AG1, A Novel Synbiotic, Demonstrates the Capability to Enhance Fermentation Using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Products
2.2. Test Gastrointestinal Tract System
2.3. Gastric Phase and Small Intestine Phase
2.4. Short-Term Colonic Batch Simulations
2.5. Statistics
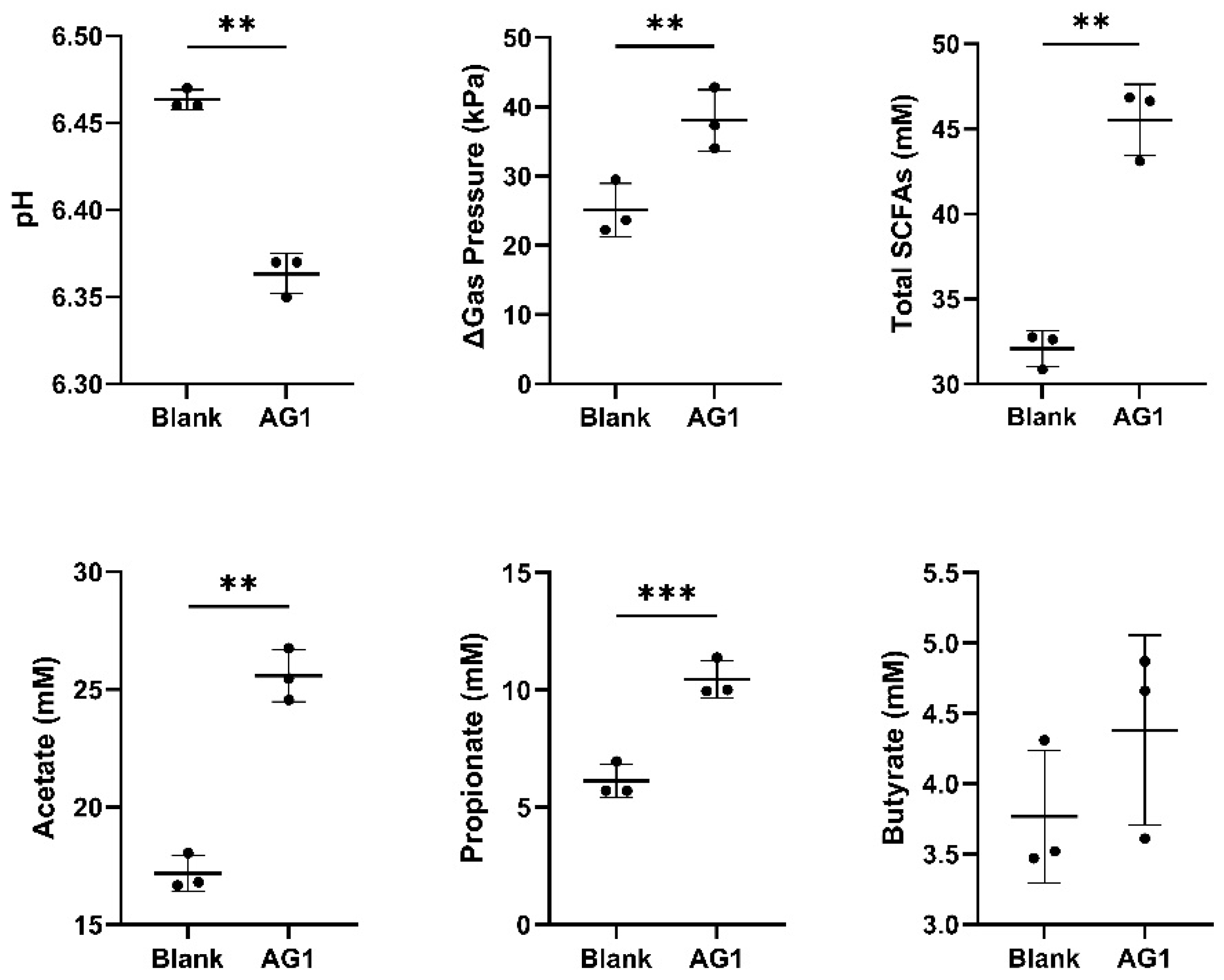
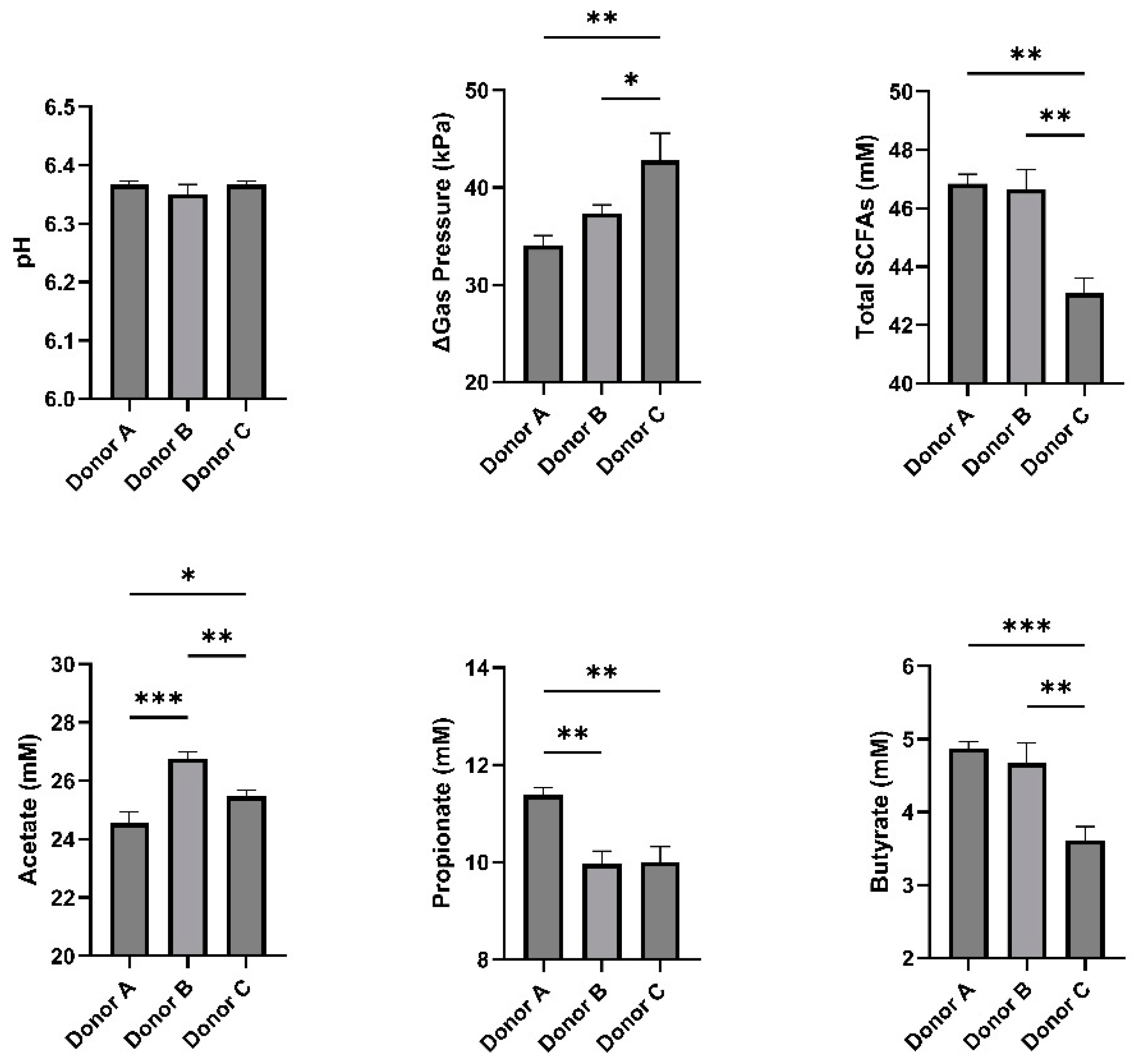
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cani, P.D.; de Hase, E.M.; Van Hul, M. Gut Microbiota and Host Metabolism: From Proof of Concept to Therapeutic Intervention. Microorganisms 2021, 9, 1302. [Google Scholar] [CrossRef]
- Pham, V.T.; Steinert, R.E.; Duysburgh, C.; Ghyselinck, J.; Marzorati, M.; Dekker, P.J.T. In Vitro Effect of Enzymes and Human Milk Oligosaccharides on FODMAP Digestion and Fecal Microbiota Composition. Nutrients 2023, 15, 1637. [Google Scholar] [CrossRef] [PubMed]
- Wiese, M.; Bashmakov, Y.; Chalyk, N.; Nielsen, D.S.; Krych, Ł.; Kot, W.; Klochkov, V.; Pristensky, D.; Bandaletova, T.; Chernyshova, M.; et al. Prebiotic Effect of Lycopene and Dark Chocolate on Gut Microbiome with Systemic Changes in Liver Metabolism, Skeletal Muscles and Skin in Moderately Obese Persons. Biomed. Res. Int. 2019, 2019, 4625279. [Google Scholar] [CrossRef] [PubMed]
- Marttinen, M.; Ala-Jaakkola, R.; Laitila, A.; Lehtinen, M.J. Gut Microbiota, Probiotics and Physical Performance in Athletes and Physically Active Individuals. Nutrients 2020, 12, 2936. [Google Scholar] [CrossRef] [PubMed]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Gadir, A.A.; Dhir, R. Probiotics: Reiterating What They Are and What They Are Not. Front. Microbiol. 2019, 10, 424. [Google Scholar] [CrossRef]
- Kumar Yadav, M.; Kumari, I.; Singh, B.; Kant Sharma, K.; Kumar Tiwari, S. Probiotics, Prebiotics and Synbiotics: Safe Options for next-Generation Therapeutics. Appl. Microbiol. Biotechnol. 2022, 106, 505–521. [Google Scholar] [CrossRef]
- Vyas, U.; Ranganathan, N. Probiotics, Prebiotics, and Synbiotics: Gut and Beyond. Gastroenterol. Res. Pract. 2012, 2012, 872716. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Roy, C.I.L. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Applied AFwrobiology Biotechnology Development of a S-Step Multi-Chamber Reactor as a Simulation of the Human Intestinal Microbial Ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- van de Wiele, T.; van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer International Publishing: Cham, Switzerland, 2015; pp. 305–317. ISBN 9783319161044. [Google Scholar]
- Carrera-Quintanar, L.; Roa, R.I.L.; Quintero-Fabián, S.; Sánchez-Sánchez, M.A.; Vizmanos, B.; Ortuño-Sahagún, D. Phytochemicals That Influence Gut Microbiota as Prophylactics and for the Treatment of Obesity and Inflammatory Diseases. Mediat. Inflamm. 2018, 2018, 9734845. [Google Scholar] [CrossRef] [PubMed]
- Dingeo, G.; Brito, A.; Samouda, H.; Iddir, M.; La Frano, M.R.; Bohn, T. Phytochemicals as Modifiers of Gut Microbial Communities. Food Funct. 2020, 11, 8444–8471. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Hoyles, L. Human Microbiome Myths and Misconceptions. Nat. Microbiol. 2023, 8, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Kriss, M.; Hazleton, K.Z.; Nusbacher, N.M.; Martin, C.G.; Lozupone, C.A. Low Diversity Gut Microbiota Dysbiosis: Drivers, Functional Implications and Recovery. Curr. Opin. Microbiol. 2018, 44, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Méric, G.; Wick, R.R.; Watts, S.C.; Holt, K.E.; Inouye, M. Correcting Index Databases Improves Metagenomic Studies. BioRxiv 2019, 712166. [Google Scholar] [CrossRef]
- Reichardt, N.; Vollmer, M.; Holtrop, G.; Farquharson, F.M.; Wefers, D.; Bunzel, M.; Duncan, S.H.; Drew, J.E.; Williams, L.M.; Milligan, G.; et al. Specific Substrate-Driven Changes in Human Faecal Microbiota Composition Contrast with Functional Redundancy in Short-Chain Fatty Acid Production. ISME J. 2018, 12, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Daniel, S.L.; Moradi, L.; Paiste, H.; Wood, K.D.; Assimos, D.G.; Holmes, R.P.; Nazzal, L.; Hatch, M.; Knight, J. Forty Years of Oxalobacter Formigenes, a Gutsy OxalateDegrading Specialist. Appl. Environ. Microbiol. 2021, 87, e00544-21. [Google Scholar] [CrossRef]
- Dronkers, T.M.G.; Ouwehand, A.C.; Rijkers, G.T. Global Analysis of Clinical Trials with Probiotics. Heliyon 2020, 6, e04467. [Google Scholar] [CrossRef] [PubMed]
- Rudrapal, M.; Khairnar, S.J.; Khan, J.; Dukhyil, A.B.; Ansari, M.A.; Alomary, M.N.; Alshabrmi, F.M.; Palai, S.; Deb, P.K.; Devi, R. Dietary Polyphenols and Their Role in Oxidative Stress-Induced Human Diseases: Insights Into Protective Effects, Antioxidant Potentials and Mechanism(s) of Action. Front. Pharmacol. 2022, 13, 283. [Google Scholar] [CrossRef]
- Makarewicz, M.; Drożdż, I.; Tarko, T.; Duda-Chodak, A. The Interactions between Polyphenols and Microorganisms, Especially Gut Microbiota. Antioxidants 2021, 10, 188. [Google Scholar] [CrossRef]
- Kan, J.; Wu, F.; Wang, F.; Zheng, J.; Cheng, J.; Li, Y.; Yang, Y.; Du, J. Phytonutrients: Sources, Bioavailability, Interaction with Gut Microbiota, and Their Impacts on Human Health. Front. Nutr. 2022, 9, 960309. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirby, T.O.; Townsend, J.R.; Sapp, P.A.; Govaert, M.; Duysburgh, C.; Marzorati, M.; Marshall, T.M.; Esposito, R. AG1, A Novel Synbiotic, Demonstrates the Capability to Enhance Fermentation Using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). Biol. Life Sci. Forum 2023, 29, 10. https://doi.org/10.3390/IECN2023-15793
Kirby TO, Townsend JR, Sapp PA, Govaert M, Duysburgh C, Marzorati M, Marshall TM, Esposito R. AG1, A Novel Synbiotic, Demonstrates the Capability to Enhance Fermentation Using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). Biology and Life Sciences Forum. 2023; 29(1):10. https://doi.org/10.3390/IECN2023-15793
Chicago/Turabian StyleKirby, Trevor O., Jeremy R. Townsend, Philip A. Sapp, Marlies Govaert, Cindy Duysburgh, Massimo Marzorati, Tess M. Marshall, and Ralph Esposito. 2023. "AG1, A Novel Synbiotic, Demonstrates the Capability to Enhance Fermentation Using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®)" Biology and Life Sciences Forum 29, no. 1: 10. https://doi.org/10.3390/IECN2023-15793
APA StyleKirby, T. O., Townsend, J. R., Sapp, P. A., Govaert, M., Duysburgh, C., Marzorati, M., Marshall, T. M., & Esposito, R. (2023). AG1, A Novel Synbiotic, Demonstrates the Capability to Enhance Fermentation Using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). Biology and Life Sciences Forum, 29(1), 10. https://doi.org/10.3390/IECN2023-15793





