Simple Summary
Long non-coding RNAs (lncRNA), which are often referred to as “Genomic Dark Matter”, are emerging as critical molecules involved in the regulation of multiple cellular events. Their aberrant expressions and activities are correlated with tumorigenesis and tumor growth in many cancers. The documented potential of lncRNAs to regulate diverse oncogenic events identifies them as promising candidates for targeted therapy in cancer. In addition, the expression profiles of lncRNAs have the potential to serve as diagnostic or prognostic markers in many cancers. Therefore, an insight into the functional role of lncRNAs in cancer could aid in the development of novel diagnostic, prognostic, and therapeutic strategies to combat cancer. This review is focused on providing a succinct treatise on the functional role of lncRNAs in the genesis, growth, and progression of different cancers.
Abstract
Cancer is one of the leading causes of death worldwide. Multifactorial etiology of cancer and tumor heterogeneity are the two most acute challenges in existing diagnostic and therapeutic strategies for cancer. An effective precision cancer medicine strategy to overcome these challenges requires a clear understanding of the transcriptomic landscape of cancer cells. Recent innovative breakthroughs in high-throughput sequencing technologies have identified the oncogenic or tumor-suppressor role of several long non-coding RNAs (lncRNAs). LncRNAs have been characterized as regulating various signaling cascades which are involved in the pathobiology of cancer. They modulate cancer cell survival, proliferation, metabolism, invasive metastasis, stemness, and therapy-resistance through their interactions with specific sets of proteins, miRNAs and other non-coding RNAs, mRNAs, or DNAs in cells. By virtue of their ability to regulate multiple sets of genes and their cognate signaling pathways, lncRNAs are emerging as potential candidates for diagnostic, prognostic, and therapeutic targets. This review is focused on providing insight into the mechanisms by which different lncRNAs play a critical role in cancer growth, and their potential role in cancer diagnosis, prognosis, and therapy.
1. Introduction
Long non-coding RNAs (lncRNAs) form a heterogenous family of non-coding RNAs (ncRNAs) that comprises almost 98% of the human transcriptome [1,2]. Linear non-coding RNAs that are greater than 200 nt in length are classified as lncRNAs. The family of lncRNAs includes long intergenic non-coding RNAs (lincRNAs), the anti-sense lncRNAs, the sense lncRNAs, the intronic lncRNAs, and the bi-directional lncRNAs [3]. Each of these subtypes carries out diverse functions, adopting different modes of action, either within the nucleus or in the cytoplasm [3]. Despite being not translated into proteins, lncRNAs actively contribute to all the major cellular processes, which span from cellular homeostasis to the cellular pathology underlying many diseases, including cancer [1,2,4,5,6]. Through their various modes of action, lncRNAs contribute significantly to the different, but synergistic, oncogenic pathways underlying cancer genesis, progression, and therapy resistance [7]. Recent observations that lncRNAs traverse through circulation, either in the form of circulating tumor cells or exosomal lncRNAs, also point to their role in long-distance signaling, including the processes involved in distant metastasis [8,9,10,11,12]. In addition, the findings that lncRNAs can be detected in blood, plasma, and urine also identify them as potential diagnostic, prognostic, or therapeutic biomarkers [13,14].
Despite the significant progress in diagnostic and therapeutic strategies over the years, cancer still ranks as the leading cause of death worldwide [15]. While increased understanding of the disease has led to the development of novel diagnostic methodologies and targeted therapies for some cancers, disease recurrence and therapy resistance contribute significantly to cancer death [16,17]. Disease recurrence and therapy resistance often arise from the evolution of rescue pathways that facilitate cancer cells to bypass treatment strategies [17]. Effective therapeutic strategies are further compounded by inter- and intra-tumoral heterogeneity in cancer [18,19]. Personalized or precision cancer medicine is emerging as an effective therapeutic strategy that can overcome the changes associated with inter- and intra-tumoral heterogeneity in cancer [20,21]. However, this requires a clear understanding of the multi-omics landscape of the cancer cells from a given patient, so that effective therapies can be launched to combat the specific aberrant or asynchronous pathway(s) presented by the patient. Thus, defining the mechanistic role of lncRNAs in the cancer transcriptome is crucial for the development of an effective targeted therapy. In this review, we present a comprehensive overview on the mechanistic role of lncRNAs in different oncogenic responses, so that the diagnostic, prognostic, and/or therapeutic potential of lncRNAs can be incorporated into precision cancer medicine strategies.
2. The Superfamily of lncRNAs and Cancer
The superfamily of lncRNAs consists of more than 70,000 annotated lncRNAs in the human genome [22]. Functionally, lncRNAs can be defined as the multifunctional regulators of gene expression. Aberrant expression of lncRNAs and the resultant dysregulated expression of oncogenes and tumor-suppressor genes are associated with tumorigenesis and tumor progression. The mechanisms by which lncRNAs execute their oncogenic function involve the modulation of gene expressions through cis- and trans-modes of epigenetic regulation, the regulation of chromatin topology, serving as RNA decoys, acting as scaffolds for proteins and RNAs, and sequestering microRNAs (miRNAs) by acting as competing endogenous RNAs (ceRNAs) [21,22,23,24,25]. They also play significant roles in enhancing the expression of neighboring genes [18]. Through their various modes of action, lncRNAs contribute to several oncogenic processes. These include cell proliferation, genomic instability, invasive migration, epithelial-to-mesenchymal transition (EMT), stem cell differentiation, maintenance of pluripotency, tumor progression and suppression, DNA damage response and repair, and metabolic reprogramming [19,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44].
Specifically, lncRNAs play a functional role in several key spatiotemporal events involved in gene expression. The functional roles of lncRNAs include the maintenance of spliceosome machinery, the inhibition of gene transcription by direct binding to the gene, regulating gene expression through the modulation of transcription factor recruitment, inducing epigenetic modifications to regulate gene expression, regulating the functions of its interacting partners such as RNA, DNA, and proteins, regulating messenger RNA (mRNA) translation, regulating mRNA decay, and preventing the miRNA-mediated mRNA inhibition as a ceRNA. However, these functional roles need not be mutually exclusive. To a certain extent, the mechanisms by which lncRNA alters gene expression are dependent on the cellular location of the lncRNAs (Figure 1).
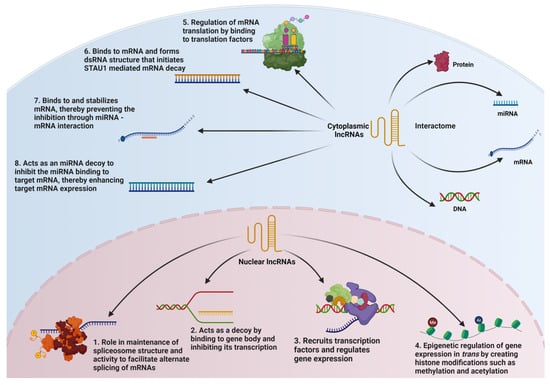
Figure 1.
Nuclear and Cytosolic Functions of lncRNAs. The nuclear functions of lncRNAs include (1) role in mRNA splicing; (2) inhibition of gene transcription by direct binding to the gene; (3) regulating gene expression through modulation of transcription factor recruitment; and (4) inducing epigenetic modifications to regulate gene expression. The cytoplasmic functions of lncRNAs include (5) regulating mRNA translation; (6) mediating mRNA decay; (7) preventing the miRNA-mediated mRNA inhibition through mRNA stabilization involving lncRNA-mRNA duplex formation; and (8) preventing miRNA-mediated mRNA inhibition by binding to the miRNA as a competing endogenous RNA though the respective interactomes.
Based on their role in oncogenic pathways, lncRNAs can be grouped into oncogenic lncRNAs and tumor-suppressor lncRNAs (TS-lncRNAs). In addition, some lncRNAs are grouped as dual-function lncRNAs due to their functional ability to be either oncogenic or tumor-suppressive in a context-specific manner.
Oncogenic, Tumor-Suppressor, and Dual-Function lncRNAs
A large number of lncRNAs have been reported to function as oncogenes in many cancers [23]. Increased expression of lncRNAs is seen in many cancers, either through gene amplification or dysregulated constitutive expression [24]. Oncogenic lncRNAs promote tumorigenic cell growth through the activation of multiple pathways involved in cell proliferation, cell survival, genomic instability, EMT, invasive metastasis, tumor angiogenesis, evading immune surveillance, cancer cell stemness, and/or radio/chemotherapy resistance (Table S1). Similarly to the multi-targeted effects of oncogenic lncRNAs, TS-lncRNAs inhibit the oncogenic growth of cells by suppressing the many different facets of oncogenic pathways (Table S2). Presumably, this involves the lncRNA-mediated suppression of expression or the activities of many of the oncogenic proteins, lncRNAs, and miRNAs.
There are also lncRNA variants that exhibit the dual functions of being either an oncogene or a tumor suppressor in a cellular or spatiotemporal context-dependent manner [25]. The plasmacytoma variant translocation 1 (PVT1), as well as the neuroblastoma-associated transcript 1 (NBAT1) locus, produce different splice variants of lncRNAs, which act as either oncogenes or as tumor suppressors, affecting the MYC levels [26,27,28,29]. Furthermore, the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) locus produces multiple splice variants of MALAT1, in addition to the full length MALAT1. These splice variants play both oncogenic and tumor-suppressor roles in a cell-type/cancer-specific manner [30,31]. In addition, H19, the NEAT1 (Nuclear Enriched Abundant Transcript 1), and BANCR (BRAF-activated non-protein coding RNA) are a few other lncRNAs that exhibit both oncogenic and tumor-suppressive functions in various tumor and cellular conditions [32,33,34,35,36,37]. Furthermore, the cancer cell type and its metabolic state serve as decisive factors to determine the exact role of the lncRNAs, be it either an oncogenic or a tumor-suppressive mode of action [23].
3. Role of lncRNAs and Cancer
Cancer can be defined as a pathophysiological state in which the cancer cells manifest an uncontrolled proliferation potential, breaking free from the cellular homeostatic regulatory mechanisms. The slow evolution of normal cells into cancer cells involves the accumulation of genetic and epigenetic changes, and various oncogenic selection processes [38,39]. The differences between normal cells and cancer cells have been defined as the hallmarks of cancer [40,41]. Major traits defined by the hallmarks of cancer include sustained cell proliferation, resistance to cell death, genomic instability, invasive metastasis, tumor angiogenesis, and metabolic reprograming. LncRNAs affect most, if not all, of the hallmarks of cancer in promoting cancer genesis and growth. A broad spectrum of oncogenic activities, regulated by different families of lncRNAs, act in concert with each other, thus cumulatively contributing to cancer genesis and progression (Figure 2).
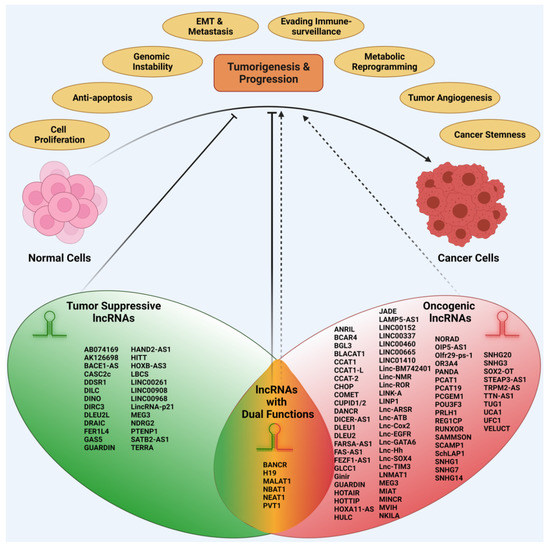
Figure 2.
Oncogenic versus Tumor Suppressive Roles of lncRNAs in cancer genesis and progression. Based on their contributory roles in cancer pathobiology, lncRNAs can be classified into three superfamilies, namely oncogenic lncRNAs, tumor-suppressor lncRNAs, and dual-function lncRNAs. Representative examples of the lncRNAs with their respective roles are categorized under the corresponding subfamilies in the figure.
3.1. lncRNAs in Cancer Cell Proliferation and Apoptosis
At normal physiological conditions, cells have stringent control over cell proliferation and apoptosis to maintain cellular, as well as organismal, homeostasis. Oncogenes have been known to disrupt cellular homeostasis and confer survival advantages to malignant cells by stimulating pro-mitotic signaling pathways and/or inhibiting apoptotic signaling pathways. Likewise, aberrant expressions of lncRNAs have been shown to provide survival advantages to cancer cells by stimulating the activation of specific pro-mitotic signaling pathways and/or inhibiting apoptotic signaling pathways. The pro-mitotic function of lncRNAs mostly involves their ability to act as a scaffold for chromatin-modifying protein complexes such as polycomb-repressive complex 2 (PRC2) and mixed-lineage leukemia 1 (MLL1), and the associated histone marks involved in gene silencing or activation. An example of this mode of action can be seen with HOTTIP (HOXA Transcript at the Distal Tip) lncRNA, which shows increased expression in many cancers (Table S1). In pancreatic cancer, the oncogenic role of HOTTIP is correlated with its ability to interact with MLL1, a H3K4 methyl transferase, and WD-repeat-containing protein 5 (WDR5), a methyl transferase adaptor [42]. Using a panel of pancreatic cancer cells, it has been shown that HOTTIP provides a scaffold for MLL1 and WDR5 that leads to the increased expression of the AURKA gene, which encodes Aurora-A kinase. Specifically, HOTTIP-MLL1 interaction leads to the increased expression of the potent oncogene AURKA [43], presumably through H3K4-trimethylation [44]. UCA1 (Urothelial Cancer Associated 1), which is overexpressed in many cancers, utilizes a different epigenetic mechanism to promote oncogenic proliferation, as evidenced in gastric cancer cells. UCA1 interacts with the enhancer of zest homologue 2 (EZH2), a histone methyl transferase, to induce repressive H3K27-trimethylation in the promoter regions of tumor-suppressor genes such as p27 and protein sprout homologue 1 (SPRY1), thereby promoting cell proliferation. Thus, the silencing of UCA1 has been shown to suppress cell proliferation, as well as the xenograft tumor growth of gastric cancer cells [45]. Similarly, EZH2-mediated suppression of the phosphatase and tensin homologue (PTEN), has been attributed to the oncogenic role of the lncRNA UFC1 (Ubiquitin Fold Modifier Conjugating Enzyme 1) in the proliferation of non-small cell lung carcinoma (NSCLC) cells, as well as xenograft tumor growth [46]. More interestingly, exosomally transmitted UFC1 could also promote the epigenetic silencing of PTEN via EZH2 in this cancer model [47]. In some instances, oncogenic lncRNAs regulate the transcription of genes by recruiting specific transcription factors and associated proteins to the promoters of the target genes. The lncRNA REG1CP (Regenerating Family Member 1 Gamma Pseudogene), which is overexpressed in colorectal cancer (CRC), forms an RNA–DNA triplex at the regenerating family member 3 alpha (REG3A) promoter region and recruits fanconi anemia J helicase to unwind the DNA. This enables the glucocorticid receptor-α mediated transactivation of REG3A expression and the subsequent REG3A-mediated proliferation of CRC cells and CRC xenograft tumor growth [48].
A primary mechanism through which lncRNAs promote cell proliferation is through the sequestration of specific miRNAs. The lncRNA PVT1 sequesters the tumor-suppressor miRNA, miR-543, to block its inhibitory effect on the expression of the oncogenic trichorhinophalangeal syndrome-1 gene. This leads to the increased proliferation and migration of breast cancer cells [49]. Similarly, UCA1 stimulates the proliferation of prostate cancer cells by sequestering miR-143. Using DU145, a prostate cancer cell line, it has been shown that the silencing of UCA1 results in the suppression of xenograft tumor growth and it is associated with the ability of UCA1 to sequester the tumor-suppressive miRNA miR-143 that targets pro-mitogenic MYO6 [50]. The mechanistic role for such a sequestration of tumor-suppressive miRNAs by lncRNAs has been shown in many other cancers by different lncRNA-miRNA combinations. The lncRNA CCAT1 sequesters miR-155, and thus upregulates c-Myc expression and the proliferation of acute myeloid leukemia (AML) cells [51]. In breast cancer cells, NEAT1 sequesters miR-141-3p to upregulate the expression of the mitogenic transcription factor, Kruppel-like factor 12 (KLF12) [52]. The lncRNAs, STEAP3-AS1 (Antisense RNA 1 to Six-Transmembrane Epithelial Antigen of Prostate-3), UFC1, and ANRIL (Antisense Non-coding RNA in the INK-4 Locus), also exhibit miRNA sequestration properties through which an array of oncogenes are upregulated, triggering cell proliferation in the cellular models of colon, pancreatic and gastric cancers, respectively [53,54,55].
In addition to the oncogenic lncRNAs, TS-lncRNAs play a critical role in cancer cell proliferation. In many cancers, the genes encoding TS-lncRNAs are deleted or their expressions are drastically reduced. SATB2-AS1 (Special AT-rich Sequence Binding protein-2 antisense RNA 1) and DIRC3 (Disrupted in Renal Carcinoma 3) are examples of lncRNAs which take part in the epigenetic regulation of genes involved in the inhibition of cell proliferation. Using a panel of CRC cell lines, it has been shown that the lncRNA, SATB2-AS1, serves as a scaffold for the recruitment of the transcriptional co-activator p300. SATB2-AS1-recruited p300 activates the expression of the tumor-suppressor gene SATB2 through the acetylation of H3K27 and H3K9 at the promoter site. However, downregulated expression of SATB2-AS1, often seen in CRC, leads to decreased levels of SATB2 with a resultant increase in cell proliferation [56]. In melanocytes, DIRC3, which is localized in the nucleus, enhances the transcription of the tumor-suppressor protein, insulin-like growth factor binding protein-5 (IGFBP5). DIRC3 acts as a decoy chromatin locus of IGFBP5 to prevent SRY box transcription factor-10 (SOX10)-mediated repression of IGFBP5. However, in melanoma cells, the melanocyte-inducing transcription factor and SOX10 complex represses the expression of DIRC3 through the repressive histone acetylation and methylation of the DIRC3 promoter. With the downregulation of DIRC3, SOX10 is able to repress the expression of IGFBP5, thus promoting the survival and growth of melanoma cells [57]. The sequestration of miRNAs by TS-lncRNAs also plays a role in tumor growth. The lncRNA MEG3 (Maternally Expressed-3) has been shown to exert its tumor-suppressive effect through the sequestration of miR-95-5p, which targets the tumor suppressor SOX11. In hepatocellular carcinoma (HCC), the downregulation of MEG3 leads to the increase in the levels of miR-9-5p. Subsequently, the expression of tumor suppressor SOX-11, which is a target of miR-9-5p, is thus suppressed, which augments cell proliferation and anti-apoptotic signals in HCC [58]. A similar role has been demonstrated with the TS-lncRNA, HAND2-AS1 (Heart and Neural Crest Derivatives Expressed Transcript 2 Antisense RNA 1), whose expression is downregulated in many cancers [59]. In vitro studies using a panel of HCC cell lines have shown that the lncRNA HAND2-AS1 sequesters miR-3118, which targets the suppressor of cytokine signaling 5 (SOCS5). The downregulation of HAND2-AS1 in HCC cells enhances the activity of miR-3118, and the subsequent suppression of its target SOCS5. SOCS5 is an inhibitor of JAK/STAT signaling. Hence, the downregulated HAND2-AS1 results in augmented JAK/STAT signaling, which contributes to enhanced proliferation, invasion, and migration in HCC cells [59]. The TS-lncRNA, GAS5 (Growth Arrest Specific-5), acts as a ceRNA to sequester miR-196a-3p in breast cancer cells. The lower levels of GAS5 in triple-negative breast cancers (TNBCs) increase the miR-196a-3p levels to inhibit the expression of the forkhead box O1 (FOXO1) gene. This activates the phosphatidyl inositol-3kinase (PI3K)-AKT kinase signaling pathway, which triggers numerous oncogenic pathways [60]. In addition to miRNA sequestration, TS-lncRNAs regulate mRNA translation by acting as scaffold in the recruitment of various translation factors. This function also plays a role in cancer cell proliferation and survival. The TS-lncRNA GAS5 interacts with the eukaryotic translation initiation factor, eIF4E and directly inhibits its recruitment to the translation site, thereby attenuating c-Myc translation and the downstream activation of c-Myc-regulated oncogenic events. This inhibition is relieved, and cell growth is promoted with the reduced expression of GAS5 in lymphoma cells [61]. In these cells, GAS5 is also found to interact directly with c-Myc mRNA to inhibit its translation. Such direct TS-lncRNA-mRNA interaction is also observed in the case of lincRNA-p21 [62]. In HeLa cells, it has been shown that lincRNA-p21 directly interacts with the mRNAs of the oncogenes JUNB and CTNNB1/β-catenin to inhibit their translation [63]. In addition, it binds to the polysomal translational machinery and the translational repressors RCK and FMRP to further suppress the translation of JUNB and CTNNB1 [62,63]. Reduced expression of lincRNA-p21 eases these constraints on JUNB and CTNNB1 expressions, which leads to oncogenic cell growth. A few other TS-lncRNAs also affect the stability of mRNAs that encode specific oncogenes. The tumor-suppressor role of NBAT1 in HCC cells has been attributed to its ability to physically associate with insulin growth factor-2 binding protein-1 to prevent its stabilizing interaction with c-Myc mRNA. Reduced expression of NBAT1, in turn, enhances the stability of c-Myc mRNA, triggering the activation of c-Myc regulated oncogenic signaling cascades [27].
The lncRNA interactome is so diverse that these interactions could determine their role in tumorigenesis and tumor progression. DRAIC (Downregulated RNA in Cancer) has been identified as a TS-lncRNA in castration-resistant prostate cancers [64]. It interacts with the IκB kinase (IKK) subunits, thereby preventing the interaction between these subunits, their phosphorylation, and the subsequent activation of nuclear factor kappa B (NF-κB). When DRAIC levels are lower, it leads to NF-κB activation and the NF-κB-pathway-mediated proliferation of prostate cancer cells [64]. A variation of this theme is seen with the NBAT1 and CASC15-003 (Cancer Susceptibility 15) lncRNAs in neuroblastoma [65]. In low-risk neuroblastoma cells, both NBAT1 and CASC15-003 show higher expressions, predicting better prognosis. In contrast, decreased expression of NBAT1 and CASC15-003 with poor prognosis was observed in high-risk neuroblastoma. A decrease in the expression levels of NBAT1 and CASC15-003 were accompanied with the increased expression levels of N-Myc (MYCN). It has been demonstrated that NBAT1 and CASC15-003 interact with ubiquitin-specific peptidase-36 (USP36), a de-ubiquitinating protein, to prevent its interaction with N-Myc. Since USP36-N-Myc interaction prevents the ubiquitin-mediated proteolytic degradation of N-Myc, increased expression of NBAT1 and CASC15-003 inhibits MYC-mediated oncogenic pathways. However, reduced levels of NBAT1 and CASC15-003 result in the enhanced expression of N-Myc and N-Myc-regulated oncogenic activities in neuroblastoma [65]. In summary, lncRNAs regulating cell proliferation and survival signals can serve as significant therapeutic targets, as well as precise biomarkers for cancers.
3.2. lncRNAs and Genomic Instability
Genome instability is a hallmark of cancer which is characterized by defects in either DNA replication fidelity, chromosome segregation, and/or DNA damage repair (DDR) mechanisms. In cancer cells, genomic instability involves the complex DNA damage response (DDR) pathways, in which lncRNAs also play a definitive role. DDR involves DNA repair enzymes, tumor suppressors, apoptotic proteins, kinases, and cell cycle checkpoint factors. Defective DDR, such as in cancers, enhance the mutational load as well as genomic instability, and adversely affects therapy with DNA-damaging agents. The prominent mammalian DDR pathways include homologous recombination repair (HRR), non-homologous end joining (NHEJ), base excision repair (BER), nucleotide excision repair (NER), and mismatch repair (MMR). The major sensors of DNA damage include ATM (ataxia telangiectasia mutated), ATR (ataxia telangiectasia and Rad3 related) and DNA-PKs (DNA-dependent protein kinase), while p53 serves as the most significant transcriptional regulator to modulate cell cycle arrest and apoptosis in case of DNA damages. LncRNAs are either induced directly owing to DNA damage, or indirectly via intermediates such as p53, which is triggered upon DNA damage sensing, thereby regulating the cell cycle and DNA repair, as well as DDR events [66,67].
DDR genes and associated DNA repair mechanisms play a major role in the maintenance of genomic stability [68]. Several lncRNAs have been documented to play a role in the genomic instability of cancer cells through diverse mechanisms. The p53-responsive lncRNA, GUARDIN, acts as a scaffold for the breast cancer type 1 susceptibility protein/BRCA1-associated ring domain 1 (BRCA1/BARD1) complex, promoting DDR in many different cancer cells, including those of CRC, osteosarcoma, and lung cancer [69]. GUARDIN also sequesters miR-23a to upregulate telomere repeat-binding factor-2 (TRF2) levels which prevent chromosomal end-to-end joining in these cells [69]. In addition, GUARDIN acts as a scaffold for the breast cancer type 1 susceptibility protein/BRCA1 associated ring domain 1 (BRCA1/BARD1) complex, promoting DDR. The reduction of GUARDIN sensitizes cancer cells to chemotherapy [70]. JADE (Gene for Apoptosis and Differentiation in Epithelia) is an ATM-induced lncRNA, which is expressed upon DNA damage in cancers. It transcriptionally activates the expression of Jade-1 protein, which forms a complex with the histone acetyltransferase enzyme, human acetylase binding to ORC1 (HBO1), to stimulate histone 4-specific acetyltanferase activity. This leads to the expression of several other DDR genes promoting genomic instability and tumorigenesis in breast cancer cells [71]. The lncRNA SNHG1 (Small Nucleolar RNA Host Gene 1) promotes genomic instability by targeting p53 stability. In doxorubicin-treated colorectal cancer cells, SNHG1 sequesters heterogenous nuclear ribonucleoprotein C (hnRNPC) from interacting with p53, which leads to the inactivation of p53 with the resultant activation of DNA repair pathways [72]. The lncRNA, ANRIL, binds to and stabilizes ATR proteins, preventing it from ubiquitination. Thus, the complex promotes DNA double-strand repair through HRR in cancer cells [73]. The lncRNA, PRLH1, (p53-regulated lncRNA for HRR 1), which is upregulated in p53-mutated HCC, modulates the HRR pathway by binding to and stabilizing the ring finger protein, RNF169, to recruit HRR factors aiding DDR [74].
Several p53-independent lncRNAs regulate DDR in cancers by acting as scaffold for DNA-repair-associated proteins. LINP1 (LncRNA in NHEJ 1) regulates NHEJ by serving as scaffold for Ku70/80 and major DNA-PKs in TNBCs. Upon DNA damage, LINP1 translocates from the cytoplasm to the nucleus, and its depletion sensitizes these cancers to radiotherapy [75]. In multiple myeloma, MALAT1 binds to poly-ADP-ribose polymerase 1 (PARP1) and DNA ligase III in regulating the alternate-NHEJ repair mechanism [76]. Single-nucleotide polymorphisms at cyclin D1 (CCND1)-promoter regions enhance the levels of lncRNAs CUPID1 and CUPID2 (CCND1 upstream intergenic DNA repair 1 and 2), which play a significant role in promoting HRR over NHEJ through interaction with p53-binding protein 1 (53BP1) and BRCA1 [77]. The lncRNA, BGL3 (beta globin locus transcript 3), promotes DDR through its interaction with PARP1 and BARD1 proteins. It acts as scaffold for the BRCA1/BARD1 complex and other DDR, as well as HRR proteins, to stimulate DNA damage repair pathways in breast, osteosarcoma, and colon cancer cells [78]. BGL3 depletion has been shown to sensitize breast cancer cells to DNA damage-mediated therapy [78]. Another oncogenic lncRNA, Ginir (Genomic Instability Inducing RNA), interferes with karyogenesis, leading to the formation of multinucleated cells, during cell division in multiple myeloma cells. This is primarily due to its regulatory role over centrosomal functions through its interaction with the centrosomal protein, Cep112. Ginir-Cep112 association impairs BRCA1-Cep112 interaction, leading to dysregulated DDR and genomic instability in multiple myeloma, both in vitro and in vivo [79]. NORAD (Non-coding RNA Activated by DNA damage) is yet another lncRNA that actively regulates DDR in cancer cells. It acts as a decoy for various RNA-binding proteins, such as PUMILIO and RBMX, to prevent them from inhibiting the expression of DDR proteins [80]. In colon cancer cells, as well as colon organoids from mouse models, it has been reported that the lncRNA, CCAT2 (colon cancer associated transcript 2), enhances the expression of genes involved in ribosomal biogenesis, including the myc-induced expression of block of proliferation 1 (BOP1). In addition, CCAT2 directly interacts with BOP1 to increase its stability. BOP1 enhances the expression and activation of Aurora Kinase B, which leads to chromosome instability and consequent tumorigenesis [81]. Overexpressed NEAT1 has been observed to promote nuclear paraspeckle formation, enhancing the ATR signaling in osteosarcoma and TNBC cell lines. This facilitates the rapid replication of cancer cells, while its silencing sensitizes TNBC cells to chemotherapy [82].
Several of the TS-lncRNAs also play a role in regulating genomic stability. LincRNA-p21 has been reported to activate p53-induced p21 expression through the interaction with hnRNP-K, which further induces the expression of various DDR and G1/S cell cycle checkpoint genes mediated via PRC2. The deficiency of lincRNA-p21 disrupts this DDR system in many cancers [83]. A greater number of lncRNAs exert their functional roles, acting as scaffold for various proteins to facilitate DDR. The lncRNA, DINO (Damage induced Non-coding), a transcriptional target of p53, binds and stabilizes p53, thus preventing the latter from ubiquitin-mediated proteolytic degradation, thereby facilitating error-proof DDR in CRC cell lines. The in vivo knockout mouse models affirmed that DINO-mediated stabilization of p53 further promotes the expressions of DDR genes such as DDB2, RRM2b, and GADD45a [84]. It has also been shown that DINO can reactivate dysfunctional p53 in human papilloma virus-positive cervical cancer cells via ATM/checkpoint kinase-2 (CHK2)-mediated DDR [85]. The lncRNA, DDSR1 (DNA damage sensitive RNA 1), an ATM-induced lncRNA upon DNA damage, acts as scaffold that binds to BRCA1 and hnRNP U-like 1 (hnRNPUL1) to facilitate the resection of DNA ends for the double-strand-break repair. The downregulation of DDSR1 results in BRCA1 accumulation at the damage site, resulting in deregulated HRR and DDR [86]. However, the seemingly tumor-suppressor function of DDSR1 is contrasted by the observation that the reduced expression of DDSR1 is correlated with the increased proliferation rate in multiple cancer cells [86]. The lncRNA, HITT (HIF-1α Inhibitor at Translational level), appears to be the first reported lncRNA that inhibits ATM activity in human cancers. HITT interacts with ATM, and prevents its association with the meiotic recombination 11 (MRE11)-RAD50-Nibrin (NBS1) complex, thereby inhibiting ATM recruitment and its activity at the damage sites, both in vitro and in vivo. Thus, HITT can sensitize colon cancer cells to chemotherapy through the inhibition of HRR [87]. The lncRNA, TERRA (Telomere Repeat Containing RNA), which is induced by tumor suppressors, such as p53, promotes genomic stability in normal cells. [88]. TERRA recruits TRF2 and associated factors at telomere ends to induce telomere stability [88]. Reduced expression of TERRA, as well as increased activity of TERRA, has been associated with the genomic instability in many cancers, including breast cancer [88,89]. From these results, it can be surmised that the modulation in TERRA levels in cancer cells leads to telomerase instability. Thus, lncRNAs play a major role in regulating various DDR components involved in genomic instability through multiple mechanisms. Further studies can reveal the precise roles in critical steps to maintain genome integrity, which can aid cancer therapy.
3.3. lncRNAs and Metabolic Reprogramming
Metabolic reprogramming is one of the major hallmarks of cancer, which involves the regulation of diverse cellular metabolic pathways, such as that of carbohydrates, lipids, amino acids, and nucleotides, so as to support cancer cell proliferation and survival. Since the cancer cells survive within a stressful tumor microenvironment, this condition can trigger various metabolic signals which can modulate the lncRNA levels. Such lncRNAs, in turn, regulate the expression and activity of various metabolic signaling pathway components to alter the cellular metabolism, termed as metabolic reprogramming, aiding tumorigenesis.
The major energy metabolism in cancer cells is through glucose utilization, whose rate is enhanced either by increasing the glucose uptake or by increasing the dependency on aerobic glycolysis rather than depending on mitochondrial oxidative phosphorylation, termed as Warburg effect [90]. To stimulate aerobic glycolysis, cancer cells increase the uptake of glucose through several lncRNAs. The lncRNAs, PCGEM1 (Prostate Cancer Gene Expression Marker 1) in prostate cancers, CRNDE (Colorectal Neoplasia Differentially Expressed Non-protein Coding Gene) in CRC, LINK-A (Long Intergenic RNA for Kinase Activation) in triple-negative breast cancers, and NRCP (lncRNA Ceruloplasmin) in ovarian as well as breast cancers, enhance the glucose uptake in respective cancer cells [91,92,93,94]. Specifically, CRNDE has been shown to stimulate glucose uptake by increasing the expression of glucose transporter-4 [92]. LncRNAs also regulate the key enzymes involved in glucose metabolism such as hexokinase 2 (HK), aldolase, pyruvate kinase, and lactate dehydrogenase (LDH). Hexokinase, which is encoded by HK1 or HK2 genes, is the first enzyme in the glycolytic pathway. In HCC cells, TUG1 (Taurine Upregulated 1) sequesters miR-455-3p, which represses the expression of the AMP-activated protein kinase β-2 (AMPKβ2). AMPKβ2, thus relieved from the inhibitory effects of miR-455-3p, stimulates HK2 expression and glycolysis [95]. In gallbladder cancer cells, PVT1 overexpression enhances glycolysis by quenching miR-143, which targets HK2, thereby increasing the expression levels of HK2 with the resultant increase in glycolysis [96]. In esophageal carcinoma, HOTAIR (HOX Transcript Antisense Intergenic RNA) has been shown to upregulate the expression of HK2 by sequestering both miR-125 and miR-143, which target HK2 [97]. Increased expression of HK2 is correlated with the overexpression of HOTAIR in pancreatic adenocarcinoma [98]. LncRNA-mediated stimulation of glycolysis also involves the increased expression of the PKM gene which encodes pyruvate kinase. In cellular as well as murine models of CRC, antisense lncRNA to FEZ family zinc finger-1 (FEZF1-AS1) increases the stability of both cytoplasmic and nuclear PKM2 by physically associating with PKM2 [99]. While the cytoplasmic PKM2 interaction increases the aerobic glycolysis of CRC cells, nuclear interaction leads to the hyperactivation of the signal transducer and the activator of transcription 3 (STAT3) signaling. Another lncRNA, PCGEM1, has been shown to increase the levels of PKM2 and lactate dehydrogenase A (LDHA) to promote prostate cancer cell survival [100]. In prostate cancer cells, it has been reported that PCGEM1 directly binds to the promoters of c-Myc and its target genes to stimulate the transactivation of PKM2 and LDHA genes. In CRC, glucose starvation induces the lncRNA, GLCC1 (glycolysis associated lncRNA of colorectal cancer 1), which directly interacts with HSP90 to stabilize c-Myc and transcriptionally activate the expression of LDHA [101]. The lncRNA, lnc-IGFBP4-1, has also been shown to promote glucose metabolism and cancer cell proliferation in lung cancers by stimulating the expression of LDHA and HK2, as well as the PDK1 gene which encodes pyruvate dehydrogenase kinase 1 [102].
In addition to glucose metabolism, lncRNAs are also involved in rewiring lipid metabolism in cancer cells. Lipid metabolism is essential for cancer cells not only due to its role in aiding proliferation and by being a cell membrane component, but by also being a source for cellular energy and a precursor to many important signaling molecules. The lncRNAs, HULC (Highly Upregulated in Liver Cancer) and TINCR (Terminal Differentiation Induced lncRNA), stimulate acyl-CoA metabolism to promote tumor growth [103,104]. Using cellular as well as murine models of HCC, it has been demonstrated that HULC increases the activity of acetyl CoA synthase (ACS), which leads to increased triacylglycerol synthesis. Specifically, HULC increases the expression of the ACS-long chain family member 1 (ACSL1) subunit of ACS through the upregulation of the transcription factor peroxisome proliferator-activated receptor alpha (PPARA), which transactivates the ACSL1 promoter. The mechanism by which HULC upregulates the expression of PPARA is through the epigenetic silencing of miR-9, which targets PPARA [103]. Using a panel of nasopharyngeal carcinoma (NPC) cell lines and a xenograft mouse model, it has been shown that TINCR interacts with adenosine triphosphate citrate lyase to protect it from ubiquitin degradation, which leads to the upregulation of acetyl CoA and an overall increase in lipid biosynthesis [104].
Several lncRNAs have been shown to converge on the regulation of fatty acid synthase (FASN), a key enzyme involved in lipid biogenesis. In the prostate cancer cell line LNCaP, the lncRNA, PCGEM1, which regulates glucose metabolism, acts in co-ordination with the androgen receptor and c-Myc to regulate fatty acid biosynthesis by increasing the expression of FASN [100]. HOXD-AS1 or HAGLR (HOXD antisense growth-associated long non-coding RNA), which is elevated in NSCLC, enhances the expression of FASN, thereby increasing the levels of free fatty acids [105]. In NPC cells, HOTAIR has been shown to induce the expression of FASN [106]. In the U2OS osteosarcoma cell line, PVT1 upregulates the expression of FASN by quenching miR-195, which targets FASN [107]. Cellular lipid metabolism is strictly regulated by the transcription factors, sterol regulatory element-binding proteins (SREBPs). In the HCC cell line HepG2, MALAT1 has been shown to associate with SREBP-1c to enhance its stability. MALAT1-stabilized SREBP-1c promotes the transcription of genes involved in lipid metabolism, such as FASN, ATP citrate synthase, stearoyl CoA desaturase 1 and acetyl-CoA carboxylase 1, leading to cellular lipid accumulation [108].
In addition to their role in reprogramming glucose and lipid metabolism in cancer cells, a host of lncRNAs target amino acid metabolism. Of the amino acids, glutamine is the most abundant and significant one which actively takes part in the bioenergetic and biosynthetic pathways in cancer cells [109]. Glutamine, taken up by the cancer cells, undergoes glutaminolysis, in which it is converted into glutamate and α-ketoglutarate. These metabolites from glutaminolysis are used to generate ATP via the tricarboxylic acid cycle, and to provide an anabolic carbon backbone for the synthesis of lipids, amino acids, and nucleotides. The key enzyme involved in glutaminolysis is glutaminase. In CRC cells, the lncRNA CCAT2 enhances the expression of GAC, a gene that encodes glutaminase-c isoform, to augment the glutamine metabolism [110]. In bladder cancer cells, UCA1 has been shown to sequester miR-16 and upregulates the expression of glutaminase 2 (GLS2). This has been correlated with a reactive oxygen species (ROS) synthesis that protects cells from oxidative toxicity, in addition to the promotion of glutaminolysis in bladder cancers [111]. HOTTIP has been shown to enhance glutamine metabolism in a panel of cancer cells by upregulating GLS1 expression to promote tumorigenesis [112]. In glioma cells, upregulated HOTAIR promotes glutamine metabolism by sequestering miR-126 and relieving its inhibition on the expression of GLS [113]. In the prostate cancer cell line, LnCAP, PCGEM1 promotes chromatin recruitment of c-Myc, for the transactivation of its target genes that regulate the glutamine metabolism [100]. LncRNAs such as TUG1 also regulate other enzymes involved in glutamine metabolism. In intrahepatic cholangiocarcinoma cells, TUG1 induces sirtuin-3 (Sirt3)-mediated glutamate dehydrogenase (GDH) levels, thereby enhancing glutaminolysis and synthesis of metabolic intermediates to promote tumor progression [114]. It is significant to note here that the lncRNA, PVT1-5, in lung cancer, has been demonstrated to promote increased amino acid uptake by promoting the expression of the neutral amino acid exchanger, solute carrier family 7 member 5 (SLC7A5). SLC7A5 mediates the transport of neutral amino acids in exchange for the intracellular glutamine. PVT1-5 increases the expression levels of SLC7A5 by sequestering miR-126 that targets SLC7A5 [115]. However, a similar lncRNA-mediated mechanism for the enhanced uptake of glutamine by cancer cells remains to be established.
The role of lncRNAs in directly targeting molecules involved in nucleotide synthesis remains poorly understood. Increased expression of the linc-NMR (long intergenic non-coding RNA–nucleotide metabolism regulator) is observed in many cancers. Its overexpression has been correlated with cell proliferation, invasion, and metastasis of the respective cancer cells. Using the HCC cell line, HLE, it has been observed that the silencing of linc-NMR expression induces G0/G1 phase cell cycle arrest, which is accompanied by a drastic reduction in the levels of all the deoxy nucleotide triphosphates. Further analyses have indicated that linc-NMR binds to and regulates Y-box-binding protein-1 (YBX1), and YBX1 in turn regulates the key enzymes in pyrimidine metabolism, such as the ribonucleotide reductase 2, thymidylate synthetase, and thymidine kinase 1 [116]. Detailed analysis of the lncRNA-regulated metabolic reprogramming would help in unraveling them as potential biomarkers and therapeutic targets in cancer.
3.4. lncRNAs and Evasion of Immune Surveillance
The human immune system includes both adaptive and innate immune responses. Adaptive immune responses in the host involve the production of antigen-responsive lymphocytes, mainly T-lymphocytes or T-cells and B-lymphocytes or B-cells, which protect the host from similar kinds of antigenic exposure or re-infection with the same pathogen. In adaptive immune responses, T-cell activation further triggers numerous T-cell subsets, such as the CD8+ cytotoxic T-lymphocytes (CTLs), which eliminate the cancer cells via the release of various cytokines such as tumor necrosis factors (TNFs) and interleukins (ILs) [117]. Several lncRNAs target CTLs to promote immune evasion by cancer cells. The lncRNA, NEAT1, which is upregulated in the PBMCs of HCC patients, enhances the apoptotic death of CTLs so that the cytolysis of cancer cells is inhibited. The paradigm that emerges from both in vitro and in vivo studies with the use of the HCC model system indicates that NEAT1 binds to miR-155, quenching its repression on the expression of T-cell immunoglobulin and mucin-domain containing-3 (Tim3), which promotes apoptosis in CTLs, thus attenuating the CTL-mediated cytolysis of cancer cells [118]. A different mechanism appears to be used by the lncRNA, lnc-TIM-3, to inhibit the cytolytic function of CTLs in HCC [119]. Using the ectopically expressed lnc-TIM3, it has been shown that it binds to and inhibits the interaction of TIM3 with the HLA-B-associated transcript 3 (BAT3) and BAT3-mediated downstream signaling. This leads to CD8+T cell exhaustion that contributes to compromised anti-tumor immunity and tumor progression [119]. Thus, the increased expression of both NEAT1 and lnc-TIM3 in the CTLs in HCC could contribute to the inactivation of functional CTLs through different mechanisms. In breast cancer, the lncRNA NKILA (NF-κB-interacting lncRNA) is involved in promoting the activation-induced cell death of CTLs and type 1 helper T-cells to facilitate immune evasion by cancer cells. NKILA has been shown to induce the tumor-antigen-activated death of T-cells by physically interacting and inhibiting the activities of IκBα and NF-κB [120]. Although the role of the lncRNA, lincMAF4 (Long-intergenic Non-coding Macrophage-activating Factor Transcriptional Regulator RNA 4) in cancers remains to be established, lincMAF4 could play a role in cancer-immune evasion through its potent inhibitory effect on T-cell differentiation [121]. It has been shown that lincMAF4 can epigenetically silence the transcriptional activation of the transcription factor MAF [121]. Since MAF is involved in the functional differentiation of regulatory T-cells (Tregs) and T-helper cells through the transcriptional activation of interleukin-4 and -10 [122], the lincMAF4-MAF link can be anticipated to have a potential role in cancer cells’ immunoevasion.
Amongst the T-cell-mediated adaptive immune response, one of the prominent tumor-infiltrating lymphocytes (TILs) that aid tumor invasion by creating immune suppressive milieu in the tumor microenvironment are the Tregs, whose maintenance and differentiation are critically regulated by lncRNAs. Studies with HCC patient-derived CD4+ T cells and xenograft mouse models indicate that lnc-EGFR (long non-coding epidermal growth factor receptor) plays a role in Treg differentiation and suppresses CTLs by binding to EGFR, so as to inhibit the signaling axis involving the transcription factors’ early growth response gene 1, and transcription factor forkhead box p3 (Foxp3) [123]. Since there are two splice variants of lnc-EGFR, namely lnc-EGFR-1 and lnc-EGFR-3, it is not clear whether both lnc-EGFRs can bind to EGFR to inhibit tumor immunoescape. A mechanism based on the ability of the lncRNA, FLICR, to epigenetically suppress the expression of FOXP3 to alleviate its tumor-suppressive functions in Tregs, has also been proposed as a potential mechanism through which cancer cells can escape immune surveillance [124]. Recent studies have shown that indolamine-2, 3-dioxygenase (IDO) is an immunoregulatory enzyme involved in tryptophan metabolism. By catabolizing tryptophan into kynurenine, IDO suppresses T-cell activities and promotes immune tolerance for cancer cells. The lncRNA, SNHG1, increases the levels of IDO in HCC cells by sequestering miR-448, which targets IDO, thus suppressing the T-cell-mediated immune response [125]. In addition, lnc-SOX5 has been reported to upregulate IDO through an unknown mechanism in CRC cells, and this has been shown to play an immuno-suppressive role promoting CRC xenograft tumor growth [126]. Immune checkpoint proteins such as programmed death ligand-1 (PD-L1), as well as cytotoxic T-lymphocyte associated protein-4 (CTLA4), are also targeted by lncRNAs to evade immune surveillance. In silico analyses have indicated a role for lncRNA RP11-571M6.8 in reducing the expression levels of PD-1, PD-L1 and CTLA4 [127]. Direct evidence for the ability to modulate the expression levels of PD-1 is shown by LINC00473, SNHG20, and MALAT1 in different cancers [128,129,130]. LINC00473 upregulates the expression of PD-L1 in pancreatic cancer cells by sequestering PD-L1-targeting miR-195-5p [128]. In esophageal squamous-cell carcinoma (ESCC), the lncRNA, SNHG20, enhances the expression as well as the activation of ATM, Janus kinase (JAK), and PD-L1, thus aiding compromised anti-tumor immunity [129]. The lncRNA, MALAT1, regulates the NF-κB-dependent innate immune response, in addition to sequestering miR-195 and upregulating PD-L1, resulting in CTL apoptosis as well as an immune escape in large B-cell lymphoma [130]. Similar to the effects of the T-cell-mediated immune response, B-cell humoral responses are also regulated by lncRNAs. The aberrant levels of soluble FAS receptors in B-cell lymphoma have been associated with the PRC2-EZH2-mediated repression of FAS gene expression by lncRNA FAS-AS1 in B-cell lymphoma cells [131]. The lncRNA, DLEU2 (deleted in lymphocytic leukemia-2), controls B-cell proliferation by sequestering the miRNA15a/16-1 axis, which targets several cyclins and cyclin-dependent kinases [132]. The lncRNA, MIAT (Myocardial Infarction-Associated Transcript), is essential for B-cell survival and proliferation through its positive regulation on the transcription factor Oct-4 (octamer-binding transcription factor-4) [133]. Thus, MIAT can play a determinant role in B-cell immunity against cancer.
The major immune cells involved in innate immune responses are natural killer cells (NKs), macrophages, and myeloid-derived suppressor cells (MDSCs). The sequestration of miRNA is one of the mechanisms through which lncRNAs modulate the differentiation and/or activities of these cells. In the case of NK cells, TS-lncRNA GAS5 has been shown to exert its inhibitory effect on NK cells through the sequestration of miR-554. GAS5 binds to miR-554, which targets the runt-related transcription factor-3 (RUNX3) and interferon-γ (IFN-γ) in liver cells. Thus, the decreased levels of GAS5 in HepG2 and Huh7 liver cancer cells result in the reduced expression of the transcription factor RUNX3 and interferon-γ levels due to increased miR-544 activity. This results in the overall reduction in the number of CD107a+ NK cells. Both of these events suppress NK cell-mediated cytotoxicity against HCC cells [134]. In the case of macrophages, lncRNAs have been shown to be involved in the pathobiology of both M1 and M2 macrophages. Activated M1 macrophages are involved in anti-tumor immune responses, whereas M2 polarization/differentiation results in the formation of tumor-associated macrophages (TAMs) that facilitate tumor progression. Using RAW264.7 macrophages, it has been shown that lincRNA-COX2 (Long Intergenic Non-coding RNA to Cyclooxygenase 2) promotes the anti-tumor functionality of M1 macrophages. In these cells, the lipopolysaccharide-induced M1 polarization and maintenance of M1-phenotype of the macrophages are very much dependent on lincRNA-COX2, and the silencing of lincRNA-COX2 results in the attenuation of M1 macrophage-mediated inhibition of HCC cell proliferation. The resultant strengthening of the functionality of M2 macrophages promotes HCC growth, both in vitro and in vivo [135]. A few of the lncRNAs are directly involved in promoting M2-polarization and the immunoescape of cancer cells. The lncRNA, LNMAT1 (Lymph Node Metastasis Associated Transcript 1), acts as scaffold and recruits hnRNPL to the promoter of C-C-motif chemokine ligand-2 (CCL2), which results in the H3K4-trimethylation-mediated transcriptional activation of CCL2, as well as CCL2-responsive genes. This promotes the M2-polarization of macrophages and upregulated expression of vascular endothelial growth factor-C (VEGF-C), which triggers lymphatic metastasis in bladder cancer [136]. The CCL2 conduit is also utilized by HOTAIR to promote M2-polarization in many different cancers [137]. The lncRNA MM2P (modulator of macrophage M2 polarization) is overexpressed in M2 macrophages. MM2P has been shown to promote the M2 polarization of macrophages by reducing the levels of phosphorylated STAT6 through a (yet to be defined) phosphatase-dependent mechanism [138]. Validating this paradigm, the silencing of MM2 negatively affects the M2-driven tumorigenesis and tumorangiogenesis of osteosarcoma xenograft tumor growth in mice [138]. In contrast to the lncRNAs that promote M2 polarization, the TS-lncrRNA, CASC2c, has been shown to inhibit the M2 polarization of macrophages through a different mechanism in glioblastoma multiforme context. CASC2c inhibits M2 polarization in glioblastoma multiforme cells by acting in concert with miR-338-3p to inhibit the expression and release of the coagulation factor X (FX), a pro-M2-polarization factor [139]. The subsequent disruption of the paracrine loop involving FX leads to a reduction in M2 polarization of macrophages.
MDSCs are another heterogeneous class of immune cells which accumulate in cancers and enhance tumor progression [140]. Upon stimulation by pro-inflammatory cytokines, such as IL-6 and granulocyte-macrophage colony-stimulating factor (GM-CSF), lncRNA olfactory receptor 29 pseudogene 1 (Olfr29-ps-1) is upregulated via JAK3/STAT3 signaling. Olfr29-ps-1 sequesters miR-214-3p and facilitates the differentiation of monocyte-MDSCs [141]. The lncRNA lnc-CHOP (Long Non-coding C/EBP Homologous Protein) interacts with the CHOP-CCAAT-enhancer-binding protein β (C/EBPβ) complex and induces the H3K4me3-mediated transcriptional activation of the genes encoding arginase 1, cyclooxygenase 2, NADPH oxygenase 2, and nitric oxide synthase 2, favoring the immunosuppression and differentiation of monocyte-MDSCs [142]. In lung cancers, the lncRNA RUNXOR (RUNX1 overlapping RNA) reduces RUNX1 expression, favoring MDSC differentiation and tumor progression [143]. The hypoxia-induced lncRNA, PVT1, was upregulated in monocyte-MDSCs as well as granulocyte-MDSCs, which increase ROS and Arg1 expression, thereby enhancing immunosuppressive functions in lung cancer cell lines as well as the murine model of lung cancer [47]. Thus, lncRNAs that regulate tumor-immune responses are emerging as precise immunotherapeutic targets for cancers.
3.5. lncRNAs in EMT and Metastasis
The advancing stages of cancers are marked by their prominent characteristics involving EMT and the invasive migration of cancer cells, as well as metastasis to distant sites, creating secondary tumors. Many different lncRNAs are critically involved in regulating the expression of genes involved in these pathological events. A host of oncogenic lncRNAs induce the EMT of cancer cells through epigenetic mechanisms. The lncRNA, HOTAIR, induces epigenetic histone marks through its interaction with either PRC2 or LSD1. Using both in vitro cellular and in vivo mouse models, it has been established that the HOTAIR-PRC2 complex induces a repressive H3K27 methylation on tumor-suppressive genes such as junctional adhesion molecule-2, protocadherin-10, PTEN, or protocadherin beta-5 [30]. On the other hand, in cellular as well as murine models of breast cancer, the HOTAIR-LSD1 complex induces H3K4 demethylation histone marks at the promoters or enhancers of oncogenic genes such as LDHA, Cyclin A, or eIF4E, to promote EMT [56]. Likewise, the MALAT1-PRC2 complex represses the expression of tumor-suppressor genes such as p21 and p27 through H3K27 trimethylation, to promote EMT and metastasis in mantle cell lymphoma cell lines [30]. Both in vitro and in vivo studies in bladder cancer have shown that H19 interacts with EZH2 to repress nucleoside diphosphate kinase 1, a Wnt-signaling antagonist, via H3K27 trimethylation, to enhance EMT through Wnt signaling [144]. The lncRNA, SChLAP1 (SWI/SNF complex antagonist associated with prostate cancer 1), binds to EZH2 and induces repressive H3K27 trimethylation on the promoters of miR-340-5p and miR-143-3p, which target the DNA methyltransferase, DNMT3a. This leads to the upregulation of DNMT3a, which methylates the promoters of tumor-suppressor genes that suppress metastasis, thus promoting cancer metastasis in prostate cancer cells [145]. The lncRNA, UFC1, interacts with EZH2 to induce H3K27me3-repressive methylation on the PTEN promoter to inhibit its expression and thereby promote the invasive metastasis of NSCLC cells, both in vitro and in vivo [46].
Many of the lncRNAs also utilize their roles as ceRNAs to modulate EMT and metastasis. HOTAIR acts through the sequestration of miR-23b-3p in HCC and the sequestration of miR-1227-5p in gastrointestinal (GI) cancer cell lines and tumor xenografts to upregulate the expressions of zinc finger E-box-binding homeobox-1 (ZEB1) and collagen type V alpha 1 chain (COL5A1), respectively, to induce EMT and metastasis [146]. MALAT1 acts as a ceRNA to sequester miR-126-5p, through which it promotes the expressions of snail family transcriptional repressor 2 (SLUG) and the transcription factor, TWIST, to induce EMT and metastasis in CRC [147]. Using HCC cell lines, it has been shown that both MALAT1 and lncRNA-ATB stimulate the expression of the ZEB family of transcription factors through the sequestration of two different miRNAs to promote EMT. While MALAT1 sequesters miR-143-3p to upregulate the expression of ZEB1, lncRNA-ATB competitively binds to miR-200 to upregulate ZEB1/2 [148]. In both the in vitro as well as the in vivo models of GI cancer, PVT1 has been shown to sequester miR-30a-5p, through which it relieves the inhibitory effect of miR-30a-5p on the expression of snail family transcriptional repressor 1 (SNAIL), ZEB1/2 and neural-cadherin (N-cadherin), to facilitate EMT and metastasis [29]. In osteosarcoma, both in vitro and in vivo studies have shown that TGF-β secreted by cancer-associated fibroblasts induce the lncRNA TUG1, which sequesters miR-143-5p to upregulate hypoxia-inducible factor-1α (HIF-1α), and promotes metastasis [149]. In cellular as well as murine models of CRC, H19 is shown to sequester miR-138 and miR-200a to upregulate ZEB1/2 to promote EMT [144]. Similar lncRNA, miRNA, and target gene nexuses are seen to promote EMT and metastasis in many other cancers. The lncRNA HULC has been shown to act through the miR-200a-3p/ZEB1 axis in HCC cell lines and mouse models [150]. LINC00460 has been observed to quench miR-433-3p to increase the expression of ANXA2, Vimentin, and N-Cadherin to induce EMT in both cellular and xenograft mouse models of colon cancers [151]. NEAT1 has been shown, both in vitro and in vivo, to restrain miR-141-3p from inhibiting the expression of KLF12 in breast cancers [152]. UFC1 has also been shown to sequester miR-34a to inhibit its repressive effect on the expression of CXCL-10 involved in invasive metastasis [153].
In addition to epigenetic and ceRNA roles, lncRNAs also act as scaffold to regulate EMT and metastasis. Increased levels of HOXA11-AS serve as scaffold for LSD1 or DNMT1, as well as ceRNA for miR-1297, which targets EZH2 to promote tumor invasion and EMT in GI cancers [154]. Using a panel of GI cancer cell lines and xenograft mouse models, the HOXA11-AS/DNMT1/EZH2 complex has been shown to repress KLF2 expression, while the HOXA11-AS/LSD1/EZH2 complex represses PRSS8 expression. The decreased expression of the tumor suppressors KLF2 and PRSS8 leads to an overall increase in the proliferation and migration of the GI cancer cells [154]. In breast cancer, the lncRNA, BCAR4 (Breast Cancer Anti-estrogen Resistance-4), is overexpressed in breast cancer cells and mediates C-C motif ligand-21 (CCL21) and C-C motif chemokine receptor-7 (CCR7)-induced EMT [28]. BCAR4 acts as scaffold for phospho-Gli-2 and protein phosphatase 1 nuclear targeting subunit (PNUTS), and Smad nuclear-interacting protein-1 (SNIP1), to regulate p300-mediated histone acetylation as well as RNA Pol II activities. Activated phospho-Gli2 stimulates the expression of the transforming growth factor beta 1 (TGFβ1), IL-6, protein patched homologue-1 (PTCH1), and mucin 5AC (MUC5AC) to promote EMT and metastasis [155]. The lncRNA CCAT2 interacts with the transcription factor TCF7L2 (Transcription Factor 7 like 2) to transactivate the expression of WNT target genes, including MYC, to promote invasive cell migration in colon cancer cells [156].
TS-lncRNAs are also involved in the regulation of invasion, migration, EMT and metastasis in cancers. In HCC cells, lincRNA-p21 has been shown to inhibit the repressive effect of miR-9 on the expression of E-cadherin to suppress the invasive migration of HCC cells. Thus, the reduced expression of lincRNA-p21 is correlated with the invasive migration of HCC cells [157]. In the context of glioma and TNBC, the expression levels of the lncRNA, GAS5, are inversely correlated with the size, staging, and metastasis of the respective cancers. GAS5 acts as a ceRNA to quench miR-196a-5p that targets FOXO1 expression, thereby increasing the expression of FOXO1 and FOXO1-mediated inhibition of PI3K/AKT activation, involved in the growth and invasive migration of glioma stem cells and TNBC cells [60,61,158].
3.6. lncRNAs and Tumor Angiogenesis
Angiogenesis is defined as the process associated with the formation of new blood vessels. Cancer growth and progression, including metastatic tumor growth, require angiogenesis to provide nutrients and oxygen for the growing tumors, in addition to the disposal of metabolic wastes. Numerous growth factors, such as vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), epidermal growth factor (EGF), transforming growth factor (TGF), and angiopoietins, as well as signaling cascades such as STAT-3, Wnt/β-Catenin, mTOR (mechanistic target of rapamycin), and NF-κB also engage in activating the pro-angiogenic mechanisms. In addition to cancer cells, numerous other cell types within the tumor microenvironment, such as TAMs, are also involved in initiating angiogenesis, thereby recruiting several cell types such as endothelial progenitor cells, haematopoietic stem cells, and mesenchymal stem cells to initiate blood vessel formation and thence, angiogenesis [159]. The hypoxic conditions in tumors activate the hypoxia-inducible factors (HIFs), which are translocated to the nucleus and escape its degradation in the cytoplasm, which further transactivates the expression of multiple genes to activate angiogenesis. A broad spectrum of lncRNAs regulate these pro-angiogenic factors, so as to regulate tumor angiogenesis.
The lncRNA, PVT1, binds to phospho-STAT3 and prevents it from ubiquitin-mediated proteolytic degradation, which then upregulates VEGFA expression, enhancing tumor angiogenesis in gastric cancer cells [160]. In cellular and murine models of lung carcinoma, LINC00665 has been shown to interact with the Y-box-binding transcription factor YB-1, to protect it from ubiquitin-mediated proteolytic degradation. YB-1, thus stabilized, binds to the promoters of angiopoietin-4, angiopoietin-like 3, and VEGFA to enhance their expressions and promote tumor angiogenesis [161]. Similarly, lncRNA olfactory receptor family 3 subfamily A member 4 pseudogene (OR3A4) promotes tumor angiogenesis by associating with the transcription factor AGGF1 (angiogenic factor with G-patch and FHA domains 1), which transactivates the expression of VEGF, FGF and Ang1 in HCC cells [162]. Tumor angiogenesis in HCC appears to also involve the lncRNA, MVIH (microvascular invasion in hepatocellular carcinoma), through a different mechanism. Using HCC tissues and cell lines, MVIH has been shown to physically interact with phosphoglycerate kinase (PGK) to block its secretion. Since the secreted PGK1 is known to inhibit angiogenesis, the MVIH-mediated block in PGK1 secretion relieves this inhibition and promotes tumor angiogenesis [163]. LncRNA-CamK-A (calcium-dependent kinase activation) in breast cancer cells activates PNCK through physical association to stimulate the expression of cytokines such as IL-6, IL-8, and VEGF through the NFκB signaling pathway. These cytokines promote tumor angiogenesis, as demonstrated by a patient-derived xenograft model using TNBC cells overexpressing lncRNA-CamK-A [164].
The lncRNA-mediated sequestration of specific miRNAs also plays a role in tumor angiogenesis. Tumor angiogenesis in gastric cancer has been shown to involve the ability of LINC01410 to sequester miR-532-5p, which represses the expression NCF-2 and NF-κB. The consequent upregulation of neutrophil cytosolic factor-2 (NCF-2)- and NF-κB-mediated signaling pathways promote angiogenesis in gastric cancers [165]. The lncRNA, TUG1, acts through the miR-299/VEGFA axis in glioblastoma, while it upregulates HIF-1α through miR-143-5p in osteosarcoma to promote angiogenesis [149]. The lncRNA, MALAT1, also promotes angiogenesis, through the miR-140/VEGFA axis in HCC, or through the miR-145/VEGFA axis in breast cancer cells [148]. Other examples of lncRNAs which act through miRNA sequestration to promote angiogenesis include DANCR (Differentiation Antagonizing Non-protein Coding RNA) through the miR-145/VEGF axis, SCAMP1 (Secretory Carrier Membrane Protein 1) through the miR-137/CXCL-12 axis, and MEG3 through the miR-421/PDGFR-A (platelet-derived growth factor receptor-A) axis, all of which are regulated in ovarian cancers [166,167,168]. In HCC, the lncRNA HULC stimulates an increase in the activation of sphingosine kinase 1 (SPHK1), a kinase that promotes tumor angiogenesis [169]. In cytosol, HULC sequesters miR-107 to upregulate the levels of the transcription factor E2F1, which targets SPHK1. In the nucleus, HULC promotes the expression of SPHK1 by recruiting E2F1 onto the SPHK1 promoter, [169].
In addition to the cancer cells, lncRNAs also influence the cells in the tumor microenvironment, as well as the cancer stem cells, to regulate angiogenesis. The lncRNA-MM2P promotes the macrophage M2-mediated angiogenesis through the direct binding and stabilization of phospho-STAT-6. The depletion of lncRNA-MM2P reduces osteosarcoma xenograft tumor growth and tumor angiogenesis [138,170]. Furthermore, the lncRNA, POU3F3 (POU class 3 homeobox 3), released through exosomes from tumor cells, is reported to be internalized by endothelial cells to enhance endothelial cell-based angiogenic signaling through the upregulation of bFGF (basic fibroblast growth factor), VEGFA, bFGFR (basic FGF receptor), and Angio in glioma cells [171]. However, the detailed mechanism behind the upregulation of these pro-angiogenic factors is yet to be elucidated. In HCC, CD90+ cancer stem cells release exosomal H19, which modulates endothelial cells by upregulating the expression of VEGF and VEGFR (VEGF receptor), thereby promoting angiogenesis [172]. Certain angiogenic lncRNAs also epigenetically regulate critical angiogenic factors. For instance, the lncRNA, SNHG14, induces cytoplasmic polyadenylate-binding protein 1 (PABPC1) expression via H3K27 acetylation. Increased levels of PABPC1 inhibit PTEN signaling, presumably through its inhibitory effect on PTEN mRNA stability, which further induces angiogenesis in both the cellular and xenograft models of HCC [127]. In CRC cells, LINC00337 recruits DNMT1 to suppress CNN1 (calponin 1) expression, thus relieving its repressive effect on VEGF-mediated angiogenesis [173,174]. In TAMs, the lncRNA, MALAT1, stimulates FGF-2 expression and associated paracrine signaling pathways to promote angiogenesis in thyroid cancers [175]. Although the mechanism by which MALAT1 upregulates the expression of FGF2 is not defined, MALAT1 has been shown to promote tumor angiogenesis in neuroblastoma via FGF2 [176].
TS-lncRNAs also contribute to the regulation of tumor-angiogenic pathways. In vitro and in vivo studies have shown that the lncRNA, GAS5, inhibits the Wnt/β-catenin pathway through the downregulation of β-catenin, Cyclin D1, and c-Myc in CRC, thereby attenuating angiogenesis [177]. The lincRNA-p21 has also been reported to regulate angiogenesis under hypoxic conditions in NSCLC through the modulation of MMP-2 (matrix metalloproteinase 2), FGF-2, VEGFA, and PDGFB (platelet-derived growth factor β) expressions [178]. MEG3 has tumor-suppressive functions and inhibits cancer cell proliferation, as well as induces apoptosis, while its knockdown promotes angiogenesis. In breast cancer cells, MDA-MB-231 and MCF-7, as well as nude mouse xenograft models, it has been shown that MEG3 suppresses AKT pathways and downstream effectors such as MMP-9 and VEGFA, to inhibit angiogenesis [179]. Furthermore, HIF-1A-AS2, an antisense lncRNA to HIF-1α, binds to and inhibits HIF-1α, to inhibit tumor angiogenesis in the OVCAR-8 ovarian cancer cell line, as well as the LNCaP prostate cancer cell line and tumor xenografts [180].
3.7. lncRNAs and Cancer Stemness
Cancer stemness plays a critical role in disease recurrence and therapy resistance [181]. Many different lncRNAs significantly contribute to cancer stemness. Cancer stemness is marked by the expression of several unique transcription factors [182]. A large number of lncRNAs have been shown to modulate the expression of one or more of these transcription factors to confer or maintain cancer stemness [183,184]. The antisense lncRNA, LAMP5-AS1 (antisense-to-lysosomal-associated membrane protein family member 5), interacts with and activates methyl transferase activity of the enzyme disruptor of telomerase-silencing-1 (DOT1L), promoting H3K79 di- and tri-methylations on the promoters of DOT1L-regulated genes to augment cancer stemness in mixed-lineage leukaemia [185]. The lncRNA-mediated sequestration of miRNA also plays a role in the regulation of cancer stemness. UCA1, transmitted exosomally from the cervical cancer cell line CaSki, has been shown to sequester miR122-5p and upregulate SOX2 expression, which promotes cancer stemness in these cells [186]. UCA1 has also been shown to promote stemness in glioma cells by inducing the expression of SLUG through the sequestering of both miR-1 and miR-203a [187]. Using CRC cells, it has been reported that H19 upregulates the expression of c-Myc, SOX-2, and Oct-4 to induce stemness. In this model, it has also been shown that the exosomal-derived H19 from the cancer-associated fibroblasts upregulates the expression of stemness-associated factors through the β-catenin signaling cascade by sequestering miR-141 [188]. Stemness in CRCs is also mediated by the lncRNA FARSA-AS1 (Phenylalanyl-tRNA Synthetase Subunit Alpha Antisense RNA1) [189]. Here, FARSA-AS1 increases the expression levels of SOX-9 by sequestering miR-18b-5p. More interestingly, SOX-9 also increases the expression levels of FARSA-AS1 by activating its transcription by binding to its promoter, thus establishing a sustained positive feedback loop to maintain cancer stemness. The lncRNA, MALAT1, has been shown to modulate cancer stemness through the quenching of miR-375 to upregulate the expression of YAP1 (Yes-associated transcriptional regulator) in liver cancer cells [190]. In gastric cancer cells, linc-ROR (long intergenic non-coding RNA—regulator of reprogramming) has been reported to enhance the expression of stemness-associated transcription factors such as Oct4, SOX-2, and Nanog through the sequestration of miRNAs [191].
In the tumor-initiating cells of renal cell carcinoma, lncARSR (lncRNA activated in renal cell carcinoma with sunitinib resistance) interacts with YAP1 and inhibits YAP1 phosphorylation, facilitating the nuclear translocation of YAP1. YAP1 upregulates the expression of Oct-4, Nanog, and SOX-2 to promote the self-renewal of CSCs and metastasis [192]. Several lncRNAs promote cancer stemness in breast cancer cells through multiple mechanisms. HOTAIR represses the expression of HoxD10 through PRC2-mediated H3K27 trimethylation, which leads to a decrease in the levels of miR-7. The resultant increased expression and activation of STAT3 promotes stemness in breast cancer cells via c-Myc and TWIST [146,193]. The lncRNA-Hh, which is regulated by TWIST, promotes stemness in MCF7 breast cancer cells by stimulating hedgehog signaling-mediated expression of Oct-4 and Sox-2 [194]. Examples of the lncRNAs acting through hedgehog signaling also include lnc-HDAC2 and lnc-HOXA10. In the liver-cancer-tumor-initiating cells, lnc-HDAC2 activates the hedgehog signaling pathway, and lnc-HOXA10 initiates the transcriptional activation of HOXA10. Together, these lncRNAs stimulate the self-renewal and tumor progression of their CSCs [195]. In the context of liver cancer, lncRNA-SOX4 has been shown to recruit STAT3 to the SOX4 promoter to enhance its expression, thereby promoting stemness [196]. In CRC cells, lncGATA6 (lncRNA GATA binding protein 6) recruits a nucleosome remodeling factor (NURF) complex to the promoter of Ehf (ETS homologous transcription factor) to induce its expression, which leads to Lgr4/5 expression, LGR4/5-mediated activation of Wnt signaling, and CSC renewal and maintenance [197].
TS-lncRNAs are also associated with cancer stemness. Reduced expression of DILC (lncRNA Downregulated in Liver Cancer Cells), a TS-lncRNA, has been correlated with cancer stemness in HCC [198]. DILC inhibits IL-6 expression by binding to its promoter and thereby attenuating the STAT3 signaling and cancer stemness. Similarly, lncRNA-LBCS (lncRNA Bladder and Prostate Cancer Suppressor) is downregulated in bladder cancer stem cells. This lncRNA, otherwise, would bind to hnRNPK and EZH2, acting as scaffold, to induce the H3K27-repressive tri-methylation of SOX-2 and repress its expression [199].
3.8. lncRNAs and Therapy Resistance
Therapy resistance contributes greatly to cancer mortality. A number of lncRNAs have been reported to play a role in either promoting or inhibiting therapy resistance in cancers. Generally, lncRNAs that promote radiosensitivity include those with tumor-suppressive activity, and the primary mechanism appears to be through the sequestration of miRNAs that target tumor suppressors. NEAT1 enhances chemosensitivity in nasopharyngeal carcinoma through the sequestering of miR-101, which targets EMP2 (epithelial membrane protein 2 [200]. Similarly, OIP5-AS1 has been shown to play a role in the chemoresistance of CRCs via its interaction with miR-369. Using the CRC cell lines LoVo and SW280, it has been shown that the sequestering of miR-369 by OPI5-AS1 leads to the increased expression of dual-specificity tyrosine phosphorylation-regulated kinase-1A (DYRK1A), which is involved in suppressing cell proliferation and therapy resistance [201]. LincRNA-p21 has been shown to promote sensitivity to radiation therapy by inhibiting the expression of β-catenin at both the mRNA and protein levels in gastric as well as CRC cells [202,203]. As can be predicted, oncogenic lncRNAs enhance the radiotherapy resistance in numerous cancers. UCA1 regulates the cell-cycle signaling molecules such as focal adhesion kinase, AKT, FGR-tyrosine kinase, AMP-activated protein kinase, and AMPKα1 to contribute to radiotherapy resistance in prostate cancer [204]. MALAT1 confers radiotherapy resistance in NPC by sequestering miR-1 to enhance the expression of SLUG, whereas LINC00963 promotes radiotherapy resistance in breast cancer by sequestering miR-324-3p to increase the expression of activated CDC42 kinase 1 [205]. In the case of resistance to chemotherapy, it could be against a single drug or to a broader spectrum of drugs. Many lncRNAs confer drug resistance, either by stimulating signaling pathways that can override drug sensitivity or by increasing the expression of proteins involved in drug efflux, such as multiple-drug resistance (MDR) or multi-drug-resistance-associated proteins (MRP) [206]. Several lncRNAs, such as MALAT1 and HOTAIR, induce drug resistance by modulating the expressions of proteins involved in the influx and efflux of therapeutic agents, or by altering specific signaling pathways. MALAT1 was first identified as overexpressed in lung cancer metastasis, and induces MDR by upregulating MRP1, as well as MDR1 proteins and STAT3 activation, to induce drug efflux [207]. MALAT1 has been associated with gefitinib resistance in lung cancer by sequestering miR-200a, thereby upregulating ZEB-1 expression, which is involved in the regulation of MDR proteins [208]. However, in gastric cancers, MALAT1 enhances ZFP91 (zinc finger protein 91 homologue) expression through the sequestration of miR-22-3p, thereby inducing oxaliplatin resistance [209]. HOTAIR has been reported to induce doxorubicin resistance in breast cancers via the activation of PI3K/AKT/mTOR pathways and upregulating the MDR1, MRP1 and ABCB1 (ATP-binding cassette subfamily B member 1) expressions [210]. In addition, HOTAIR also induces cisplatin resistance in NSCLC via the upregulation of MDR1, MRP1, and Wnt/β-catenin signaling [211]. HOTAIR induces paclitaxel and doxorubicin resistance in gastric cancers by sequestering miR-217, targeting glypican 5 (GPC5) and protein tyrosine phosphatase non-receptor type 14 (PTPN14) [212]. It also induces cisplatin resistance in gastric cancers by targeting miR-126, which targets and activates PI3K/AKT/MRP1 proteins [213]. It has been reported that the natural product, curcumin, inhibited the HOTAIR/miR-20a-5p/WT-1 transcription factor signaling axis to reduce the adriamycin resistance in acute myeloid leukemia (AML), thus suggesting the potential role of the HOTAIR/miR-20a-5p/WT-1 axis in conferring adriamycin resistance in AML [214].
In gastric cancers, UCA1 sequesters miR-27b, which upregulates cyclin G1 (CCNG1). CCNG1 enhances p53 expression, as well as downregulates miR-508-5p, thereby inducing MDR [215,216]. Several other lncRNAs, such as TRPM2-AS (antisense lncRNA to transient receptor potential cation channel subfamily M member 2), which acts through the miR-138-5p/EGFR/PI3K/AKT axis in NSCLC; lncRNA OIP5-AS1, which acts through miR-340-5p/LPAATβ (lysophosphatidic acid β)/PI3K/AKT/mTOR axis in osteosarcoma; lncRNA H19, which acts through the miR-107/HMGB1 (high mobility group box 1) axis in laryngeal squamous cell carcinoma; lncRNA SNHG7, which acts through miR-34a in breast cancers; lncRNA BLACAT1 (bladder cancer-associated transcript 1), which acts through the miR-519d-3p/CREB1 (cAMP-responsive element-binding protein 1) axis in CRCs; lncRNA TTN-AS1 (antisense lncRNA to titin), which acts through miR-16-5p/Cyclin E1 in HCC, lncRNA DICER-AS1 (antisense lncRNA to DICER ribonuclease), which acts through miR-34a-5p in osteosarcoma; and lncRNA PCGEM1, which acts through the miR-129-5p/EVT1 (ETS variant gene 1) axis, alter the MDR proteins to confer drug resistance [206].
In addition, some of these lncRNAs overcome drug resistance by stimulating the expression of signaling proteins that can overcome drug sensitivity. The exosomal lncRNA, SOX2-OT (SOX-2 overlapping transcript), sequesters miR-627-3p, which targets Smad2/3/4, and activates Smad signaling. This promotes macrophage M2 polarization, resulting in the production of TAMs to enhance resistance to EGFR-TKIs (EGFR-tyrosine kinase inhibitors) in NSCLC [217]. The extra-vesicular NEAT1 sequesters miR-141-3p and upregulates KLF12 to induce resistance to paclitaxel, cisplatin, and 5-fluorouracil in breast cancer model [152]. The exosomal HOTTIP inhibits miR-214, thereby upregulating karyopherin subunit alpha 3 (KPNA3) expression to promote mitomycin resistance in CRC cancers [218]. The lncRNA, UCA1, induces tamoxifen resistance in breast cancers by associating with EZH2 to generate repressive H3K27me3 methylation on the p21 promoter, thereby downregulating p21 expression and activating PI3K/AKT signaling [219]. UCA1 induces cisplatin resistance in gastric cancers through its association with EZH2 and the activation of PI3K/AKT signaling through upregulation of p-PI3K and p-Akt [220]. UCA1 has also been reported to activate Wnt/β-catenin signaling, resulting in both tamoxifen and cisplatin resistance in breast and bladder cancers, respectively [221,222]. HOTAIR upregulates DNMT3b levels and induces the promoter hypermethylation of PTEN, which downregulates PTEN expression, leading to drug resistance in AML [223]. LINC00665 interacts with EZH2 and induces Akt expression to activate the PI3K/AKT pathway, thereby leading to the gefitinib resistance in NSCLC [224]. LINC00152 interacts with EZH2 to induce ZEB1 expression, thereby leading to EMT and oxaliplatin resistance in esophageal cancers [225]. In gliomas, the lncRNA DLEU1 (Deleted in Lymphocytic Leukemia 1) generates temozolomide resistance by regulating autophagy [226]. Similarly, HOTAIR has been reported to induce autophagy and imatinib resistance in GI cancers through the inhibition of miR-130a and the subsequent upregulation of ATG2B (autophagy-related 2B) levels, which promotes autophagy-mediated cell survival [227]. The lncRNA, PCAT-1 (Prostate Cancer Associated Transcript-1), activates AKT and GSK-3 (glycogen synthase kinase-3) phosphorylation, resulting in signaling which augments gefitinib resistance [228]. PVT1 inhibits kelch-like ECH-associated protein 1 (Keap1)–nuclear factor erythroid 2-related factor 2 (Nrf-2) association and stabilizes the latter, resulting in doxorubicin resistance in TNBCs [229]. Though these studies have shown an increased drug sensitivity through the knockdown of the lncRNAs, the exact mechanism has yet to be defined.
In addition to the oncogenic lncRNAs described above, TS-lncRNAs play a role in regulating drug resistance in cancers. The downregulated expression of GAS5 is inversely correlated with ABCB1 expression, as shown through silencing studies. GAS5, otherwise, targets miR-221-3p and upregulates DKK2 (Dickkopf Wnt Signaling Pathway Inhibitor 2) expression to inhibit the Wnt/β-catenin pathway, to promote adriamycin resistance in breast cancers [230]. In HCC, the downregulated GAS5 levels contribute to doxorubicin resistance via the activation of miR-222-3p and downregulation of PTEN [231]. In NSCLC, it has been demonstrated that the lncRNA AC078883.3 levels are downregulated, which increases miR-19a activity that targets PTEN. Similarly, downregulation of lncRNA TP53TG1 (TP53 Target 1), which otherwise suppresses miR-18a, reduces PTEN levels. Both these lncRNAs have been reported to contribute to cisplatin resistance in NSCLC through the regulation of PTEN [232,233]. The lncRNA FER1L4 (Fer-1-like Family Member 4 Pseudogene) levels are downregulated in ovarian cancers, which induces paclitaxel resistance through the activation of mitogen-activated protein kinase (MAPK) signaling [234]. The lncRNA, LINC00968 is downregulated in breast cancers and contributes to drug resistance through its inhibitory effects on Wnt signaling. LINC00968 interacts with the transcription factor HEY1 and inhibits Wnt2, thereby leading to Wnt/β-catenin signaling [235]. In pancreatic cancers, the lncRNA DLEU2L targets miR-210-3p and upregulates BRCA2 to reduce the gemcitabine resistance [236]. Targeting these oncogenic—as well as tumor-suppressive—lncRNAs, and their associated axes, will be an ideal therapeutic strategy to combat the therapy resistance in cancers.
4. Perspective and Conclusions
LncRNAs form a diverse class of non-coding RNAs which regulate the cellular processes, both at the physiological as well as pathophysiological states in the human body. They regulate cellular homeostasis through the modulation of gene expressions and protein levels, as well as their stability and interactions. As discussed here, lncRNAs critically participate in diverse cell signaling cascades, bringing out their multitudinous roles, ranging from embryonic development to disease biogenesis, and most importantly, cancer genesis and progression. From the initial oncogenic signals triggering the transformation of normal cells to the tumorous state, through the regulation of diverse functional attributes that promote tumor progression and metastasis, to the response to anti-cancer therapies, lncRNAs perform a cardinal role (Figure 3).
Figure 3.
Multifacted role of lncRNAs in tumorigenesis and tumor progression. The diverse classes of lncRNAs play a role in tumor progression by variably regulating the major arms of tumorigenesis, either by promoting cell proliferation, invasion, migration, metastasis, EMT, angiogenesis, drug resistance, cancer stemness, and metabolic reprogramming, or by inhibiting apoptosis, immune surveillance, and efficient DNA damage response within the cancer cells.
Non-coding RNAs were considered to be the dark matter of the genome, as they could not be translated into proteins and carry out cellular functions. The advent of the existence of functional non-coding RNAs such as that of ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), lncRNAs, small interfering RNAs (siRNAs), miRNAs, guide RNAs (gRNAs), piwi-interacting RNAs (piRNAs), and small nucleolar RNAs (snRNAs) has revolutionized the significance of transcriptomes. Deeper understanding of transcriptomes has transfigured the whole paradigm of cancer pathobiology. lncRNAs are now recognized as an active component of complex cellular regulatory networks, playing significant roles in cancer genesis, progression, and therapy resistance. Since lncRNAs lack sequence conservation, multiple factors such as secondary, tertiary, and quaternary structure–function relationships in their respective interactomes must be taken into account, in addition to their linear primary sequence. Thus, targeting lncRNAs in precision cancer medicine requires a clear understanding of their functional roles and underlying mechanisms in relation to their interactomes in specific cancers, as well as in normal cells. The present review has discussed the salient functional features of lncRNAs in cancer biology. However, it should be noted that further analyses of the expression profiles and functional mechanisms of lncRNAs could unravel them as effective diagnostic, prognostic, and therapeutic targets for cancers. This calls for conjoint research demanding various omics technologies such as genomics, transcriptomics, proteomics, and metabolomics, that would help towards deciphering the precise upstream and downstream events underlying the aberrant expression of lncRNAs in different cancers. With the advent of newer high-throughput technologies, the wait is not long for when tailored therapy involving lncRNAs will translate into clinics.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/onco1020014/s1, Table S1: Functional role of oncogenic lncRNAs in cancer; Table S2: Functional role of tumor-suppressor lncRNAs in cancer.
Author Contributions
Conceptualization: D.N.D.; Data curation, R.N.; Writing—original draft preparation, R.N.; Writing, revising, and editing, D.N.D. and R.N.; Supervision, D.N.D.; Project administration, D.N.D.; Funding acquisition, D.N.D. All authors have read and agreed to the published version of the manuscript.
Funding
The present research was supported by the Department of Defense Ovarian Cancer Research Program Award (grant no. W81XWH-18-1-0066) and The National Institutes of Health grant (grant no. GM103639). Computational services were supported by the National Institute of General Medical Sciences P20 (grant no. GM103639) and The National Cancer Institute of the National Institutes of Health (grant no. P30CA225520).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [Green Version]
- Tsagakis, I.; Douka, K.; Birds, I.; Aspden, J.L. Long non-coding RNAs in development and disease: Conservation to mechanisms. J. Pathol. 2020, 250, 480–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mole. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef]
- Taft, R.J.; Pheasant, M.; Mattick, J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 2007, 29, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [Green Version]
- Jeon, Y.; Lee, J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2011, 146, 119–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, C.; Brex, D.; Caponnetto, A.; Cirnigliaro, M.; Scalia, M.; Magnano, A.; Caltabiano, R.; Barbagallo, D.; Biondi, A.; Cappellani, A.; et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol. Therapy Nucleic Acids 2018, 12, 229–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rueff, J.; Rodrigues, A.S. Cancer Drug Resistance: A Brief Overview from a Genetic Viewpoint. Methods Mol. Biol. 2016, 1395, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, L.; Kriner, M.; Coleman, I.; Morrissey, C.; Roudier, M.; True, L.D.; Gulati, R.; Plymate, S.R.; Zhou, Z.; Birditt, B.; et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 2021, 12, 1426. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dang, H.; Wang, X.W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 2018, 50, e416. [Google Scholar] [CrossRef]
- Ramón y Cajal, S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Rosa, S.; Rubbia-Brandt, L.; Scoazec, J.-Y.; Weber, A. Editorial: Tumor Heterogeneity. Front. Med. 2019, 6, 156. [Google Scholar] [CrossRef] [Green Version]
- Seifuddin, F.; Singh, K.; Suresh, A.; Judy, J.T.; Chen, Y.C.; Chaitankar, V.; Tunc, I.; Ruan, X.; Li, P.; Chen, Y.; et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci. Data 2020, 7, 326. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Akimitsu, N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int. J. Mol. Sci. 2021, 22, 632. [Google Scholar] [CrossRef]
- Hua, J.T.; Ahmed, M.; Guo, H.; Zhang, Y.; Chen, S.; Soares, F.; Lu, J.; Zhou, S.; Wang, M.; Li, H.; et al. Risk SNP-Mediated Promoter-Enhancer Switching Drives Prostate Cancer through lncRNA PCAT19. Cell 2018, 174, 564–575.e518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aprile, M.; Katopodi, V.; Leucci, E.; Costa, V. LncRNAs in Cancer: From garbage to Junk. Cancers 2020, 12, 3220. [Google Scholar] [CrossRef]
- Wang, D.L.; Yuan, P.; Tian, J.Y. Expression of long noncoding RNA NBAT1 is associated with the outcome of patients with non-small cell lung cancer. Rev. Assoc. Med. Bras. 2020, 66, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ling, M.; Yang, S.; Xie, Y.; Liu, C.; Yi, W. Long noncoding RNA NBAT1 suppresses hepatocellular carcinoma progression via competitively associating with IGF2BP1 and decreasing c-Myc expression. Hum. Cell 2021, 34, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Olivero, C.E.; Martínez-Terroba, E.; Zimmer, J.; Liao, C.; Tesfaye, E.; Hooshdaran, N.; Schofield, J.A.; Bendor, J.; Fang, D.; Simon, M.D.; et al. p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cell 2020, 77, 761–774.e768. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xiao, B.; Yu, T.; Gong, L.; Wang, Y.; Zhang, X.; Zou, Q.; Zuo, Q. lncRNA PVT1 promotes the migration of gastric cancer by functioning as ceRNA of miR-30a and regulating Snail. J. Cell. Physiol. 2021, 236, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Piao, H.L.; Kim, B.J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Weidle, U.H.; Birzele, F.; Kollmorgen, G.; Rüger, R. Long Non-coding RNAs and their Role in Metastasis. Cancer Genom. Proteom. 2017, 14, 143–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.; Crenshaw, T.; Moulton, T.; Newcomb, E.; Tycko, B. Tumour-suppressor activity of H19 RNA. Nature 1993, 365, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Liu, X.H.; Wang, K.M.; Nie, F.Q.; Kong, R.; Yang, J.S.; Xia, R.; Xu, T.P.; Jin, F.Y.; Liu, Z.J.; et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol. Cancer 2014, 13, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Zhang, L.; Jia, L.; Duan, Y.; Li, Y.; Bao, L.; Sha, N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS ONE 2014, 9, e100893. [Google Scholar] [CrossRef] [Green Version]
- Ewald, P.W.; Swain Ewald, H.A. Toward a general evolutionary theory of oncogenesis. Evol. Appl. 2013, 6, 70–81. [Google Scholar] [CrossRef]
- Aktipis, C.A.; Boddy, A.M.; Jansen, G.; Hibner, U.; Hochberg, M.E.; Maley, C.C.; Wilkinson, G.S. Cancer across the tree of life: Cooperation and cheating in multicellularity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar]
- Cheng, Y.; Jutooru, I.; Chadalapaka, G.; Corton, J.C.; Safe, S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 2015, 6, 10840–10852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Wang, Y.; Tang, C.; Jones, L.; Ye, H.; Zhang, G.; Cao, W.; Li, J.; Liu, L.; Liu, Z.; et al. TP53, PIK3CA, FBXW7 and KRAS Mutations in Esophageal Cancer Identified by Targeted Sequencing. Cancer Genom. Proteom. 2016, 13, 231–238. [Google Scholar]
- Lu, K.; Tao, H.; Si, X.; Chen, Q. The Histone H3 Lysine 4 Presenter WDR5 as an Oncogenic Protein and Novel Epigenetic Target in Cancer. Front. Oncol. 2018, 8, 502. [Google Scholar] [CrossRef]
- He, X.; Wang, J.; Chen, J.; Han, L.; Lu, X.; Miao, D.; Yin, D.; Geng, Q.; Zhang, E. lncRNA UCA1 Predicts a Poor Prognosis and Regulates Cell Proliferation and Migration by Repressing p21 and SPRY1 Expression in GC. Mol. Therapy Nucleic Acids 2019, 18, 605–616. [Google Scholar] [CrossRef] [Green Version]
- Zang, X.; Gu, J.; Zhang, J.; Shi, H.; Hou, S.; Xu, X.; Chen, Y.; Zhang, Y.; Mao, F.; Qian, H.; et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020, 11, 215. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Tian, X.; Wang, T.; Xia, X.; Cao, F.; Tian, J.; Xu, P.; Ma, J.; Xu, H.; Wang, S. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol. Cancer 2019, 18, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yari, H.; Jin, L.; Teng, L.; Wang, Y.; Wu, Y.; Liu, G.Z.; Gao, W.; Liang, J.; Xi, Y.; Feng, Y.C.; et al. LncRNA REG1CP promotes tumorigenesis through an enhancer complex to recruit FANCJ helicase for REG3A transcription. Nat. Commun. 2019, 10, 5334. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, W.; Lian, J.; Zhang, H.; Yu, B.; Zhang, M.; Wei, F.; Wu, J.; Jiang, J.; Jia, Y.; et al. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2020, 27, 695–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Wang, Y.; Yuan, S.; Wen, F.; Liu, J.; Zou, L.; Zhang, J. Regulatory role of long non-coding RNA UCA1 in signaling pathways and its clinical applications. Oncol. Lett. 2021, 21, 404. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; Cao, L.; Li, Z.; Wang, X. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to Regulate Cell Growth and Differentiation in Acute Myeloid Leukemia. Mol. Cells 2016, 39, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Yao, T.; Zhang, Y.; Li, W.; Wang, Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: Role, mechanism and therapeutic potential. Int. J. Biol. Sci. 2021, 17, 3428–3440. [Google Scholar] [CrossRef] [PubMed]
- Na, H.; Li, X.; Zhang, X.; Xu, Y.; Sun, Y.; Cui, J.; Chen, Z.; Shi, X.; Ren, S.; Zuo, Y. lncRNA STEAP3-AS1 Modulates Cell Cycle Progression via Affecting CDKN1C Expression through STEAP3 in Colon Cancer. Mol. Ther. Nucleic Acids 2020, 21, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Sun, Q.Q.; Liu, T.X.; Lu, K.; Zhang, N.; Zhu, Y.; Chen, M. Serum lncRNA-UFC1 as a potential biomarker for diagnosis and prognosis of pancreatic cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 4125–4129. [Google Scholar]
- Lou, N.; Liu, G.; Pan, Y. Long noncoding RNA ANRIL as a novel biomarker in human cancer. Future Oncol. 2020, 16, 2981–2995. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Jiang, D.M.; Hu, S.S.; Zhao, L.; Wang, L.; Yang, M.H.; Ai, M.L.; Jiang, H.J.; Han, Y.; Ding, Y.Q.; et al. SATB2-AS1 Suppresses Colorectal Carcinoma Aggressiveness by Inhibiting SATB2-Dependent Snail Transcription and Epithelial-Mesenchymal Transition. Cancer Res. 2019, 79, 3542–3556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coe, E.A.; Tan, J.Y.; Shapiro, M.; Louphrasitthiphol, P.; Bassett, A.R.; Marques, A.C.; Goding, C.R.; Vance, K.W. The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor. PLoS Genet. 2019, 15, e1008501. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Wang, X.; Gu, Y.; Wang, F.; Li, L.; Qiu, X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch. Biochem. Biophys. 2019, 661, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zheng, Q.; Chu, Q.; Zhu, H. HAND2-AS1: A functional cancer-related long non-coding RNA. Biomed. Pharm. 2021, 137, 111317. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, J.; Wang, Z.; Wang, P.; Gao, X.; Wang, Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed. Pharm. 2018, 104, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, Z.; Lei, X.; Gan, R. Long non-coding RNA GAS5 in human cancer. Oncol. Lett. 2020, 20, 2587–2594. [Google Scholar] [CrossRef] [PubMed]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020, 10, 82. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Kiran, M.; Kuscu, C.; Chatrath, A.; Wotton, D.; Mayo, M.W.; Dutta, A. Long Noncoding RNA DRAIC Inhibits Prostate Cancer Progression by Interacting with IKK to Inhibit NF-κB Activation. Cancer Res. 2020, 80, 950–963. [Google Scholar] [CrossRef]
- Juvvuna, P.K.; Mondal, T.; Di Marco, M.; Kosalai, S.T.; Kanduri, M.; Kanduri, C. NBAT1/CASC15-003/USP36 control MYCN expression and its downstream pathway genes in neuroblastoma. Neuro-Oncol. Adv. 2021, 3, vdab056. [Google Scholar] [CrossRef]
- Rajan, A.; Nadhan, R.; Latha, N.R.; Krishnan, N.; Warrier, A.V.; Srinivas, P. Deregulated estrogen receptor signaling and DNA damage response in breast tumorigenesis. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188482. [Google Scholar] [CrossRef] [PubMed]
- Guiducci, G.; Stojic, L. Long Noncoding RNAs at the Crossroads of Cell Cycle and Genome Integrity. Trends Genet. TIG 2021, 37, 528–546. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.L.; Jin, L.; Xu, A.; Wang, Y.F.; Thorne, R.F.; Zhang, X.D.; Wu, M. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 2018, 20, 492–502. [Google Scholar] [CrossRef]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013, 32, 2833–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Liu, S.; Fan, J.; Jin, Y.; Tian, B.; Zheng, X.; Fu, H. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep. 2017, 18, 536–548. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Chen, Y.; Huang, Y.; Cao, K.; Liu, T.; Shen, H.; Cui, J.; Li, B.; Cai, J.; Gao, F.; et al. Long non-coding RNA ANRIL promotes homologous recombination-mediated DNA repair by maintaining ATR protein stability to enhance cancer resistance. Mol. Cancer 2021, 20, 94. [Google Scholar] [CrossRef]
- Deng, B.; Xu, W.; Wang, Z.; Liu, C.; Lin, P.; Li, B.; Huang, Q.; Yang, J.; Zhou, H.; Qu, L. An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination. EMBO Rep. 2019, 20, e47650. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Shi, L.; Yu, X.; Gu, Y.; Sun, X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018, 17, 439–447. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, L.; Mei, Z.; Jiang, Y.; Yi, Y.; Liu, L.; Meng, Y.; Zhou, L.; Zeng, J.; Wu, H. Interaction of E3 ubiquitin ligase MARCH7 with long noncoding RNA MALAT1 and autophagy-related protein ATG7 promotes autophagy and invasion in ovarian cancer. Cell. Physiol. Biochem. 2018, 47, 654–666. [Google Scholar] [CrossRef]
- Betts, J.A.; Moradi Marjaneh, M.; Al-Ejeh, F.; Lim, Y.C.; Shi, W.; Sivakumaran, H.; Tropée, R.; Patch, A.M.; Clark, M.B.; Bartonicek, N.; et al. Long Noncoding RNAs CUPID1 and CUPID2 Mediate Breast Cancer Risk at 11q13 by Modulating the Response to DNA Damage. Am. J. Hum. Genet. 2017, 101, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Mi, S.; Zhao, T.; Peng, C.; Peng, Y.; Chen, L.; Zhu, W.; Yao, Y.; Song, Q.; Li, X.; et al. BGL3 lncRNA mediates retention of the BRCA1/BARD1 complex at DNA damage sites. EMBO J. 2020, 39, e104133. [Google Scholar] [CrossRef]
- Panda, S.; Setia, M.; Kaur, N.; Shepal, V.; Arora, V.; Singh, D.K.; Mondal, A.; Teli, A.; Tathode, M.; Gajula, R.; et al. Noncoding RNA Ginir functions as an oncogene by associating with centrosomal proteins. PLoS Biol. 2018, 16, e2004204. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Azimi, T.; Hussen, B.M.; Abak, A.; Taheri, M.; Dilmaghani, N.A. Non-coding RNA Activated by DNA Damage: Review of Its Roles in the Carcinogenesis. Front. Cell Dev. Biol. 2021, 9, 714787. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Dragomir, M.P.; Fabris, L.; Bayraktar, R.; Knutsen, E.; Liu, X.; Tang, C.; Li, Y.; Shimura, T.; Ivkovic, T.C.; et al. The Long Noncoding RNA CCAT2 Induces Chromosomal Instability Through BOP1-AURKB Signaling. Gastroenterology 2020, 159, 2146–2162.e2133. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.; Siu, M.T.; Ho, C.W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef] [Green Version]
- Dimitrova, N.; Zamudio, J.R.; Jong, R.M.; Soukup, D.; Resnick, R.; Sarma, K.; Ward, A.J.; Raj, A.; Lee, J.T.; Sharp, P.A.; et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 2014, 54, 777–790. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Mol. Cell 2014, 48, 1370–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Munger, K.; Shenk, T. Expression of the Long Noncoding RNA DINO in Human Papillomavirus-Positive Cervical Cancer Cells Reactivates the Dormant TP53 Tumor Suppressor through ATM/CHK2 Signaling. MBio 2020, 11, e01190-20. [Google Scholar] [CrossRef]
- Sharma, V.; Khurana, S.; Kubben, N.; Abdelmohsen, K.; Oberdoerffer, P.; Gorospe, M.; Misteli, T. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 2015, 16, 1520–1534. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Wang, X.; Xue, X.; Li, L.; Hu, Y. A long noncoding RNA sensitizes genotoxic treatment by attenuating ATM activation and homologous recombination repair in cancers. PLoS Biol. 2020, 18, e3000666. [Google Scholar] [CrossRef] [PubMed]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Vohhodina, J.; Goehring, L.J.; Liu, B.; Kong, Q.; Botchkarev, V.V., Jr.; Huynh, M.; Liu, Z.; Abderazzaq, F.O.; Clark, A.P.; Ficarro, S.B.; et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat. Commun. 2021, 12, 3542. [Google Scholar] [CrossRef] [PubMed]
- Kamarajugadda, S.; Stemboroski, L.; Cai, Q.; Simpson, N.E.; Nayak, S.; Tan, M.; Lu, J. Glucose oxidation modulates anoikis and tumor metastasis. Mol. Cell Biol. 2012, 32, 1893–1907. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Duxbury, M.; Zinner, M.J.; Ashley, S.W.; Whang, E.E. Glucose transporter-1 gene expression is associated with pancreatic cancer invasiveness and MMP-2 activity. Surgery 2004, 136, 548–556. [Google Scholar] [CrossRef]
- Ellis, B.C.; Graham, L.D.; Molloy, P.L. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta BBA Mol. Cell Res. 2014, 1843, 372–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltrán-Anaya, F.O.; Cedro-Tanda, A.; Hidalgo-Miranda, A.; Romero-Cordoba, S.L. Insights into the Regulatory Role of Non-coding RNAs in Cancer Metabolism. Front. Physiol. 2016, 7, 342. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Li, C.; Xing, Z.; Hu, Q.; Liang, K.; Han, L.; Wang, C.; Hawke, D.H.; Wang, S.; Zhang, Y.; et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 2016, 18, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Wu, M.H.; Huang, Y.H.; Yeh, C.T.; Cheng, M.L.; Chi, H.C.; Tsai, C.Y.; Chung, I.H.; Chen, C.Y.; Lin, K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology 2018, 67, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Fan, Y.; Feng, T.; Chen, F.; Xu, Z.; Li, S.; Lin, Q.; He, X.; Shi, W.; Liu, Y.; et al. HOTAIR regulates HK2 expression by binding endogenous miR-125 and miR-143 in oesophageal squamous cell carcinoma progression. Oncotarget 2017, 8, 86410–86422. [Google Scholar] [CrossRef]
- Ma, Y.; Hu, M.; Zhou, L.; Ling, S.; Li, Y.; Kong, B.; Huang, P. Long non-coding RNA HOTAIR promotes cancer cell energy metabolism in pancreatic adenocarcinoma by upregulating hexokinase-2. Oncol. Lett. 2019, 18, 2212–2219. [Google Scholar] [CrossRef]
- Bian, Z.; Zhang, J.; Li, M.; Feng, Y.; Wang, X.; Zhang, J.; Yao, S.; Jin, G.; Du, J.; Han, W.; et al. LncRNA-FEZF1-AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin. Cancer Res. 2018, 24, 4808–4819. [Google Scholar] [CrossRef] [Green Version]
- Hung, C.L.; Wang, L.Y.; Yu, Y.L.; Chen, H.W.; Srivastava, S.; Petrovics, G.; Kung, H.J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 18697–18702. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Yan, T.; Bao, Y.; Shen, C.; Yu, C.; Zhu, X.; Tian, X.; Guo, F.; Liang, Q.; Liu, Q.; et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat. Commun. 2019, 10, 3499. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.B.; Bai, Y.J.; Tang, Y.; Liu, Z.H.; Wei, Q.; Han, P. Detection of Circulating Tumor Cells in Bladder Cancer by Subtraction Enrichment and Immunostaining-fluorescence in situ Hybridization. Sichuan Da Xue Xue Bao Yi Xue Ban 2017, 48, 605–609. [Google Scholar]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Z.Q.; Li, Z.X.; Guan, J.L.; Liu, X.; Li, J.Y.; Chen, Y.; Lin, L.; Kou, J.; Lv, J.W.; Zhang, L.L.; et al. Long Noncoding RNA TINCR-Mediated Regulation of Acetyl-CoA Metabolism Promotes Nasopharyngeal Carcinoma Progression and Chemoresistance. Cancer Res. 2020, 80, 5174–5188. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ma, J.; Cai, D. Increased HAGLR expression promotes non–small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumor Biol. 2017, 39, 1010428317697574. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.D.; Yuan, L.L.; Lin, L.Q. LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5143–5152. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, F.; Zhao, J.; Li, B.; Liang, Y.; Pan, W.; Zhang, S.; Wang, X.; Zheng, D. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget 2016, 7, 82620. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Taheri, M. Colon Cancer-Associated Transcripts 1 and 2: Roles and functions in human cancers. J. Cell. Physiol. 2019, 234, 14581–14600. [Google Scholar] [CrossRef]
- Wei, R.; DeVilbiss, F.T.; Liu, W. Genetic Polymorphism, Telomere Biology and Non-Small Lung Cancer Risk. J. Genet. Genom. 2015, 42, 549–561. [Google Scholar] [CrossRef]
- Ge, Y.; Yan, X.; Jin, Y.; Yang, X.; Yu, X.; Zhou, L.; Han, S.; Yuan, Q.; Yang, M. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015, 11, e1005726. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cui, S.; Wan, T.; Li, X.; Tian, W.; Zhang, R.; Luo, L.; Shi, Y. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126-5p. J. Cell Physiol. 2018, 233, 6822–6831. [Google Scholar] [CrossRef]
- Zeng, B.; Ye, H.; Chen, J.; Cheng, D.; Cai, C.; Chen, G.; Chen, X.; Xin, H.; Tang, C.; Zeng, J. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget 2017, 8, 113650–113661. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Chen, S.; Liu, J.; Guo, X.; Xiang, X.; Dong, T.; Ran, P.; Li, Q.; Zhu, B.; Zhang, X.; et al. Long non-coding RNA PVT1-5 promotes cell proliferation by regulating miR-126/SLC7A5 axis in lung cancer. Biochem. Biophys. Res. Commun. 2018, 495, 2350–2355. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Groß, M.; Holler, J.M.; Coggins, S.A.; Patil, N.; Leupold, J.H.; Munschauer, M.; Schenone, M.; Hartigan, C.R.; Allgayer, H.; et al. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1-RRM2 axis in cancer. Nat. Commun. 2020, 11, 3214. [Google Scholar] [CrossRef]
- Varier, K.M.; Dhandapani, H.; Liu, W.; Song, J.; Wang, C.; Hu, A.; Ben-David, Y.; Shen, X.; Li, Y.; Gajendran, B. An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells. J. Exp. Clin. Cancer Res. CR 2021, 40, 242. [Google Scholar] [CrossRef]
- Yan, K.; Fu, Y.; Zhu, N.; Wang, Z.; Hong, J.L.; Li, Y.; Li, W.J.; Zhang, H.B.; Song, J.H. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int. J. Biochem. Cell Biol. 2019, 110, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yin, Y.; Ju, H.; Xu, X.; Liu, W.; Fu, Q.; Hu, J.; Zhang, X.; Sun, B. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018, 9, 478. [Google Scholar] [CrossRef]
- Huang, D.; Chen, J.; Yang, L.; Ouyang, Q.; Li, J.; Lao, L.; Zhao, J.; Liu, J.; Lu, Y.; Xing, Y.; et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat. Immunol. 2018, 19, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Ranzani, V.; Rossetti, G.; Panzeri, I.; Arrigoni, A.; Bonnal, R.J.; Curti, S.; Gruarin, P.; Provasi, E.; Sugliano, E.; Marconi, M.; et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015, 16, 318–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Guo, Z.M. Multiple functions of Maf in the regulation of cellular development and differentiation. Diabetes Metab. Res. Rev. 2015, 31, 773–778. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.; Tang, J.; Chen, Y.; Deng, L.; Ji, J.; Xie, Y.; Wang, K.; Jia, W.; Chu, W.M.; Sun, B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 2017, 8, 15129. [Google Scholar] [CrossRef] [Green Version]
- Zemmour, D.; Pratama, A.; Loughhead, S.M.; Mathis, D.; Benoist, C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, e3472–e3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, X.; Wang, X.; Li, H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int. J. Biol. Macromol. 2018, 118, 24–30. [Google Scholar] [CrossRef]
- Wu, K.; Zhao, Z.; Liu, K.; Zhang, J.; Li, G.; Wang, L. Long noncoding RNA lnc-sox5 modulates CRC tumorigenesis by unbalancing tumor microenvironment. Cell Cycle 2017, 16, 1295–1301. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liao, G.; Bai, J.; Zhang, X.; Xu, L.; Deng, C.; Yan, M.; Xie, A.; Luo, T.; Long, Z.; et al. Identifying Cancer Driver lncRNAs Bridged by Functional Effectors through Integrating Multi-omics Data in Human Cancers. Mol. Therapy Nucleic Acids 2019, 17, 362–373. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.Y.; Zhang, M.M.; Liu, C.; Kang, Y.; Wang, J.O.; Yang, X.H. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J. Cell Physiol. 2019, 234, 23176–23189. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, F.; Su, C.; Xie, P.; Xu, L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J. Cell Biochem. 2019, 120, 11642–11650. [Google Scholar] [CrossRef]
- Wang, Q.M.; Lian, G.Y.; Song, Y.; Huang, Y.F.; Gong, Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019, 231, 116335. [Google Scholar] [CrossRef]
- Sehgal, L.; Mathur, R.; Braun, F.K.; Wise, J.F.; Berkova, Z.; Neelapu, S.; Kwak, L.W.; Samaniego, F. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 2014, 28, 2376–2387. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar] [CrossRef] [Green Version]
- Sattari, A.; Siddiqui, H.; Moshiri, F.; Ngankeu, A.; Nakamura, T.; Kipps, T.J.; Croce, C.M. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget 2016, 7, 54174–54182. [Google Scholar] [CrossRef]
- Fang, P.; Xiang, L.; Chen, W.; Li, S.; Huang, S.; Li, J.; Zhuge, L.; Jin, L.; Feng, W.; Chen, Y.; et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. 2019, 25, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Xu, Y.; Lai, Y.; He, W.; Li, Y.; Wang, R.; Luo, X.; Chen, R.; Chen, T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J. Cell. Biochem. 2018, 119, 2951–2963. [Google Scholar] [CrossRef]
- Chen, C.; He, W.; Huang, J.; Wang, B.; Li, H.; Cai, Q.; Su, F.; Bi, J.; Liu, H.; Zhang, B.; et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun. 2018, 9, 3826. [Google Scholar] [CrossRef] [Green Version]
- Leija Montoya, G.; González Ramírez, J.; Sandoval Basilio, J.; Serafín Higuera, I.; Isiordia Espinoza, M.; González González, R.; Serafín Higuera, N. Long Non-coding RNAs: Regulators of the Activity of Myeloid-Derived Suppressor Cells. Front. Immunol. 2019, 10, 1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Dong, R.; Jiang, L.; Gong, Y.; Yuan, M.; You, J.; Meng, W.; Chen, Z.; Zhang, N.; Weng, Q.; et al. LncRNA-MM2P Identified as a Modulator of Macrophage M2 Polarization. Cancer Immunol. Res. 2019, 7, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, J.; Fu, H.; Liu, C.; Yu, Z.; Sun, Y.; She, X.; Li, P.; Zhao, C.; Liu, Y.; et al. Coagulation Factor X Regulated by CASC2c Recruited Macrophages and Induced M2 Polarization in Glioblastoma Multiforme. Front. Immunol. 2018, 9, 1557. [Google Scholar] [CrossRef] [Green Version]
- Veglia, F.; Sanseviero, E.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021, 21, 485–498. [Google Scholar] [CrossRef]
- Shang, W.; Gao, Y.; Tang, Z.; Zhang, Y.; Yang, R. The Pseudogene Olfr29-ps1 Promotes the Suppressive Function and Differentiation of Monocytic MDSCs. Cancer Immunol. Res. 2019, 7, 813–827. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, T.; Li, Y.; Zhang, Y.; Yang, R. Lnc-chop Promotes Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor and Inflammatory Environments. J. Immunol. 2018, 200, 2603–2614. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Ma, J.; Wang, T.; Tian, J.; Zheng, Y.; Peng, R.; Wang, Y.; Zhang, Y.; Mao, L.; Xu, H.; et al. Long non-coding RNA RUNXOR accelerates MDSC-mediated immunosuppression in lung cancer. BMC Cancer 2018, 18, 660. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. LncRNA H19: A novel oncogene in multiple cancers. Int. J. Biol. Sci. 2021, 17, 3188–3208. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Tang, Y. SChLAP1 promotes prostate cancer development through interacting with EZH2 to mediate promoter methylation modification of multiple miRNAs of chromosome 5 with a DNMT3a-feedback loop. Cell Death Dis. 2021, 12, 188. [Google Scholar] [CrossRef]
- Xin, X.; Li, Q.; Fang, J.; Zhao, T. LncRNA HOTAIR: A Potential Prognostic Factor and Therapeutic Target in Human Cancers. Front. Oncol. 2021, 11, 679244. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ou, C.; Liu, J.; Chen, C.; Zhou, Q.; Yang, S.; Li, G.; Wang, G.; Song, J.; Li, Z.; et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 2019, 38, 2627–2644. [Google Scholar] [CrossRef] [Green Version]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta. Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef]
- Da, M.; Zhuang, J.; Zhou, Y.; Qi, Q.; Han, S. Role of long noncoding RNA taurine-upregulated gene 1 in cancers. Mol. Med. 2021, 27, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, X.; Wang, X.; Hu, J.; Jiang, L. Silencing of LncRNA HULC Enhances Chemotherapy Induced Apoptosis in Human Gastric Cancer. J. Med. Biochem. 2016, 35, 137–143. [Google Scholar] [CrossRef]
- Hong, W.; Ying, H.; Lin, F.; Ding, R.; Wang, W.; Zhang, M. lncRNA LINC00460 Silencing Represses EMT in Colon Cancer through Downregulation of ANXA2 via Upregulating miR-433-3p. Mol. Ther. Nucleic Acids 2020, 19, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Gu, J.; Wang, Y.; Wu, H.; Cheng, W.; Wang, Q.; Zheng, G.; Wang, X. Long non-coding RNA NEAT1 transported by extracellular vesicles contributes to breast cancer development by sponging microRNA-141-3p and regulating KLF12. Cell Biosci. 2021, 11, 68. [Google Scholar] [CrossRef]
- Xie, R.; Wang, M.; Zhou, W.; Wang, D.; Yuan, Y.; Shi, H.; Wu, L. Long Non-Coding RNA (LncRNA) UFC1/miR-34a Contributes to Proliferation and Migration in Breast Cancer. Med. Sci. Monit. 2019, 25, 7149–7157. [Google Scholar] [CrossRef]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bei, W.; Wen, X.; Yuxuan, C.; Kai, L.; Jiacheng, W.; Chong, G.; Chengfu, Y. The Functional Role of Oncogenic LncRNA BCAR4 for Cancer Outcome. Curr. Pharm. Des. 2021, 27, 4107–4113. [Google Scholar] [CrossRef]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, G.; Peng, Z.; Shang, J.; Kang, Y.; Ning, H.; Mao, C. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through miR-9/E-cadherin cascade signaling pathway molecular mechanism. OncoTargets Ther. 2017, 10, 3241–3247. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Liu, Y.; Zheng, J.; Liu, X.; Chen, J.; Liu, L.; Wang, P.; Xue, Y. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1605–1617. [Google Scholar] [CrossRef]
- Deryugina, E.I.; Quigley, J.P. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol. 2015, 44–46, 94–112. [Google Scholar] [CrossRef]
- Zhao, J.; Du, P.; Cui, P.; Qin, Y.; Wu, J.; Zhou, Z.; Zhang, W.; Qin, L.; Huang, G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018, 37, 4094–4109. [Google Scholar] [CrossRef] [PubMed]
- Cong, Z.; Diao, Y.; Li, X.; Jiang, Z.; Xu, Y.; Zhou, H.; Qiang, Y.; Wu, H.; Shen, Y. Long non-coding RNA linc00665 interacts with YB-1 and promotes angiogenesis in lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2020, 527, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Fu, Q.; Man, W.; Guo, H.; Yang, P. LncRNA OR3A4 participates in the angiogenesis of hepatocellular carcinoma through modulating AGGF1/akt/mTOR pathway. Eur. J. Pharmacol. 2019, 849, 106–114. [Google Scholar] [CrossRef]
- Yuan, S.X.; Yang, F.; Yang, Y.; Tao, Q.F.; Zhang, J.; Huang, G.; Yang, Y.; Wang, R.Y.; Yang, S.; Huo, X.S.; et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012, 56, 2231–2241. [Google Scholar] [CrossRef] [PubMed]
- Sang, L.J.; Ju, H.Q.; Liu, G.P.; Tian, T.; Ma, G.L.; Lu, Y.X.; Liu, Z.X.; Pan, R.L.; Li, R.H.; Piao, H.L.; et al. LncRNA CamK-A Regulates Ca(2+)-Signaling-Mediated Tumor Microenvironment Remodeling. Mol. Cell 2018, 72, 71–83.e77. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.X.; Chen, Z.H.; Chen, D.L.; Tian, X.P.; Wang, C.Y.; Zhou, Z.W.; Gao, Y.; Xu, Y.; Chen, C.; Zheng, Z.S.; et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene 2018, 37, 2660–2675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Yang, F.; Qi, X.; Li, Q.; Wang, D.; Yi, T.; Yin, R.; Zhao, X.; Zhong, X.; Bian, C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol. Carcinog. 2019, 58, 2286–2296. [Google Scholar] [CrossRef]
- Song, R.; Liu, Z.; Lu, L.; Liu, F.; Zhang, B. Long Noncoding RNA SCAMP1 Targets miR-137/CXCL12 Axis to Boost Cell Invasion and Angiogenesis in Ovarian Cancer. DNA Cell Biol. 2020, 39, 1041–1050. [Google Scholar] [CrossRef]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNA-Meg3/miR-421/PDGFRA axis. Int. J. Oncol. 2019, 55, 1296–1312. [Google Scholar] [CrossRef]
- Lu, Z.; Xiao, Z.; Liu, F.; Cui, M.; Li, W.; Yang, Z.; Li, J.; Ye, L.; Zhang, X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget 2016, 7, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, L.; Han, Z.Y.; Wang, Z.X.; Qin, L.X. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am. J. Cancer Res. 2019, 9, 1367–1381. [Google Scholar]
- Lang, H.L.; Hu, G.W.; Chen, Y.; Liu, Y.; Tu, W.; Lu, Y.M.; Wu, L.; Xu, G.H. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 959–972. [Google Scholar]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, H.B.; Kurban, E.; Luo, H.W. LncRNA SNHG14 promotes hepatocellular carcinoma progression via H3K27 acetylation activated PABPC1 by PTEN signaling. Cell Death Dis. 2020, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Nie, J.; Lu, L.; Du, C.; Meng, F.; Song, D. LINC00337 promotes tumor angiogenesis in colorectal cancer by recruiting DNMT1, which suppresses the expression of CNN1. Cancer Gene Ther. 2021, 28, 1285–1297. [Google Scholar] [CrossRef]
- Huang, J.K.; Ma, L.; Song, W.H.; Lu, B.Y.; Huang, Y.B.; Dong, H.M.; Ma, X.K.; Zhu, Z.Z.; Zhou, R. LncRNA-MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor-Associated Macrophage FGF2 Protein Secretion. J. Cell Biochem. 2017, 118, 4821–4830. [Google Scholar] [CrossRef] [PubMed]
- Tee, A.E.; Liu, B.; Song, R.; Li, J.; Pasquier, E.; Cheung, B.B.; Jiang, C.; Marshall, G.M.; Haber, M.; Norris, M.D.; et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget 2016, 7, 8663–8675. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shu, H.; Zhang, L.; Xiong, J. Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wnt/β-catenin signaling pathway. J. Cell Biochem. 2019, 120, 6937–6951. [Google Scholar] [CrossRef]
- Castellano, J.J.; Navarro, A.; Viñolas, N.; Marrades, R.M.; Moises, J.; Cordeiro, A.; Saco, A.; Muñoz, C.; Fuster, D.; Molins, L.; et al. LincRNA-p21 Impacts Prognosis in Resected Non-Small Cell Lung Cancer Patients through Angiogenesis Regulation. J. Thorac. Oncol. 2016, 11, 2173–2182. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Taheri, M. Maternally expressed gene 3 (MEG3): A tumor suppressor long non coding RNA. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 118, 109129. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.R.; Wu, J.S.; Tang, Y.L.; Liang, X.H. Long noncoding RNAs: Emerging regulators of tumor angiogenesis. Future Oncol. 2017, 13, 1551–1562. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, Y.; Zhang, X. Stemness-Related Markers in Cancer. Cancer Transl. Med. 2017, 3, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhu, J.; Wang, F.; Guan, Z.; Ge, Y.; Yang, X.; Cai, J. LncRNAs and their role in cancer stem cells. Oncotarget 2017, 8, 110685–110692. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, H. Regulation of Chromatin Organization in Cell Stemness: The Emerging Role of Long Non-coding RNAs. Stem Cell Rev. Rep. 2021, 17, 2042–2053. [Google Scholar] [CrossRef]
- Wang, W.T.; Chen, T.Q.; Zeng, Z.C.; Pan, Q.; Huang, W.; Han, C.; Fang, K.; Sun, L.Y.; Yang, Q.Q.; Wang, D.; et al. The lncRNA LAMP5-AS1 drives leukemia cell stemness by directly modulating DOT1L methyltransferase activity in MLL leukemia. J. Hematol. Oncol. 2020, 13, 78. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Wang, Q.; Ji, M.; Guo, X.; Li, L.; Su, X. Exosomal lncRNA UCA1 modulates cervical cancer stem cell self-renewal and differentiation through microRNA-122-5p/SOX2 axis. J. Transl. Med. 2021, 19, 229. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; Zhong, Q.; Wu, J.; Tang, Z. LncRNA UCA1 is necessary for TGF-β-induced epithelial–mesenchymal transition and stemness via acting as a ceRNA for Slug in glioma cells. FEBS Open Bio 2018, 8, 1855–1865. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef]
- Zhou, T.; Wu, L.; Ma, N.; Tang, F.; Yu, Z.; Jiang, Z.; Li, Y.; Zong, Z.; Hu, K. SOX9-activated FARSA-AS1 predetermines cell growth, stemness, and metastasis in colorectal cancer through upregulating FARSA and SOX9. Cell Death Dis. 2020, 11, 1071. [Google Scholar] [CrossRef]
- Zhao, L.; Lou, G.; Li, A.; Liu, Y. lncRNA MALAT1 modulates cancer stem cell properties of liver cancer cells by regulating YAP1 expression via miR375 sponging. Mol. Med. Rep. 2020, 22, 1449–1457. [Google Scholar] [CrossRef]
- Wang, S.; Liu, F.; Deng, J.; Cai, X.; Han, J.; Liu, Q. Long Noncoding RNA ROR Regulates Proliferation, Invasion, and Stemness of Gastric Cancer Stem Cell. Cell. Reprogram. 2016, 18, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Wu, Z.; Li, Y.; Xu, Z.; Liu, B.; Liu, F.; Bao, Y.; Wu, D.; Liu, J.; Wang, A.; et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat. Commun. 2016, 7, 12692. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, M.; Yang, Q.; Zhu, W.; Jin, H.; Wang, J.; Song, J.; Kong, Y.; Lv, X. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol. Cancer 2018, 17, 173. [Google Scholar] [CrossRef]
- Chen, Z.Z.; Huang, L.; Wu, Y.H.; Zhai, W.J.; Zhu, P.P.; Gao, Y.F. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat. Commun. 2016, 7, 12598. [Google Scholar] [CrossRef]
- Zhu, P.; Wu, J.; Wang, Y.; Zhu, X.; Lu, T.; Liu, B.; He, L.; Ye, B.; Wang, S.; Meng, S.; et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat. Cell Biol. 2018, 20, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, W.; Shen, W.; Xia, M.; Chen, C.; Xiang, D.; Ning, B.; Cui, X.; Li, H.; Li, X.; et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016, 64, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xie, R.; Gu, P.; Huang, M.; Han, J.; Dong, W.; Xie, W.; Wang, B.; He, W.; Zhong, G.; et al. Long Noncoding RNA LBCS Inhibits Self-Renewal and Chemoresistance of Bladder Cancer Stem Cells through Epigenetic Silencing of SOX2. Clin. Cancer Res. 2019, 25, 1389–1403. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, C.; Chen, C.; Wu, F.; Shen, P.; Zhang, P.; He, G.; Li, X. Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism. Oncotarget 2017, 8, 70156–70171. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Yao, S.; Chen, X.; Liu, D.; Wang, J.; Yuan, X.; Rao, J.; Xiong, H.; Yu, S.; Yuan, X.; et al. LncRNA OIP5-AS1 regulates radioresistance by targeting DYRK1A through miR-369-3p in colorectal cancer cells. Eur. J. Cell Biol. 2018, 97, 369–378. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Li, Z.; Zhao, Q.; Zhu, Y.; Zhao, C.; Li, X.; Ma, Z.; Li, X.; Zhang, Y. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/beta-catenin signaling pathway. Oncol. Rep. 2014, 31, 1839–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Yuan, D.; Yang, Y.; Ren, M. LincRNA-p21 enhances the sensitivity of radiotherapy for gastric cancer by targeting the beta-catenin signaling pathway. J. Cell Biochem. 2019, 120, 6178–6187. [Google Scholar] [CrossRef]
- Fotouhi Ghiam, A.; Taeb, S.; Huang, X.; Huang, V.; Ray, J.; Scarcello, S.; Hoey, C.; Jahangiri, S.; Fokas, E.; Loblaw, A.; et al. Long non-coding RNA urothelial carcinoma associated 1 (UCA1) mediates radiation response in prostate cancer. Oncotarget 2017, 8, 4668–4689. [Google Scholar] [CrossRef]
- Podralska, M.; Ciesielska, S.; Kluiver, J.; van den Berg, A.; Dzikiewicz-Krawczyk, A.; Slezak-Prochazka, I. Non-Coding RNAs in Cancer Radiosensitivity: MicroRNAs and lncRNAs as Regulators of Radiation-Induced Signaling Pathways. Cancers 2020, 12, 1662. [Google Scholar] [CrossRef]
- He, J.; Zhu, S.; Liang, X.; Zhang, Q.; Luo, X.; Liu, C.; Song, L. LncRNA as a multifunctional regulator in cancer multi-drug resistance. Mol. Biol. Rep. 2021, 48, 6151–6165. [Google Scholar] [CrossRef]
- Fang, Z.; Chen, W.; Yuan, Z.; Liu, X.; Jiang, H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed. Pharmacother. 2018, 101, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhao, Y.; Li, Y.; Zhang, T.; Ma, Y.; Liu, Y. LncRNA MALAT1 Promotes Lung Cancer Proliferation and Gefitinib Resistance by Acting as a miR-200a Sponge. Arch. Bronconeumol. 2019, 55, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, M.; Zhang, Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. OncoTargets Ther. 2020, 13, 1343–1354. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Qian, J.; Li, J.; Zhu, C. Knockdown of lncRNA-HOTAIR downregulates the drug-resistance of breast cancer cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp. Ther. Med. 2019, 18, 435–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Cao, Z.; Guo, H.; Li, S. The action mechanism of lncRNA-HOTAIR on the drug resistance of non-small cell lung cancer by regulating Wnt signaling pathway. Exp. Ther. Med. 2018, 15, 4885–4889. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Qin, R.; Guan, A.; Yao, Y.; Huang, Y.; Jia, H.; Huang, W.; Gao, J. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J. Cell Biochem. 2018, 119, 7226–7234. [Google Scholar] [CrossRef]
- Yan, J.; Dang, Y.; Liu, S.; Zhang, Y.; Zhang, G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol. 2016, 37, 16345–16355. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Li, M.; Luo, W.; Sun, H.B. Curcumin attenuates Adriamycin-resistance of acute myeloid leukemia by inhibiting the lncRNA HOTAIR/miR-20a-5p/WT1 axis. Lab. Investig. 2021, 101, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Chen, X.; Zhi, X. Long Non-Coding RNA (LncRNA) Urothelial Carcinoma Associated 1 (UCA1) Increases Multi-Drug Resistance of Gastric Cancer via Downregulating miR-27b. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 3506–3513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Y.; Feng, B.; Zhou, L.; Ren, G.; Zhang, Z.; Fan, X.; Sun, Y.; Luo, G.; Liang, J.; Wu, K.; et al. The miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget 2016, 7, 538–549. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Xia, Z.; Xie, M.; Gao, Y.; Yu, Q.; He, B. Exosomal long non-coding RNA SOX2 overlapping transcript enhances the resistance to EGFR-TKIs in non-small cell lung cancer cell line H1975. Hum. Cell 2021, 34, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Liu, J.; Gen, H. Exosomal Long Non-coding RNA HOTTIP Increases Resistance of Colorectal Cancer Cells to Mitomycin via Impairing MiR-214-Mediated Degradation of KPNA3. Front. Cell Dev. Biol. 2020, 8, 582723. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, D.; Li, H.; Lv, Y.; Li, S. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int. J. Oncol. 2019, 54, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Zhang, T.; Pan, J.; Li, C. LncRNA UCA1 promotes cisplatin resistance in gastric cancer via recruiting EZH2 and activating PI3K/AKT pathway. J. Cancer 2020, 11, 3882–3892. [Google Scholar] [CrossRef]
- Liu, H.; Wang, G.; Yang, L.; Qu, J.; Yang, Z.; Zhou, X. Knockdown of Long Non-Coding RNA UCA1 Increases the Tamoxifen Sensitivity of Breast Cancer Cells through Inhibition of Wnt/β-Catenin Pathway. PLoS ONE 2016, 11, e0168406. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014, 281, 1750–1758. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, S.; Chen, X.; Wang, C. HOTAIR suppresses PTEN via DNMT3b and confers drug resistance in acute myeloid leukemia. Hematology 2021, 26, 170–178. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Zhen, F.; Jin, S.; Yu, T.; Zhu, Q.; Wang, W.; Xu, K.; Yao, J.; Guo, R. LINC00665 Induces Acquired Resistance to Gefitinib through Recruiting EZH2 and Activating PI3K/AKT Pathway in NSCLC. Molecular therapy. Nucleic Acids 2019, 16, 155–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Liao, W.; Wu, Q.; Huang, X.; Pan, Z.; Chen, W.; Gu, S.; Huang, Z.; Wang, Y.; Tang, X.; et al. LINC00152 upregulates ZEB1 expression and enhances epithelial-mesenchymal transition and oxaliplatin resistance in esophageal cancer by interacting with EZH2. Cancer Cell Int. 2020, 20, 569. [Google Scholar] [CrossRef]
- Lv, Q.L.; Wang, L.C.; Li, D.C.; Lin, Q.X.; Shen, X.L.; Liu, H.Y.; Li, M.; Ji, Y.L.; Qin, C.Z.; Chen, S.H. Knockdown lncRNA DLEU1 Inhibits Gliomas Progression and Promotes Temozolomide Chemosensitivity by Regulating Autophagy. Front. Pharmacol. 2020, 11, 560543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, K.; Tang, Y.; Luan, X.; Zheng, X.; Lu, X.; Mao, J.; Hu, L.; Zhang, S.; Zhang, X.; et al. LncRNA-HOTAIR activates autophagy and promotes the imatinib resistance of gastrointestinal stromal tumor cells through a mechanism involving the miR-130a/ATG2B pathway. Cell Death Dis. 2021, 12, 367. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, C.; Lei, Q.; Wu, Z.; Miao, X.; Zhu, D.; Yang, X.; Li, N.; Tang, M.; Chen, Y.; et al. Relationship between long non-coding RNA PCAT-1 expression and gefitinib resistance in non-small-cell lung cancer cells. Respir. Res. 2021, 22, 146. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, W.; Xu, L.; Chen, Y.; Xu, Y.; Yuan, L. Long Non-Coding RNA PVT1 Regulates the Resistance of the Breast Cancer Cell Line MDA-MB-231 to Doxorubicin via Nrf2. Technol. Cancer Res. Treat. 2020, 19, 1533033820980763. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Pan, T.; Jiang, D.; Jin, L.; Geng, Y.; Feng, X.; Shen, A.; Zhang, L. The lncRNA-GAS5/miR-221-3p/DKK2 Axis Modulates ABCB1-Mediated Adriamycin Resistance of Breast Cancer via the Wnt/β-Catenin Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 19, 1434–1448. [Google Scholar] [CrossRef]
- Wang, C.; Ke, S.; Li, M.; Lin, C.; Liu, X.; Pan, Q. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol. Genet. Genom. 2020, 295, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Qu, Y.; Li, C.; Huang, A.; Tong, S.; Wu, C.; Fan, K. Deregulation of lncRNA-AC078883.3 and microRNA-19a is involved in the development of chemoresistance to cisplatin via modulating signaling pathway of PTEN/AKT. J. Cell Physiol. 2019, 234, 22657–22665. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Liu, Y.; Liang, P.; Wang, B.; Tan, H.; Zhang, Y.; Gao, X.; Gao, J. TP53TG1 enhances cisplatin sensitivity of non-small cell lung cancer cells through regulating miR-18a/PTEN axis. Cell Biosci. 2018, 8, 23. [Google Scholar] [CrossRef]
- Liu, S.; Zou, B.; Tian, T.; Luo, X.; Mao, B.; Zhang, X.; Lei, H. Overexpression of the lncRNA FER1L4 inhibits paclitaxel tolerance of ovarian cancer cells via the regulation of the MAPK signaling pathway. J. Cell. Biochem. 2018, 120, 7581–7589. [Google Scholar] [CrossRef] [PubMed]
- Xiu, D.-H.; Liu, G.-F.; Yu, S.-N.; Li, L.-Y.; Zhao, G.-Q.; Liu, L.; Li, X.-F. Long non-coding RNA LINC00968 attenuates drug resistance of breast cancer cells through inhibiting the Wnt2/β-catenin signaling pathway by regulating WNT2. J. Exp. Clin. Cancer Res. 2019, 38, 94. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Wu, H.; Xiong, J.; Peng, T. Long Non-coding RNA DLEU2L Targets miR-210-3p to Suppress Gemcitabine Resistance in Pancreatic Cancer Cells via BRCA2 Regulation. Front. Mol. Biosci. 2021, 8, 645365. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).