Abstract
To gain insight into two different plant strategies (hyperaccumulation and phytostabilization) for managing heavy metals, we conducted a network based functional enrichment analysis. Protein-protein interactions of A. halleri root and shoot were derived by weighted gene co-expression analysis. While, in the case of A. thaliana, protein-protein interactions characterizing the organs were derived from STRING database based on genes known to be expressed in root and shoot. Protein-protein interaction clusters of root and shoot networks of both species were analyzed to identify enriched pathways. Thus, we provide a first clear analysis of the biological peculiarities of different organs of both species.
1. Introduction
Phytoremediation is one of the auspicious strategies to manage heavy-metal polluted soils [1]. Several plants act differently to accommodate this issue rather by uptake (phytoextraction), emit in the atmosphere (phytovolatilization), or stabilize heavy metals in the root system (phytostabilization) [2]. Among these different processes, phytostabilization is of particular interest to be a low-cost and effective strategy. Moreover, this process is characteristic of a wide range of plants which include also commercial species and model organisms such as Arabidopsis thaliana [3]. On the other hand, some plants developed a rare adaptation to extract heavy metals from the soil and hyperaccumulate them into the shoot, such in the case as Arabidopsis halleri [4]. Within the genus Arabidopsis, A. thaliana and A. halleri possesses an unique global distributions, mating systems, life histories and adaptations providing opportunities to study them under different growth conditions. The recently long-scaffold assembly of the A. halleri permits now to identify long-range patterns of polymorphism and diversity and perform further genotype–phenotype association studies in different fields [5]. The major area of research in A. halleri focused on the study of heavy metal tolerance and hyperaccumulation, a constitutive phenotype in all tested genotypes. However, up to now, molecular mechanisms underpinning this ability are not completely understood; thus, in the present study, the prediction of all protein-protein interactions (PPIs), followed by network based functional enrichment analysis, were used to identify all pathways characterizing A. halleri, followed by comparing the uniqueness and similarities with A. thaliana.
2. Methodology
A weighted gene co-expression network analysis (WGCNA) was applied on RNA-seq data of A. halleri root and shoot [6] to identify and describe the PPI for the first time by WGCNA, a free accessible R package [7]. A soft threshold power was identified for root and shoot, separately, and used it to construct adjacency matrix based on scale-free topology. The adjacency matrix was converted to topological overlap matrix (TOM) and related dissimilarity of TOM (1-TOM) was computed to classify genes, having same expression pattern, into same module. Clustering height cut-off was set to 0.25 in order to merge likely modules with minimum module size of 30.
For A. thaliana, taking advantage of already available data, a list of genes expressed in the root and shoot was identified by reviewing the recent published papers, and organ-specific PPI was reconstructed and, successively, clustered according to their topological position into the networks. The gene lists were used to filter a high-confidence set of interactions of A. thaliana derived from STRING database [8]. The resulting network was subjected to MCODE plugin of Cytoscape to identify the highly connected regions in the PPI network which represent molecular clusters (modules) [9].
After identifying and filtering the clusters/modules in the two species, we performed KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis by using g:Profiler to have a view of biological pathways in common and dissimilar and involvement of genes [10].
3. Results and Discussion
3.1. Network Construction of A. halleri
The WGCNA was carried out on RNA-seq data of A. halleri root and shoot. A soft threshold power of β = 14 for root, and β = 8 in case of shoot was used to construct adjacency matrix based on scale-free topology (R2 = 0.97) and, successively, all genes having similar expression patterns were categorized into 14 modules in root and shoot (Figure 1).
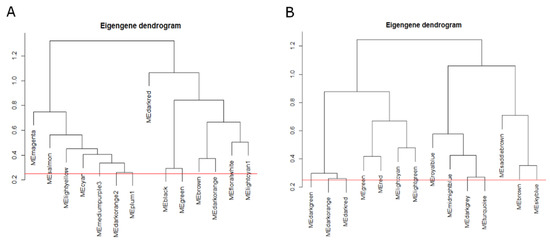
Figure 1.
Dendrograms representing the tree-based clustering of module eigengenes in root (A) and shoot (B). Red line represents the merging threshold.
3.2. Network Reconstruction of A. thaliana
Among all STRING interactions available for A. thaliana, we considered only those with a score higher than 700 composing a network made of a high-confidence set of interactions and only if both of its node IDs were present in the list of genes expressed in the two organs [11]. The modules (clusters) within each organ-specific networks were identified by applying a molecular complex detection algorithm available as MCODE plugin of Cytoscape, with the following criteria: degree cut-off equal to 2, node score cut-off equal to 0.2, K-core score equal to 2 and maximum depth from the seed equal to 100. A final score was calculated for the i-th module according to the formula:
where Di is the density of the module and Ni the number of nodes of the module. Modules were ranked and retained for following analysis if their final score was higher than 10. By this procedure we identified 15 modules including 1023 genes from root network and 18 modules including 1804 genes form shoot.
Final scorei = Di × Ni
3.3. KEGG Enrichment Analysis
With the purpose of getting insights of the biological pathways characterizing the biology of root and shoot of A. halleri and A. thaliana, KEGG pathway enrichment analysis was performed and identified the top 3 KEGG pathways significantly enriched (p < 0.05) in each module of root and shoot in A. halleri and A. thaliana (Figure 2).
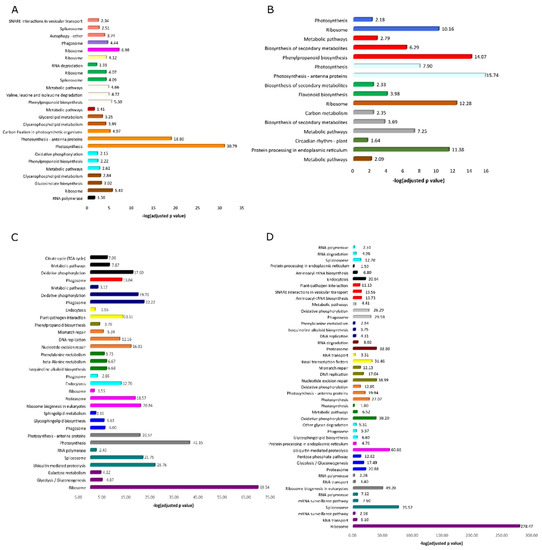
Figure 2.
Bars are the three most representative enriched pathways identified in each module of root (A) and shoot (B) of A. halleri, and of root (C) and shoot (D) of A. thaliana. The x-axis represents -log10(adjusted p-value) and bar colors distinguish modules.
In case of A. halleri, the most enriched pathway in both root and shoot was “photosynthesis”. While, in case of A. thaliana, the pathway related to “ribosome” was most enriched in both organs. Figure 3 is showing a Venn diagram representing the number of common and unique pathways identified in the root and shoot of both plants. In case of root, there are 8 unique pathways for A. halleri (24.2% of the total pathways identified), whereas for A. thaliana, there were 16 unique pathways (48.5% of total). On the other hand, in shoot, there were 5 unique pathways in A. halleri (14.7% of the total) and 24 pathways in case of A. thaliana (70.6% of the total).
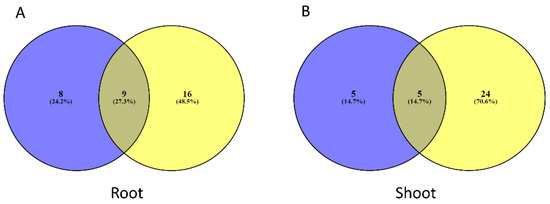
Figure 3.
Venn diagram illustrating the number of pathways overlapping in root (A) and shoot (B) in both species. Blue color represents A. halleri, and yellow color represents A. thaliana.
Glucosinolate biosynthesis was one of the enriched unique pathways identified in the in root of A. halleri. Glucosinolates comprise of diverse range of organic compounds involved in interactions between plant and insects [12]. Few studies reported the role of glucosinolates in anti-herbivory in A. thaliana and A. halleri [13,14]. Cadmium exposure to A. halleri results in the increased accumulation of glucosinolates in leaves and the result was opposite in case of A. thaliana with the decreased deposition of some glucosinolates in leaves and roots with Cd exposure [13,15]. These results can be relatable to our results where glucosinolate pathway was found to be more enriched in root of A. halleri. On the other hand, in A. thaliana root, citrate cycle (TCA cycle) was found to be among the unique pathways. TCA cycle is necessary to maintain the normal growth and development of plant under stress environment [16]. A study revealed that Cd exposure caused an increased accumulation of enzymes involved in TCA cycle, including citrate synthase (At2g44350) [17], which is also identified to be involved in this pathway in our study. The significance of this pathway in A. thaliana is well defined also in literature, as knockout of one of its components resulted in dwarfing phenotype accompanying with more prone to oxidative stress [18]. Till date, there is no prominent study regarding the component of this pathway in A. halleri. However, a study found more citrate (produced in TCA cycle) in the roots of A. halleri grown in contaminated soil with Zn [5,19]. But here we cannot conclude based on a single result.
Phenylpropanoids biosynthesis, pointed out as unique in case of A. halleri shoot, is a pathway which consists of a sequence of enzyme regulated reactions leading to different aromatic end products [20]. A study summarized the involvement of this pathway in increased tolerance against several abiotic stresses including heavy metals [21]. However, there is a lack of knowledge regarding the relation between metal stress and phenylpropanoid biosynthesis pathway in A. halleri and A. thaliana. But it should be considered that there is a chance of possible crosstalk between phenylpropanoid and glucosinolate metabolic pathways [22,23]. Based on this view, we can expect the same trend of phenylpropanoid metabolic pathway in accordance with the previous mentioned review in terms of glucosinolate metabolic pathway. For A. thaliana, pentose phosphate pathway and SNARE interactions were among the most enriched unique pathways in shoot. The pentose phosphate pathway is mandatory for metabolism as it produces components essential for nucleotide synthesis and for aromatic amino acids [24]. However, according to our knowledge, no study has been reported regarding the role of this pathway in metal stress in both plants. On the other side, several studies mentioned the role of SNAREs in abiotic tolerance in plants. The decrease in expression of v-SNAREs proteins in A. thaliana resulted in improved salt tolerance, leading to efficient functioning of tonoplast and vacuole [25]. This aspect can also be considered to regulate redox activity in relation to heavy metal stress, as the heavy metals have the ability to generate reactive oxygen species [26]. However, there is a lack in knowledge of these proteins in A. halleri.
4. Conclusions
Our study presented, for the first time, the prediction of PPI in A. halleri and, followed by the functional enrichment analysis, we provide new insights into the biology of this recently sequenced organism. In addition, a simple methodology was established to gain knowledge about similar or contrasting biological behaviors of A. halleri and A. thaliana.
A. halleri is a close relative of A. thaliana, but the main mechanisms for tolerating metal stress may be specific to one or the other species and therefore may be based on the possession of some different and particular biological processes. Here, we pointed out few organ-specific biological pathways identified as unique to A. halleri and/or A. thaliana, which can be suggested as possible targets to do future research. Moreover, this study will also help the researchers in order to pinpoint the gaps in research and to explore more of these two Arabidopsis species.
Author Contributions
Conceptualization, G.S.S., G.S., D.T. and D.M.; methodology, S.H.H. and G.S.; software, S.H.H., G.S. and D.F.; validation, G.S., D.T. and G.S.S.; formal analysis, S.H.H., G.S. and D.F.; investigation, S.H.H., G.S. and M.S.; writing—original draft preparation, review and editing, S.H.H., G.S., M.S., G.S.S., D.M. and D.T.; supervision, G.S.S., G.S. and D.T.; project administration, G.S.S., D.M. and D.T.; funding acquisition, G.S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data underlying this article will be shared on reasonable request 236 from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shah, V.; Daverey, A. Phytoremediation: A multidisciplinary approach to clean up heavy metal contaminated soil. Environ. Technol. Innov. 2020, 18, 100774. [Google Scholar] [CrossRef]
- Kushwaha, A.; Rani, R.; Kumar, S.; Gautam, A. Heavy metal detoxification and tolerance mechanisms in plants: Implications for phytoremediation. Environ. Rev. 2015, 24, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Simiele, M.; Sferra, G.; Lebrun, M.; Renzone, G.; Bourgerie, S.; Scippa, G.S.; Morabito, D.; Scaloni, A.; Trupiano, D.J.E.; Botany, E. In-depth study to decipher mechanisms underlying Arabidopsis thaliana tolerance to metal (loid) soil contamination in association with biochar and/or bacteria. Environ. Exp. Bot. 2021, 182, 104335. [Google Scholar] [CrossRef]
- Sarret, G.; Saumitou-Laprade, P.; Bert, V.; Proux, O.; Hazemann, J.-L.; Traverse, A.; Marcus, M.A.; Manceau, A. Forms of zinc accumulated in the hyperaccumulator Arabidopsis halleri. Plant Physiol. 2002, 130, 1815–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briskine, R.V.; Paape, T.; Shimizu-Inatsugi, R.; Nishiyama, T.; Akama, S.; Sese Shimizu, K.K. Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol. Ecol. Resour. 2017, 17, 1025–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corso, M.; An, X.; Jones, C.Y.; Gonzalez-Doblas, V.; Schvartzman, M.S.; Malkowski, E.; Willats, W.G.; Hanikenne, M.; Verbruggen, N. Adaptation of Arabidopsis halleri to extreme metal pollution through limited metal accumulation involves changes in cell wall composition and metal homeostasis. New Phytol. 2021, 230, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 8, 49. [Google Scholar]
- Bader, G.D.; Houge, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 13, 2. [Google Scholar]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Bozhilova, L.V.; Whitmore, A.V.; Wray, J.; Reinert, G.; Deane, C. Measuring rank robustness in scored protein interaction networks. BMC Bioinform. 2019, 20, 446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P.J.P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
- Stolpe, C.; Krämer, U.; Müller, C. Heavy metal (hyper) accumulation in leaves of Arabidopsis halleri is accompanied by a reduced performance of herbivores and shifts in leaf glucosinolate and element concentrations. Environ. Exp. Bot. 2017, 133, 78–86. [Google Scholar] [CrossRef]
- Kim, J.H.; Jander, G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 2007, 49, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, J.; Zhang, H.; Zhang, Q.; Ni, Y.; Chen, J.; Guan, Y. Glucosinolate profiles of Arabidopsis thaliana in response to cadmium exposure. Water Air Soil Pollut. 2009, 200, 109–117. [Google Scholar] [CrossRef]
- Hossain, Z.; Komatsu, S. Contribution of proteomic studies towards understanding plant heavy metal stress response. Front. Plant Sci. 2013, 3, 310. [Google Scholar] [CrossRef] [Green Version]
- Sarry, J.E.; Kuhn, L.; Ducruix, C.; Lafaye, A.; Junot, C.; Hugouvieux, V.; Jourdain, A.; Bastien, O.; Fievet, J.B.; Vailhen, D.J.P. The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 2006, 6, 2180–2198. [Google Scholar] [CrossRef]
- Bouché, N.; Fait, A.; Bouchez, D.; Møller, S.G.; Fromm, H. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. USA 2003, 100, 6843–6848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, N.C.; O’Neill, L. A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Gou, M.; Liu, C.-J. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B.J.M. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemm, M.R.; Ruegger, M.O.; Chapple, C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 2003, 15, 179–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.I.; Dolan, W.L.; Anderson, N.A.; Chapple, C. Indole glucosinolate biosynthesis limits phenylpropanoid accumulation in Arabidopsis thaliana. Plant Cell 2015, 27, 1529–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussell, J.D.; Keech, O.; Fenske, R.; Smith, S. Requirement for the plastidial oxidative pentose phosphate pathway for nitrate assimilation in Arabidopsis. Plant J. 2013, 75, 578–591. [Google Scholar] [CrossRef] [PubMed]
- Leshem, Y.; Melamed-Book, N.; Cagnac, O.; Ronen, G.; Nishri, Y.; Solomon, M.; Cohen, G.; Levine, A. Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 2006, 103, 18008–18013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.S.; Dietz, K.-J. The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci. 2009, 14, 43–50. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).