Genetic Diversity of Oat Genotypes Using SCoT Markers †
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. PCR Conditions
2.3. Electrophoresis of DNA
2.4. Data Analysis
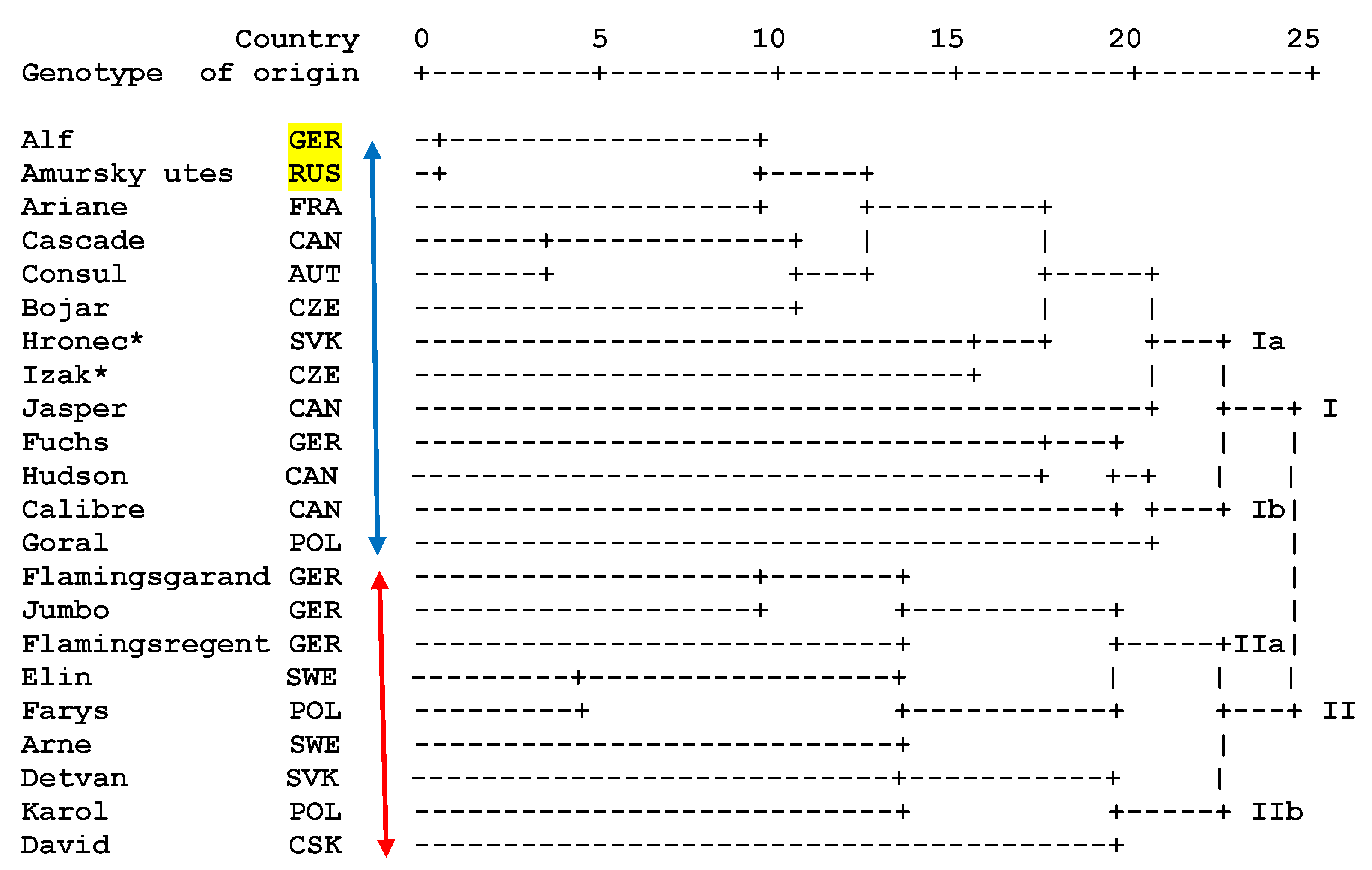
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daou, C.; Zhang, H. Oat Beta-Glucan: Its Role in Health Promotion and Prevention of Diseases. Compr. Rev. Food Sci. Food Saf. 2012, 11, 355–365. [Google Scholar] [CrossRef]
- Boczkowska, M.; Tarczyk, E. Genetic diversity among Polish landrances of common oat (Avena sativa L.). Gen. Res. Crop Rev. 2013, 60, 2157–2169. [Google Scholar]
- Gálová, Z.; Palenčáková, E.; Chňapek, M.; Balážová, Ž. Use of Cereals, Pseudo-Cereals and Legumes in a Gluten-Free Diet, 1st ed.; Slovak University of Agriculture: Nitra, Slovakia, 2011; pp. 38–39. [Google Scholar]
- Gaborská, D. Can celiacs consume oats—A still solved and unresolved issue. Nutr. Food. 2007, 62, 162–163. [Google Scholar]
- Gálová, Z.; Balážová, Ž.; Chňapek, M.; Vivodík, M.; Oslovičová, V. Protein and DNA Markers of Wheat, 1st ed.; Slovak University of Agriculture: Nitra, Slovakia, 2011; pp. 86–96. [Google Scholar]
- Gregáňová, Ž. Molecular Markers in the Identification and Characterization of Summer Wheat. Dissertation Work, University of Agriculture, Nitra, Slovakia, 2005. [Google Scholar]
- Vivodík, M.; Balážová, Ž.; Gálová, Z.; Kuťka Hlozáková, T. Differentiation of ricin using RAPD markers. Pak. J. Bot. 2015, 47, 1341–1345. [Google Scholar]
- Vyhnánek, T.; Trojan, V.; Štiasna, K.; Presinszká, M.; Hřivna, L.; Mrkvicová, E.; Havel, L. Testing of DNA isolation for the identification of Hemp. Potravin. Slovak J. Food Sci. 2015, 9, 393–397. [Google Scholar]
- Žiarovská, J.; Senková, S.; Beţo, M.; Ražná, K.; Masnica, M.; Labajová, M. ISSR markers as a tool to distinguish Idt and SSS populations of Zea mays L. J. Cent. Eur. Agric. 2013, 14, 489–499. [Google Scholar] [CrossRef][Green Version]
- Collard, B.Y.; Mackill, D. Start codon targeted (SCoT) polymorphism: Asimple, novel DNA marker technique for generating gene-targeted markers inplants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Satya, P.; Karan, M.; Jana, S.; Mitra, S.; Sharma, A.; Karmakar, P.G.; Rayb, D.P. Start codon targeted (SCoT) polymorphism reveals genetic diversity in wild and domesticated populations of ramie (Boehmeria nivea L. Gaudich.), a premium textile fiberproducing species. Meta Gene 2015, 3, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Kallamadi, P.R.; Nadigatlab, V.P.R.; Mulpuriba, S. Molecular diversity in castor (Ricinus communis). Ind. Crop. Prod. 2015, 66, 271–281. [Google Scholar] [CrossRef]
- Tsaballa, A.; Ganopoulos, I.; Timplalexi, A.; Aliki, X.; Bosmali, I.; Irini, N.O.; Athanasios, T.; Madesis, P. Molecular characterization of Greek pepper (Capsicumannuum L.) landraces with neutral (ISSR) and gene-based (SCoT and EST-SSR) molecular markers. Biochem. Syst. Ecol. 2015, 59, 257–259. [Google Scholar] [CrossRef]
- Vivodík, M.; Saadaoui, E.; Balážová, Ž.; Gálová, Z.; Petrovičová, L. Genetic diversity and population structure in Tunisian castor genotypes (Ricinus communis L.) detected using SCoT markers. Potravin. Slovak J. Food Sci. 2018, 12, 143–149. [Google Scholar] [CrossRef]
- Etminan, A.; Pour-Aboughadareh, A.; Mohammadi, R.; Ahmadi-Rad, A.; Noori, A.; Mahdavian, Z.; Moradi, Z. Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol. Biotechnol. Equip. 2016, 30, 1075–1081. [Google Scholar] [CrossRef]
- Vivodík, M.; Gálová, Z.; Balážová, Ž.; Petrovičová, L. Start codon targeted (SCoT) polymorphism reveals genetic diversity in European old maize (Zea mays L.) genotypes. Potravin. Slovak J. Food Sci. 2016, 10, 563–569. [Google Scholar]
- Petrovičová, L.; Balážová, Ž.; Vivodík, M.; Gálová, Z. Detection genetic variability of Secale cereale L. by scot markers. Potravin. Slovak J. Food Sci. 2017, 11, 197–202. [Google Scholar] [CrossRef][Green Version]
- Balážová, Ž.; Gálová, Z.; Vivodík, M.; Petrovičová, L.; Hornyák Gregáňová, R. Molecular variability of oat based on gene specific markers. Potravin. Slovak J. Food Sci. 2017, 11, 332–337. [Google Scholar] [CrossRef]
- Cieplak, M.; Okoń, S.; Werwińska, K. Genetic Similarity of Avena sativa L. Varieties as an Example of a Narrow Genetic Pool of Contemporary Cereal Species. Plants 2021, 10, 1424. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.L. Informativeness of human (dC-dA)n x (dG-dT)n polymorphism. Genomics 1990, 7, 524–530. [Google Scholar] [CrossRef]
| SCoT Primer | Sequence of Primers (5′-3′) | Anealing Temperature [°C] |
|---|---|---|
| SCoT 8 | CAACAATGGCTACCACGT | 50 °C |
| SCoT 9 | CAACAATGGCTACCAGCA | 50 °C |
| SCoT 12 | ACGACATGGCGACCAACG | 50 °C |
| SCoT 23 | CACCATGGCTACCACCAG | 50 °C |
| SCoT 26 | ACCATGGCTACCACCGTC | 50 °C |
| SCoT 28 | CCATGGCTACCACCGCCA | 50 °C |
| SCoT 29 | CCATGGCTACCACCGGCC | 50 °C |
| SCoT Marker | Number of All Fragments | Number of Polymorphic Fragments | Percentage of Polymorphic Bands (%) | PIC |
|---|---|---|---|---|
| SCoT8 | 7 | 5 | 71.42 | 0.674 |
| SCoT9 | 6 | 4 | 66.66 | 0.537 |
| SCoT12 | 6 | 2 | 33.33 | 0.305 |
| SCoT23 | 5 | 4 | 80.00 | 0.390 |
| SCoT26 | 6 | 4 | 66.66 | 0.547 |
| SCoT28 | 4 | 3 | 75.00 | 0.519 |
| SCoT29 | 6 | 4 | 66.66 | 0.567 |
| Average | 5.71 | 3.71 | 65.67 | 0.506 |
| Total | 40 | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chňapek, M.; Mikolášova, L.; Vivodík, M.; Gálová, Z.; Hromadová, Z.; Ražná, K.; Balážová, Ž. Genetic Diversity of Oat Genotypes Using SCoT Markers. Biol. Life Sci. Forum 2022, 11, 29. https://doi.org/10.3390/IECPS2021-11926
Chňapek M, Mikolášova L, Vivodík M, Gálová Z, Hromadová Z, Ražná K, Balážová Ž. Genetic Diversity of Oat Genotypes Using SCoT Markers. Biology and Life Sciences Forum. 2022; 11(1):29. https://doi.org/10.3390/IECPS2021-11926
Chicago/Turabian StyleChňapek, Milan, Lucia Mikolášova, Martin Vivodík, Zdenka Gálová, Zuzana Hromadová, Katarína Ražná, and Želmíra Balážová. 2022. "Genetic Diversity of Oat Genotypes Using SCoT Markers" Biology and Life Sciences Forum 11, no. 1: 29. https://doi.org/10.3390/IECPS2021-11926
APA StyleChňapek, M., Mikolášova, L., Vivodík, M., Gálová, Z., Hromadová, Z., Ražná, K., & Balážová, Ž. (2022). Genetic Diversity of Oat Genotypes Using SCoT Markers. Biology and Life Sciences Forum, 11(1), 29. https://doi.org/10.3390/IECPS2021-11926






