; Venkatesan, M.; Mani, D.; Thirunarayanan, M.A.; Veeraraghavan, B. Detection of Chromosomal and Plasmid-Mediated Mechanisms of Colistin Resistance in Escherichia coli and Klebsiella pneumoniae from Indian Food Samples. J. Glob. Antimicrob. Resist. 2019, 16, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K. Central Government Bans Use of Colistin in Livestock to Tackle Anti-Microbial Resistance–Food, Drugs, Healthcare, Life Sciences–India. Available online: https://www.mondaq.com/india/food-and-drugs-law/853840/central-government-bans-use-of-colistin-in-livestock-to-tackle-anti-microbial-resistance (accessed on 18 February 2022).
- Wadaskar, B.H. Detection of Colistin Resistance in Escherichia coli and Salmonella Species Isolated from Flies Trapped at Farm Premises of Food Animal and Poultry. Master’s Thesis, Maharashtra Animal and Fishery Sciences University, Nagpur, India, 2019. [Google Scholar]
- Das, A.K.; Anjaneyulu, A.S.R.; Verma, A.K.; Biswas, S. Scenario of Indian Livestock and Meat Marketing. Indian Food Ind. 2006, 25, 58. [Google Scholar]
- Mehta, R.; Nambiar, R.G. The Poultry Industry in India. In Proceedings of the FAO Conference on Poultry in the 21st Century, Bangkok, Thailand, 5–7 November 2007. [Google Scholar]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The Application of Antibiotics in Broiler Production and the Resulting Antibiotic Resistance in Escherichia coli: A Global Overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Griffiths, M.W. Cloning and Sequencing of the Gene Encoding Universal Stress Protein from Escherichia coli O157:H7 Isolated from Jack-in-a-Box Outbreak. Lett. Appl. Microbiol. 1999, 29, 103–107. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Table for Interpretation of MICs and Zone Diameters; The European Committee on Antimicrobial Susceptibility Testing: Copenhagen, Denmark, 2020. [Google Scholar]
- EFSA. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2019–2020. EFSA J. 2022, 20, e07209. [Google Scholar] [CrossRef] [PubMed]
- ISO 20776-2:2021 (En). Clinical Laboratory Testing and In Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 2: Evaluation of Performance of Antimicrobial Susceptibility Test Devices Against Reference Broth Micro-Dilution. Available online: https://www.iso.org/obp/ui/en/#iso:std:iso:20776:-2:ed-2:v1:en (accessed on 9 October 2025).
- Handayani, K.S.; Setiyono, A.; Lukman, D.W.; Pisestyani, H.; Rahayu, P. Distribution of Extended-Spectrum β-Lactamase Producing Escherichia coli Genes in an Integrated Poultry-Fish Farming System in Bogor, Indonesia. Vet. World 2024, 17, 1596–1602. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. CLSI Document VET01, 5th ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2018. [Google Scholar]
- Boyd, D.A.; Tyler, S.; Christianson, S.; McGeer, A.; Muller, M.P.; Willey, B.M.; Bryce, E.; Gardam, M.; Nordmann, P.; Mulvey, M.R. Complete Nucleotide Sequence of a 92-Kilobase Plasmid Harboring the CTX-M-15 Extended-Spectrum Beta-Lactamase Involved in an Outbreak in Long-Term-Care Facilities in Toronto, Canada. Antimicrob. Agents Chemother. 2004, 48, 3758–3764. [Google Scholar] [CrossRef]
- Buelow, E.; Bayjanov, J.R.; Majoor, E.; Willems, R.J.L.; Bonten, M.J.M.; Schmitt, H.; van Schaik, W. Limited Influence of Hospital Wastewater on the Microbiome and Resistome of Wastewater in a Community Sewerage System. FEMS Microbiol. Ecol. 2018, 94, fiy087. [Google Scholar] [CrossRef] [PubMed]
- Ciesielczuk, H.; Hornsey, M.; Choi, V.; Woodford, N.; Wareham, D.W. Development and Evaluation of a Multiplex PCR for Eight Plasmid-Mediated Quinolone-Resistance Determinants. J. Med. Microbiol. 2013, 62, 1823–1827. [Google Scholar] [CrossRef]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Jaja, I.F.; Oguttu, J.; Jaja, C.J.I.; Green, E. Prevalence and Distribution of Antimicrobial Resistance Determinants of Escherichia coli Isolates Obtained from Meat in South Africa. PLoS ONE 2020, 15, e0216914. [Google Scholar] [CrossRef]
- Lescat, M.; Poirel, L.; Nordmann, P. Rapid Multiplex Polymerase Chain Reaction for Detection of Mcr-1 to Mcr-5 Genes. Diagn. Microbiol. Infect. Dis. 2018, 92, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, N.; Beidokhti, H.; Jamehdar, A.S.; Ghahraman, M. Cpgenetic Properties of blaCTX-M and blaPER β-Lactamase Genes in Clinical Isolates of Enterobacteriaceae by Polymerase Chain Reaction. Iran. J. Basic Med. Sci. 2014, 17, 378–383. [Google Scholar]
- Tsukamoto, N.; Ohkoshi, Y.; Okubo, T.; Sato, T.; Kuwahara, O.; Fujii, N.; Tamura, Y.; Yokota, S.I. High Prevalence of Cross-Resistance to Aminoglycosides in Fluoroquinolone-Resistant Escherichia coli Clinical Isolates. Chemotherapy 2014, 59, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Mezhoud, H.; Chantziaras, I.; Iguer-Ouada, M.; Moula, N.; Garmyn, A.; Martel, A.; Touati, A.; Smet, A.; Haesebrouck, F.; Boyen, F. Presence of Antimicrobial Resistance in Coliform Bacteria from Hatching Broiler Eggs with Emphasis on ESBL/AmpC-Producing Bacteria. Avian Pathol. 2016, 45, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Patel, A.C.; Mohapatra, S.K.; Chauhan, H.C.; Sharma, K.K.; Shrimali, M.D.; Raval, S.H.; Prajapati, B.I. Antibiotic Resistance and Virulence Gene Patterns Associated with Multi Drug Resistant Avian Pathogenic Escherichia coli (APEC) Isolated from Broiler Chickens in India. Indian J. Microbiol. 2024, 64, 917–926. [Google Scholar] [CrossRef]
- Sarker, M.S.; Mannan, M.S.; Ali, M.Y.; Bayzid, M.; Ahad, A.; Bupasha, Z.B. Antibiotic Resistance of Escherichia coli Isolated from Broilers Sold at Live Bird Markets in Chattogram, Bangladesh. J. Adv. Vet. Anim. Res. 2019, 6, 272. [Google Scholar] [CrossRef]
- Bhuvaneswa, M.; Shanmughap, S.; Natarajase, K. Prevalence of Multidrug-Resistant (MDR) Salmonella enteritidis in Poultry and Backyard Chicken from Tiruchirappalli, India. Microbiol. J. 2015, 5, 28–35. [Google Scholar] [CrossRef]
- Khurana, A.; Sinha, R.; Nagaraju, M. Antibiotic Resistance in Poultry Environment. Spread of Resistance from Poultry Farm to Agricultural Field. Cent. Sci. Environ. 2017, 1–36. Available online: https://cdn.cseindia.org/userfiles/report-antibiotic-resistance-poultry-environment.pdf.
- Samanta, I.; Joardar, S.N.; Das, P.K.; Sar, T.K.; Bandyopadhyay, S.; Dutta, T.K.; Sarkar, U. Prevalence and Antibiotic Resistance Profiles of Salmonella Serotypes Isolated from Backyard Poultry Flocks in West Bengal, India. J. Appl. Poult. Res. 2014, 23, 536–545. [Google Scholar] [CrossRef]
- Petersen, A.; Christensen, J.P.; Kuhnert, P.; Bisgaard, M.; Olsen, J.E. Vertical Transmission of a Fluoroquinolone-Resistant Escherichia coli within an Integrated Broiler Operation. Vet. Microbiol. 2006, 116, 120–128. [Google Scholar] [CrossRef]
- Schwaiger, K.; Huther, S.; Hölzel, C.; Kämpf, P.; Bauer, J. Prevalence of Antibiotic-Resistant Enterobacteriaceae Isolated from Chicken and Pork Meat Purchased at the Slaughterhouse and at Retail in Bavaria, Germany. Int. J. Food Microbiol. 2012, 154, 206–211. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Antimicrobial Resistance in Bacteria of Animal Origin; Wiley: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Perreten, V. Resistance in the Food Chain and in Bacteria from Animals: Relevance to Human Infections. In Frontiers in Antimicrobial Resistance; American Society for Microbiology: Washington, DC, USA, 2014; pp. 446–464. [Google Scholar] [CrossRef]
- Chen, H.M.; Wang, Y.; Su, L.H.; Chiu, C.-H. Nontyphoid Salmonella Infection: Microbiology, Clinical Features, and Antimicrobial Therapy. Pediatr. Neonatol. 2013, 54, 147–152. [Google Scholar] [CrossRef]
- Vuthy, Y.; Lay, K.S.; Seiha, H.; Kerleguer, A.; Aidara-Kane, A. Antibiotic Susceptibility and Molecular Characterization of Resistance Genes among Escherichia coli and among Salmonella Subsp. in Chicken Food Chains. Asian Pac. J. Trop. Biomed. 2017, 7, 670–674. [Google Scholar] [CrossRef]
- Osman, A.Y.; Elmi, S.A.; Simons, D.; Elton, L.; Haider, N.; Khan, A.M.; Othman, I.; Zumla, A.; McCoy, D.; Kock, R. Antimicrobial Resistance Patterns and Risk Factors Associated with Salmonella Spp. Isolates from Poultry Farms in the East Coast of Peninsular Malaysia: A Cross-Sectional Study. Pathogens 2021, 10, 1160. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of Sulfonamide Resistance Genes (Sul1, Sul2, and Sul3) in Portuguese Salmonella enterica Strains and Relation with Integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef]
- Sin, M.; Yoon, S.; Kim, Y.B.; Noh, E.B.; Seo, K.W.; Lee, Y.J. Molecular Characteristics of Antimicrobial Resistance Determinants and Integrons in Salmonella Isolated from Chicken Meat in Korea. J. Appl. Poult. Res. 2020, 29, 502–514. [Google Scholar] [CrossRef]
- MoFL Ministry of Fisheries and Livestock. Fish Feed and Animal Feed Act, 2010; Bangladesh Parliament: Dhaka, Bangladesh, 2010. [Google Scholar]
- Acharya, K.P.; Wilson, R.T. Antimicrobial Resistance in Nepal. Front. Med. 2019, 6, 105. [Google Scholar] [CrossRef]
- Dawadi, P.; Bista, S.; Bista, S. Prevalence of Colistin-Resistant Escherichia coli from Poultry in South Asian Developing Countries. Vet. Med. Int. 2021, 2021, 6398838. [Google Scholar] [CrossRef]
- Xu, F.; Zeng, X.; Hinenoya, A.; Lin, J. MCR-1 Confers Cross-Resistance to Bacitracin, a Widely Used In-Feed Antibiotic. mSphere 2018, 3, e00411-18. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, M.; Huang, J.; Shah, J.M.; Ali, I.; Rahman, S.U.; Wang, L. Characterization and Resistant Determinants Linked to Mobile Elements of ESBL-Producing and Mcr-1-Positive Escherichia coli Recovered from the Chicken Origin. Microb. Pathog. 2021, 150, 104722. [Google Scholar] [CrossRef] [PubMed]
- Shang, K.; Wei, B.; Cha, S.Y.; Zhang, J.F.; Park, J.Y.; Lee, Y.J.; Jang, H.K.; Kang, M. The Occurrence of Antimicrobial-Resistant Salmonella enterica in Hatcheries and Dissemination in an Integrated Broiler Chicken Operation in Korea. Animals 2021, 11, 154. [Google Scholar] [CrossRef]
- Apostolakos, I.; Mughini-Gras, L.; Fasolato, L.; Piccirillo, A. Assessing the Occurrence and Transfer Dynamics of ESBL/pAmpC-Producing Escherichia coli across the Broiler Production Pyramid. PLoS ONE 2019, 14, e0217174. [Google Scholar] [CrossRef]
- Bista, S.; Shrestha, U.T.; Dhungel, B.; Koirala, P.; Gompo, T.R.; Shrestha, N.; Adhikari, N.; Joshi, D.R.; Banjara, M.R.; Adhikari, N.; et al. Detection of Plasmid-Mediated Colistin Resistant Mcr-1 Gene in Escherichia coli Isolated from Infected Chicken Livers in Nepal. Animals 2020, 10, 2060. [Google Scholar] [CrossRef] [PubMed]
- Dierikx, C.M.; van der Goot, J.A.; Smith, H.E.; Kant, A.; Mevius, D.J. Presence of ESBL/AmpC -Producing Escherichia coli in the Broiler Production Pyramid: A Descriptive Study. PLoS ONE 2013, 8, e79005. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.M.; Capik, S.F.; Giebel, S.; Nickodem, C.; Piñeiro, J.M.; Scott, H.M.; Vinasco, J.; Norman, K.N. Prevalence and Profiles of Antibiotic Resistance Genes Mph(A) and qnrB in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli Isolated from Dairy Calf Feces. Microorganisms 2022, 10, 411. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, P.; Umadevi, S.; Kumar, S.; Stephen, S. Determination of Correlation between Biofilm and Extended Spectrum β Lactamases Producers of Enterobacteriaceae. Sch. Res. J. 2012, 2, 2. [Google Scholar] [CrossRef]
- García-Tello, A.; Gimbernat, H.; Redondo, C.; Arana, D.M.; Cacho, J.; Angulo, J.C. Extended-Spectrum Beta-Lactamases in Urinary Tract Infections Caused by Enterobacteria: Understanding and Guidelines for Action. Actas Urológicas Españolas 2014, 38, 678–684. [Google Scholar] [CrossRef]



| No. | Types of Samples Collected | No. of Samples |
|---|---|---|
| Broiler breeder farms (BBFs) | 81 | |
| 1 | Water from the water tank of the farm | 9 |
| 2 | Water from nipples from 30 nipples pooled into one/shed) | 12 |
| 3 | Feed sample from feed bags (10 bags pooled into one sample/shed) | 12 |
| 4 | Feed sample from feeders (From 30 areas/feeders pooled/shed) | 12 |
| 5 | Cloacal swabs (30 birds/shed and pooled) | 12 |
| 6 | Egg surface (30 eggs/shed and pooled) | 12 |
| 7 | Swabs from Egg Yolk (30 eggs/shed pooled) | 8 |
| 8 | Environmental soil sample (Boot socks): one pair per farm | 4 |
| Hatcheries | 36 | |
| 1 | Swabs from egg setting room (10 swabs pooled to one per Hatchery) | 4 |
| 2 | Swabs from the Incubator (3 swabs from different areas of the incubator) | 4 |
| 3 | Swabs from air tunnels and fans of incubators | 4 |
| 4 | Swabs from hatchers | 3 |
| 5 | Swabs from Hatcher Egg Tray | 3 |
| 6 | Meconium | 3 |
| 7 | Yolk sac swabs of dead chicks (Ten dead chicks and samples were pooled into one/hatchery) | 3 |
| 8 | Hand swabs from hatchery workers | 5 |
| 9 | Boot socks from the hatchery floor | 7 |
| Commercial broiler farms, Day 1, Day 18, Day 36 | 160 | |
| 1 | Water from the water tank | 25 |
| 2 | Water from nipples/drinkers | 25 |
| 3 | Feed samples from feed bags | 25 |
| 4 | Feed feeders | 25 |
| 5 | Faecal swabs | 25 |
| 6 | Internal (inside the shed) environment samples using sterile boot socks | 25 |
| 7 | External (outside the shed) environment samples using sterile boot socks | 10 |
| Retail meat shops | 57 | |
| 1 | Swabs from the surface of the cutting/chopping board (100 cm2) | 6 |
| 2 | Swabs from the cutter/knife | 3 |
| 3 | Meat rinsing water | 3 |
| 4 | Chicken carcasses (5 Carcasses/shop). | 15 |
| 5 | Ileal contents from five carcasses | 15 |
| 6 | Caecal contents from five carcasses | 15 |
| Total number of samples | 332 | |
| Antimicrobial | Proportion of AMR in the Broiler Production Chain | Proportion of AMR in the Commercial Broiler Farm Crop Cycle | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Drug A | BBF (N = 38) B | Hatchery (N = 41) B | CBF (N = 117) B | RMS (N = 95) B | Total (N = 291) | 95% CI C | Day 1 (N = 36) B | Day 18 (N = 36) B | Day 36 (N = 45) B | Total (N = 117) B | 95% CI C |
| n (%) D | n (%) D | n (%) D | n (%) D | n (%) D | n (%) D | n (%) D | n (%) D | n (%) D | ||||
| Aminoglycosides | GEN | 21 (55.26) | 13 (31.71) | 70 (59.83) | 59 (62.11) | 163 (56.01) | 50.1–61.8 | 23 (63.89) | 18 (50.00) | 29 (64.44) | 70 (59.83) | 50.36–68.78 |
| AMK | 12 (31.58) | 10 (24.39) | 37 (31.62) | 42 (44.21) | 101 (34.71) | 29.25–34.71 | 10 (27.78) | 14 (38.89) | 13 (28.89) | 37 (31.62) | 23.34–40.87 | |
| NEO | 10 (26.31) | 6 (14.63) | 37 (31.62) | 13 (13.68) | 66 (22.68) | 18–27.93 | 14 (38.89) | 9 (25.00) | 13 (28.89) | 36 (30.77) | 22.57–39.97 | |
| Fluoroquinolones | CIP | 28 (73.68) | 29 (70.73) | 100 (85.47) | 74 (77.89) | 231 (79.73) | 74.64–84.19 | 32 (88.89) | 32 (88.89) | 36 (80.00) | 100 (85.47) | 77.76–91.3 |
| Tetracycline | DOX | 35 (92.11) | 41 (100.00) | 114 (97.44) | 92 (96.84) | 282 (96.91) | 94.21–98.58 | 36 (100.00) | 35 (97.22) | 43 (95.56) | 114 (97.44) | 92.69–99.47 |
| Folate pathway antagonists | COT | 17 (44.74) | 18 (43.9) | 57 (48.72) | 46 (48.42) | 138 (47.72) | 41.57–53.33 | 19 (52.78) | 15 (41.67) | 23 (51.11) | 57 (48.72) | 39.37–58.13 |
| Phenicols | CHL | 10 (26.32) | 8 (19.51) | 34 (29.06) | 15 (15.79) | 67 (23.02) | 18.31–28–29 | 10 (27.78) | 9 (25.00) | 15 (33.33) | 34 (29.06) | 21.04–38.17 |
| Polymyxins | COL | 3 (7.89) | 8 (19.51) | 31 (26.50) | 18 (18.95) | 60 (20.61) | 16.12–27.73 | 1(2.78) | 15 (41.67) | 15 (41.67) | 31 (26.5) | 18.77–35.45 |
| Penicillin | AMP | 20 (52.63) | 25 (60.98) | 88 (75.21) | 68 (71.58) | 201 (69.07) | 63.41–74.34 | 30 (83.33) | 28 (77.78) | 30 (66.67) | 88 (75.21) | 66.36–82.73 |
| β-Lactamase combination agents | AMC | 22 (57.89) | 19 (46.34) | 41 (35.04) | 36 (37.89) | 118 (40.55) | 34.86–46.44 | 9 (25.00) | 11 (30.56) | 20 (44.44) | 40 (34.19) | 25.67–43.53 |
| Third-generation cephalosporins (3CGR) | CTX | 19 (50.00) | 21 (51.22) | 63 (53.85) | 61 (64.21) | 164 (56.36) | 50.45–62.14 | 19 (52.78) | 16 (44.44) | 28 (62.22) | 63 (53.85) | 44.39–63.1 |
| CAZ | 15 (39.47) | 22 (53.66) | 57 (48.71) | 63 (66.32) | 157 (53.95) | 48.04–59.78 | 16 (44.44) | 11 (30.56) | 29 (64.44) | 56 (47.86) | 38.54–57.29 | |
| CPD | 33 (86.84) | 39 (95.12) | 99 (84.62) | 79 (83.16) | 250 (85.91) | 81.38–89.7 | 32 (88.89) | 30 (83.33) | 37 (82.22) | 99 (84.62) | 76.78–90.62 | |
| 3CGR average | - | 58.77 | (66.66) | (62.40) | (71.23) | (65.41) | 62.25–68.56 | (62.03) | (52.77) | (69.63) | (62.11) | 53.24–70.34 |
| ESBL E Producer | 17(44.74) | 18(43.90) | 40 (34.19) | 46(48.42) | 121 (41.58) | 35.86–47.48 | 14 (38.89) | 17 (47.22) | 17(37.78) | 48 (41.03) | 32.02–50.5 | |
| MDR F | 37 (97.37) | 40 (97.56) | 113 (96.58) | 92 (96.84) | 282 (96.84) | 94.21–98.58 | 35 (97.22) | 35 (97.22) | 43 (95.56) | 113 (97.44) | 91.48–99.06 | |
| Sampling Source | MDR-ESBL-Producer | Fluoroquinolones-ESBL-Producer | Colistin-ESBL-Producer | ||
|---|---|---|---|---|---|
| Proportion of AMR in the broiler production chain | BBFs (N A = 13) | n B (%) | 13 (100) | 7 (53.85) | 1 (7.69) |
| Hatchery (N = 11) | n (%) | 11 (100) | 7 (63.64) | 3 (27.27) | |
| CBFs (N = 49) | n (%) | 49 (100) | 46 (93.38) | 16 (32.65) | |
| RMSs (N = 48) | n (%) | 46 (95.85) | 37 (77.08) | 10 (20.83) | |
| Total (N = 121) | n (%) | 119 (98.35) | 97 (80.17) | 30 (24.79) | |
| 95% CI C | 64.16–99.8 | 71.94–86.86 | 17.4–33.46 | ||
| Proportion of AMR in the commercial broiler farm crop cycle | Day 1 (N = 14) | n (%) | 14 (100) | 14 (100) | 1 (7.14) |
| Day 18 (N = 12) | n (%) | 12 (100) | 12 (100) | 4 (33.33) | |
| Day 36 (N = 23) | n (%) | 23 (100) | 20 (86.96) | 11 (47.83) | |
| Total | n (%) | 49 (100) | 46 (93.38) | 16 (32.65) | |
| 95% CI C | 92.75–100 | 83.13–98.72 | 19.95–47.54 | ||
| Class of Antimicrobials | Target Genes A | Proportion of ARGs in the Broiler Production Chain | Proportion of ARGs in the Broiler Crop Cycle | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBF (N = 38)A | Hatchery (N = 41) A | CBF (N = 117) A | RMS (N = 95) A | Total (N = 291) A | 95% CI C | Day 1 (N = 36) A | Day 18 (N = 36) A | Day 36 (N = 45) A | Total (N = 117) A | 95% CI C | ||
| n (%) B | n (%) B | n (%) B | n (%) B | n (%) B | n (%) B | n (%) B | n (%) B | n (%) B | ||||
| β Lactams | blaTEM | 5 (13.16) | 20 (48.78) | 59 (50.43) | 36 (37.89) | 120 (41.24) | 35.52–47.13 | 22 (61.11) | 21 (58.33) | 16 (55.56) | 59 (50.43) | 41.03–59.8 |
| blaSHV | 0(0) | 0(0) | 8 (6.84) | 13 (13.68) | 21 (7.22) | 4.52–10.82 | 1 (2.78) | 0 (0) | 7 (15.56) | 8 (6.84) | 3–13.03 | |
| blaOXA | 0(0) | 1 (2.44) | 4 (3.42) | 5 (5.26) | 10 (3.44) | 1.66–6.23 | 1 (2.78) | 1 (2.78) | 2 (4.44) | 4 (3.42) | 0.94–8.52 | |
| blaCTX-M | 5 (13.16) | 13 (31.71) | 32 (27.08) | 32 (32.63) | 82 (28.18) | 23.08–33.72 | 10 (27.8) | 8 (22.22) | 14 (31.11) | 32 (27.35) | 19.52–36.36 | |
| blaCTX-M Group 1 | 5 (13.16) | 9 (21.95) | 27 (23.08) | 31 (32.63) | 72 (24.74) | 19.89–30.11 | 7 (19.44) | 8 (22.22) | 12 (26.67) | 27 (23.08) | 15.79–31.77 | |
| blaCTX-M Group 2 | 0(0) | 2 (4.88) | 2 (1.71) | 2 (2.11) | 6 (2.06) | 0.76–4.43 | 1 (2.78) | 0 (0) | 1 (22.22) | 2 (1.71) | 0.21–6.04 | |
| blaCTX-M Group 9 | 0(0) | 2 (4.88) | 1 (0.85) | 1 (1.05) | 4 (1.37) | 0.38–3.48 | 0 (0) | 0 (0) | 1 (22.22) | 1 (0.85) | 0.02–4.67 | |
| blaCTX-M Group 8/25 | 0(0) | 2 (4.88) | 1 (0.85) | 1 (1.05) | 4 (1.37) | 0.38–3.48 | 1 (2.78) | 0 (0) | 0 (00) | 1 (0.85) | 0.02–4.67 | |
| Quinolone | qnrB | 12 (31.58) | 22 (53.66) | 71 (60.68) | 83 (87.37) | 188 (64.6) | 58.81–70.1 | 13 (36.11) | 28 (77.78) | 30 (66.67) | 71 (60.68) | 51.27–69.59 |
| qepA | 9 (23.68) | 15 (36.59) | 25 (21.37) | 7 (7.37) | 56 (19.24) | 14.88–24.25 | 14 (38.89) | 4 (11.11) | 7 (15.56) | 25 (21.37) | 14.33–29.91 | |
| qnrD | 0(0) | 0(0) | 14 (11.97) | 4 (4.21) | 18 (6.19) | 3.71–9.6 | 2 (5.55) | 1 (2.780 | 11 (24.44) | 14 (11.97) | 6.7–19.26 | |
| qnrS | 12 (31.58) | 16 (39.02) | 74 (63.25) | 52 (54.74) | 154 (52.92) | 47.01–58.77 | 16 (44.44) | 22 (61.11) | 36 (80.00) | 74 (63.25) | 53.84–71.97 | |
| aac(6′)-Ib-cr | 11 (28.95) | 8 (19.51) | 35 (29.91) | 35 (36.84) | 89 (30.58) | 25.34–36.23 | 9 (25) | 6 (16.67) | 20 (44.44) | 35 (29.91) | 21.8–39.07 | |
| Polymyxins | mcr-1 | 0(0) | 15 (36.59) | 42 (35.9) | 25 (26.32) | 82 (28.18) | 23.08–33.72 | 5 (13.89) | 21 (58.33) | 16 (35.56) | 42 (35.9) | 27.24–45.29 |
| mcr-2 | 3 (7.89) | 3 (7.32) | 29 (24.79) | 30 (31.58) | 65 (22.34) | 17.68–27.56 | 9 (25) | 9 (25) | 11 (24.44) | 29 (24.79) | 17.27–33.62 | |
| mcr-3 | 2 (5.26) | 4 (9.76) | 25 (21.37) | 33 (34.74) | 64 (21.99) | 17.37–27.2 | 9 (25) | 9 (25) | 7 (15.56) | 25 (21.37) | 14.33–29.91 | |
| mcr-4 | 1 (2.63) | 0(0) | 24 (20.51) | 28 (29.47) | 53 (18.21) | 13.95–23.14 | 9 (25) | 9 (25) | 6 (13.33) | 24 (20.51) | 13.61–28.97 | |
| mcr-5 | 0 (0) | 3 (7.32) | 27 (23.08) | 27 (28.42) | 57 (19.59) | 15.19–24.62 | 8 (22.22) | 12 (33.33) | 7 (15.56) | 27 (23.08) | 15.79–31.77 | |
| Tetracycline | tetA | 10 (26.32) | 32 (78.05) | 88 (75.21) | 72 (75.79) | 202 (69.42) | 63.77–74.66 | 23 (63.89) | 28 (77.7) | 37 (82.22) | 88 (75.21) | 66.38–82.73 |
| Phenicol | catA1 | 0 (0) | 8 (19.51) | 24 (20.51) | 6 (6.32) | 38 (13.06) | 9.41–17.48 | 2 (5.55) | 7 (9.44) | 15 (33.33) | 24 (20.51) | 13.61–28.97 |
| catA2 | 0 (0) | 7 (17.07) | 11 (9.4) | 3 (3.16) | 21 (7.22) | 4.52–10.82 | 2 (5.55) | 5 (13.89) | 4 (8.89) | 11 (9.4) | 4.79–16.2 | |
| cmlA1 | 3 (7.89) | 12 (29.27) | 20 (17.09) | 12 (12.63) | 47 (16.15) | 12.12–20.89 | 10 (27.78) | 5 (13.89) | 5 (11.11) | 20 (17.09) | 10.77–25.16 | |
| Folate pathway antagonists | sul1 | 2 (5.26) | 4 (9.76) | 31 (26.5) | 9 (9.47) | 46 (15.81) | 11.81–20.52 | 5 (13.89) | 8 (22.22) | 18 (40.00) | 31 (26.5) | 18.77–35.45 |
| sul2 | 1 (2.63) | 2 (4.88) | 7 (5.98) | 0(0) | 10 (3.44) | 1.66–6.23 | 0 (0) | 2 (5.55) | 5 (11.11) | 7 (5.98) | 2.44–11.94 | |
| Total ARGs D | 14/35 (40.00) | 21/35 (60.00) | 24/35 (68.57) | 23/35 (65.71) | 24/35 (68.57) | 50.71–83.15 | 22/35 (62.86) | 20/35 (57.14) | 23/35 (65.71) | 24/35 (68.57) | 50.71–83.15 | |
| No. | Type of Samples | AMR Genes Detected (%) |
|---|---|---|
| BBFs | ||
| 1 | External environment | qnrB, aac(6′)-Ib-cr, qnrS, and tetA (33) |
| 2 | Cloacal swabs | mcr-2 & cmlA1 (29), qnrB, aac(6′)-Ib-cr, qnrS & (14), |
| 3 | Egg yolk | blaTEM, qnrB & tetA (14). |
| 4 | Feed from feed bags | qnrS and aac(6′)-Ib-cr (14). |
| 5 | Feed from feeders | qnrB, qepA, aac(6′)-Ib-cr, qnrS, tetA, cmlA1 and sul1 (14) |
| Hatcheries | ||
| 1 | Hatcheries internal environments | tetA (100), blaTEM, qnrS, qepA (67), qnrB, aac(6′)-Ib-cr, mcr-2, and cmlA1 (33), |
| 2 | Chick trays | tetA (100), qepA, qnrS, aac(6′)-Ib-cr (66.67), blaTEM, blaCTX-M, blaCTX-M group1, qnrB, mcr-1, cmlA2 and sul2 (33) |
| 3 | Hatchers samples | tetA (100), qnrB and sul1 (67), blaTEM, blaCTX-M, qnrS, aac(6′)-Ib-cr, mcr-1, mcr-2 and cmlA1(33) |
| 4 | Incubator air tunnels | tetA, qnrB and qnrS (67), blaTEM, blaCTX-M, group, qepA, aac(6′)-Ib-cr and cmlA1 (33) |
| 5 | Incubators | blaTEM, qnrB, tetA, and mcr-1 (33). |
| 6 | Meconium dropping | tetA (100), qnrB & qepA (67) and blaTEM, qnrS, cmlA1 & mcr-1 (33) |
| 7 | Eggs setting room | mcr-5 & catA1 1/3(33) |
| 8 | In the workers’ hand swabs | qnrS and tetA (60), qnrB and qepA mcr-3 catA1, cmlA1 (40), blaTEM, aac(6′)-Ib-cr and mcr-1 (20). |
| CBFs | ||
| 1 | Farm internal environment samples (Boot socks inside the farm) | tetA (80), qnrS (67), qnrB (60), mcr-1 (47), blaTEM, qepA, mcr-2,(27), catA1 (33), aac(6′)-Ib-cr, mcr2-5, and, sul1 (20), catA2, cmlA1 (13) |
| 2 | farm external environment (Boot socks outside the farm) | tetA (67), blaTEM, qnrS (56), aac(6′)-Ib-cr 4/9(44), qepA, mcr-2-5 and catA1 (33), qnrD and, cmlA1 (22), mcr-1, sul2 (11) |
| 3 | Fecal swabs | tetA (87), qnrS (73), blaTEM (60), qnrB (53), aac(6′)-Ib-cr (47), sul1, mcr-1 mcr-2, and mcr-5 and cmalA1 (27), mcr-3 and mcr-4 (20), sul2 and blaCTX-M (13), blaSHV, bla group 1, qepA and catA1 (7) |
| 4 | Feed samples from feeders | tetA (80), qnrB & qnrS (67), blaTEM (53), catA1 6/15 (40), mcr-1 (33), sul1 and aac(6′)-Ib-cr (27), qepA, mcr-2 mcr-3, mcr-4 and mcr-5 and cmalA1 (20) and blaCTX-M (Universal & bla group (7) |
| 5 | Feed from feed bags | blaCTX-M, cmlA1, mcr-2 and mcr-3 and mcr-4 (7), blaTEM, mcr-5, sul1 and sul2 (13), qnrS, qepA mcr-1 and tetA (27) |
| 6 | Water nipple samples | qnrS (47) tetA (40), blaTEM (13), mcr-2 (33), qnrB, mcr-3-5 (20), blaSHV, blaCTX-M and bal group1, catA2, sul1 and aac(6′)-Ib-cr \(13) and blaOXA (7) |
| RMSs | ||
| 1 | Meat rinsing water | qnrB, qnrS, mcr-3, catA1 and tetA (67), blaTEM, aac(6′)-Ib-cr, catA1, mcr-2 mcr-4 and mcr-5 (33) |
| 2 | Ileal contents | tetA (100), qnrB (93%), qnrS (73), & blaTEM (40), mcr2-4 (33), mcr-5 (27), mcr-1 (20), qepA, catA1, cmlA1 (13), blaCTX-M blaSHV, bla group 1 and sul1 1/15 (67) |
| 3 | Caecal contents | tetA (100), qnrB (93.33), mcr-1 (53), cmlA1 (47), blaTEM, aac(6′)-Ib-cr & qnrS (33), mcr2-4 (27), mcr-5 (20), blaCTX-M, blaSHV, qepA and sul1 (13), blaOXA, catA1 and blaTEM group1 (7) |
| 4 | chicken carcass | qnrD (80%), tetA (73), qnrS (67), blaTEM (47), aac(6′)-Ib-cr 5/20(33), blaSHV, mcr1-4 (27), mcr-5, cmlA1 and sul2(20), qepA and catA1 (13), blaCTX-M, group1, catA2 group8/25 (7) |
| 5 | Cutting knife | qnrB & tetA 2/3 (67), blaTEM, qepA, qnrS, mcr-3, 1/mcr-5 (33), |
| 6 | Cutting board | blaCTX-M & bla group1, bla group2, qnrB, qnrS, aac(6′)-Ib-cr, mcr-2 and mcr-3 (33) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohail, M.N.; Isloor, S.; Rathnamma, D.; Chandra Priya, S.; Veeregowda, B.M.; Hegde, N.R.; Varga, C.; Williams, N.J. Phenotypic and Genotypic Characterization of Colistin, ESBL, and Multidrug Resistance in Escherichia coli Across the Broiler Production Chain in Karnataka, India. Poultry 2025, 4, 51. https://doi.org/10.3390/poultry4040051
Sohail MN, Isloor S, Rathnamma D, Chandra Priya S, Veeregowda BM, Hegde NR, Varga C, Williams NJ. Phenotypic and Genotypic Characterization of Colistin, ESBL, and Multidrug Resistance in Escherichia coli Across the Broiler Production Chain in Karnataka, India. Poultry. 2025; 4(4):51. https://doi.org/10.3390/poultry4040051
Chicago/Turabian StyleSohail, Mohammad Nasim, Srikrishna Isloor, Doddamane Rathnamma, S. Chandra Priya, Belamaranahally M. Veeregowda, Nagendra R. Hegde, Csaba Varga, and Nicola J. Williams. 2025. "Phenotypic and Genotypic Characterization of Colistin, ESBL, and Multidrug Resistance in Escherichia coli Across the Broiler Production Chain in Karnataka, India" Poultry 4, no. 4: 51. https://doi.org/10.3390/poultry4040051
APA StyleSohail, M. N., Isloor, S., Rathnamma, D., Chandra Priya, S., Veeregowda, B. M., Hegde, N. R., Varga, C., & Williams, N. J. (2025). Phenotypic and Genotypic Characterization of Colistin, ESBL, and Multidrug Resistance in Escherichia coli Across the Broiler Production Chain in Karnataka, India. Poultry, 4(4), 51. https://doi.org/10.3390/poultry4040051








