Fungal Communities across an Edaphic Gradient in Central Borneo †
Abstract
:1. Introduction
2. Methods
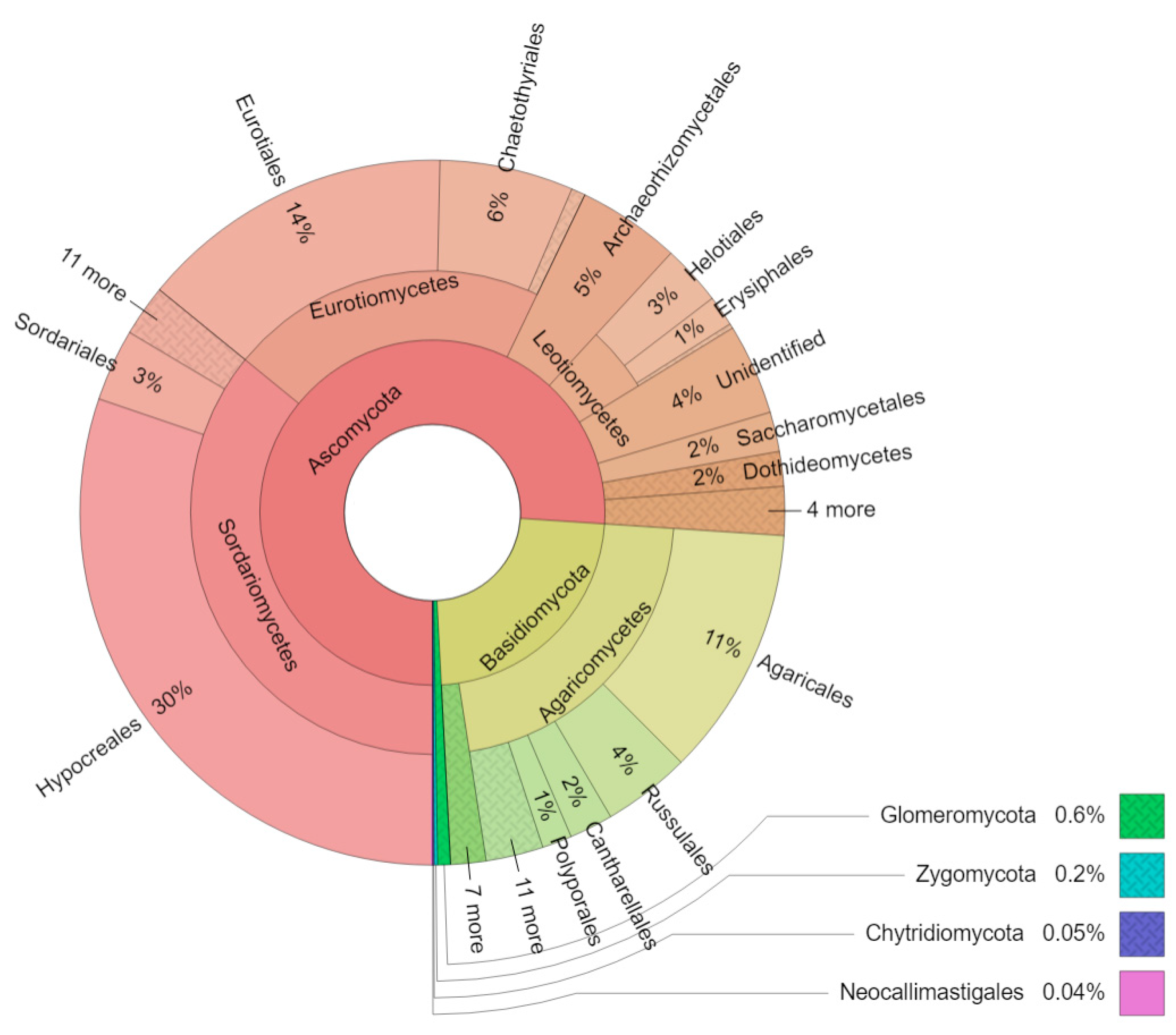
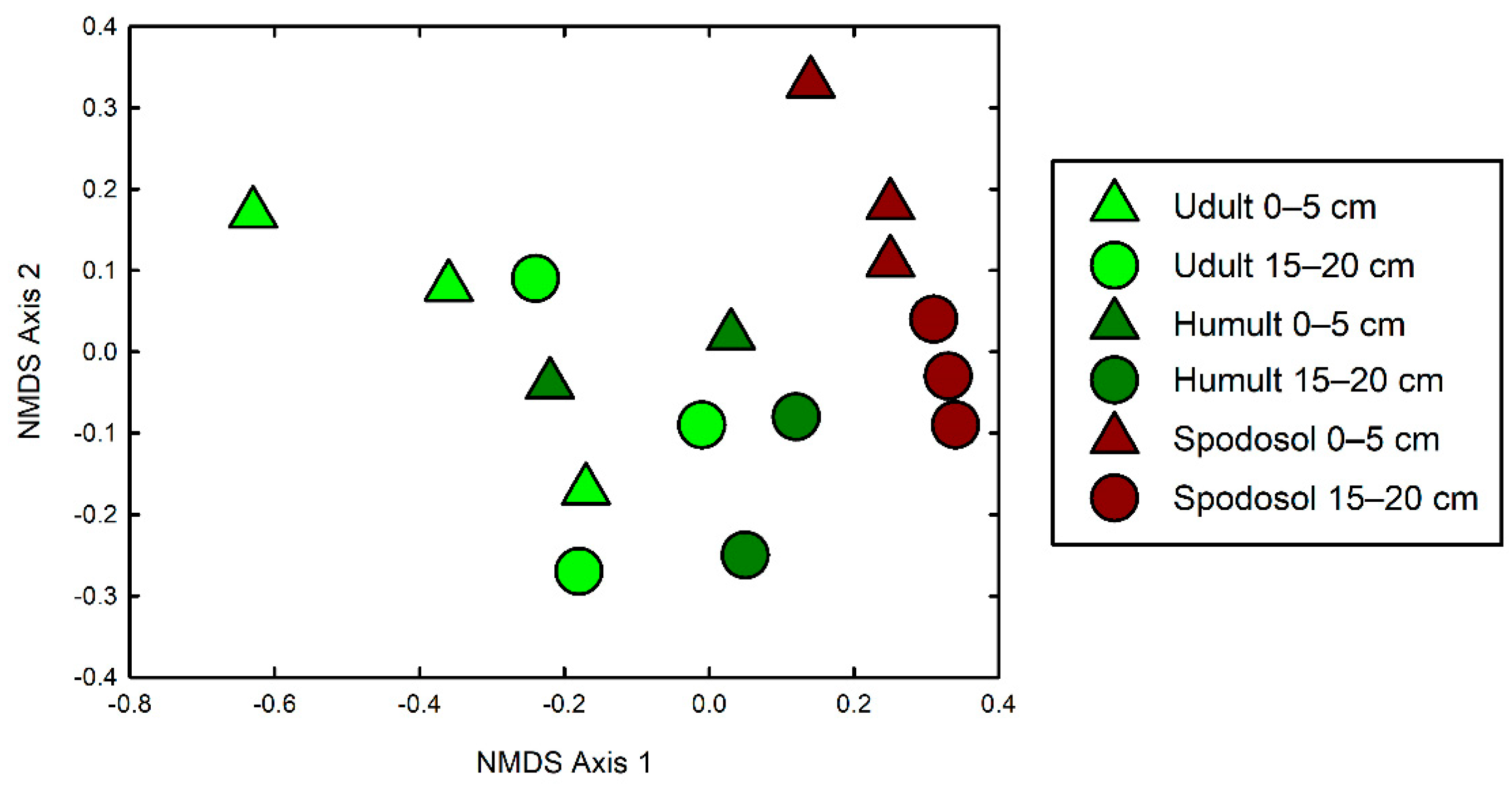
3. Results
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dighton, J. Fungi in Ecosystem Processes, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Aime, M.C.; Brearley, F.Q. Tropical fungal diversity: Closing the gap between species estimates and species discovery. Biodivers. Conserv. 2012, 21, 2177–2180. [Google Scholar] [CrossRef]
- McGuire, K.L.; Allison, S.D.; Fierer, N.; Treseder, K.K. Ectomycorrhizal-dominated boreal and tropical forests have distinct fungal communities, but analogous spatial patterns across soil horizons. PLoS ONE 2013, 8, e68278. [Google Scholar]
- Peay, K.G.; Baraloto, C.; Fine, P.V.A. Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J. 2013, 7, 1852–1861. [Google Scholar] [CrossRef] [PubMed]
- Schappe, T.; Albornoz, F.E.; Turner, B.L.; Neat, A.; Condit, R.; Jones, F.A. The role of soil chemistry and plant neighbourhoods in structuring fungal communities in three Panamanian rainforests. J. Ecol. 2017, 105, 569–579. [Google Scholar] [CrossRef]
- Ritter, C.D.; Dunthorn, M.; Anslan, S.; de Lima, V.X.; Tedersoo, L.; Nilsson, R.H.; Antonelli, A. Advancing biodiversity assessments with environmental DNA: Long-read technologies help reveal drivers of Amazonian fungal diversity. Ecol. Evol. 2020, 10, 7509–7524. [Google Scholar] [CrossRef]
- Vasco-Palacios, A.M.; Bahram, M.; Boekhout, T.; Tedersoo, L. Carbon content and pH as important drivers of fungal community structure in three Amazon forests. Plant Soil 2020, 450, 111–131. [Google Scholar] [CrossRef]
- McGuire, K.L.; D’Angelo, H.; Brearley, F.Q.; Gedallovich, S.M.; Babar, N.; Yang, N.; Gillikin, C.M.; Gradoville, R.; Bateman, C.; Turner, B.L.; et al. Responses of soil fungi to logging and oil palm agriculture in Southeast Asian tropical forests. Microb. Ecol. 2015, 69, 733–747. [Google Scholar] [CrossRef]
- Geml, J.; Morgado, L.N.; Semenova-Nelsen, T.A.; Schilthuizen, M. Changes in richness and community composition of ectomycorrhizal fungi among altitudinal vegetation types on Mount Kinabalu in Borneo. New Phytol. 2017, 215, 454–468. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Voitk, A.; Villarreal Ruiz, L.; Thu, P.Q.; Dang, T.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Ihrmark, K.; Boberg, J.; Trumbore, S.E.; Högberg, P.; Stenlid, J.; Finlay, R.D. Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol. 2017, 173, 611–620. [Google Scholar] [CrossRef]
- Davies, S.J.; Tan, S.; LaFrankie, J.V.; Potts, M.D. Soil-related floristic variation in a hyperdiverse dipterocarp forest. In Pollination Ecology and the Rain Forest: Sarawak Studies; Roubik, D., Sakai, S., Hamid, A.A., Eds.; Springer: New York, NY, USA, 2005; pp. 22–34. [Google Scholar]
- Sellan, G.; Brearley, F.Q.; Nilus, R.; Titin, J.; Majalap-Lee, N. Differences in soil properties among contrasting soil types in northern Borneo. J. Trop. For. Sci. 2021, 33, 105–115. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Song, W.; Slik, J.W.F.; Sukri, R.S.; Jaafar, S.; Dong, K.; Adams, J.M. Distinctive tropical forest variants have unique soil microbial communities, but not always low microbial diversity. Front. Microbiol. 2016, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Brearley, F.Q.; Proctor, J.; Suriantata; Nagy, L.; Dalrymple, G.; Voysey, B.C. Reproductive phenology over a 10-year period in a lowland evergreen rain forest of central Borneo. J. Ecol. 2007, 95, 828–839. [Google Scholar] [CrossRef]
- Proctor, J. Heath forests and acid soils. Bot. J. Scotl. 1999, 51, 1–14. [Google Scholar] [CrossRef]
- Brearley, F.Q. Microbial functioning in response to a simulated drought in Malaysian rain forest and oil palm soils. In Land-Use Change Impacts on Soil Processes: Tropical and Savannah Environments; Brearley, F.Q., Thomas, A.D., Eds.; CABI: Wallingford, UK, 2015; pp. 31–40. [Google Scholar]
- Epp, L.S.; Boessenkool, S.; Bellemain, E.P.; Haile, J.; Esposito, A.; Riaz, T.; Erséus, C.; Gusarov, V.I.; Edwards, M.E.; Johnsen, A.; et al. New environmental metabarcodes for analysing soil DNA: Potential for studying past and present ecosystems. Mol. Ecol. 2012, 8, 1821–1833. [Google Scholar] [CrossRef]
- Brearley, F.Q. Sequence data describing the fungal community in a tropical quartzite soil. Data Br. 2020, 29, 105112. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files, Version 1.33. 2011. Available online: https://github.com/najoshi/sickle (accessed on 29 September 2016).
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi—Recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomics visualisation in a web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan Community Ecology Package, Version 2.5-7. 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 9 November 2021).
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef]
- Floudas, D.; Bentzer, J.; Ahrén, D.; Johansson, T.; Persson, P.; Tunlid, A. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 2020, 14, 2046–2059. [Google Scholar] [CrossRef]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef]

| Parameter | Soil Type | Depth (cm) | |
|---|---|---|---|
| 0–5 | 15–20 | ||
| pH S ***, D ***, S × D ** | Udult | 3.36 ± 0.08 | 3.55 ± 0.09 |
| Humult | 2.81 ± 0.04 | 3.63 ± 0.05 | |
| Spodosol | 2.76 ± 0.07 | 3.14 ± 0.07 | |
| N (%) S ***, D ***, S × D n.s. | Udult | 0.27 ± 0.10 | 0.09 ± 0.02 |
| Humult | 1.11 ± 0.08 | 0.15 ± 0.02 | |
| Spodosol | 1.25 ± 0.09 | 0.17 ± 0.04 | |
| C (%) S ***, D ***, S × D ** | Udult | 4.09 ± 0.69 | 1.07 ± 0.10 |
| Humult | 39.8 ± 4.95 | 2.31 ± 0.29 | |
| Spodosol | 48.9 ± 1.05 | 4.71 ± 1.36 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brearley, F.Q. Fungal Communities across an Edaphic Gradient in Central Borneo. Biol. Life Sci. Forum 2022, 15, 31. https://doi.org/10.3390/IECD2022-12350
Brearley FQ. Fungal Communities across an Edaphic Gradient in Central Borneo. Biology and Life Sciences Forum. 2022; 15(1):31. https://doi.org/10.3390/IECD2022-12350
Chicago/Turabian StyleBrearley, Francis Q. 2022. "Fungal Communities across an Edaphic Gradient in Central Borneo" Biology and Life Sciences Forum 15, no. 1: 31. https://doi.org/10.3390/IECD2022-12350
APA StyleBrearley, F. Q. (2022). Fungal Communities across an Edaphic Gradient in Central Borneo. Biology and Life Sciences Forum, 15(1), 31. https://doi.org/10.3390/IECD2022-12350






