Molecular Docking as a Key Driver of Biocontrol for Agri-Food Security
Abstract
1. Introduction
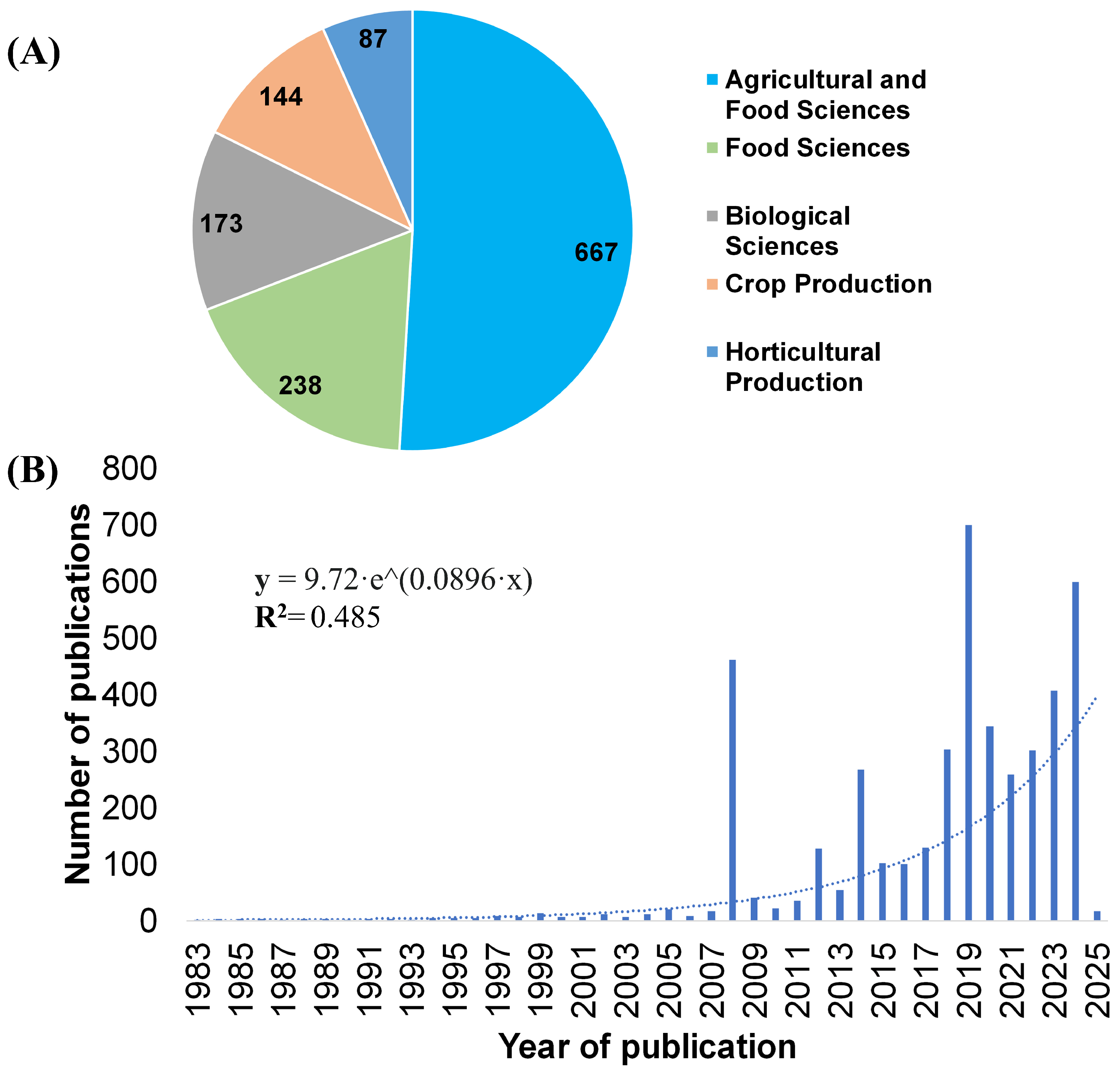
2. Search Strategy, Literature Sources and Selection Criteria
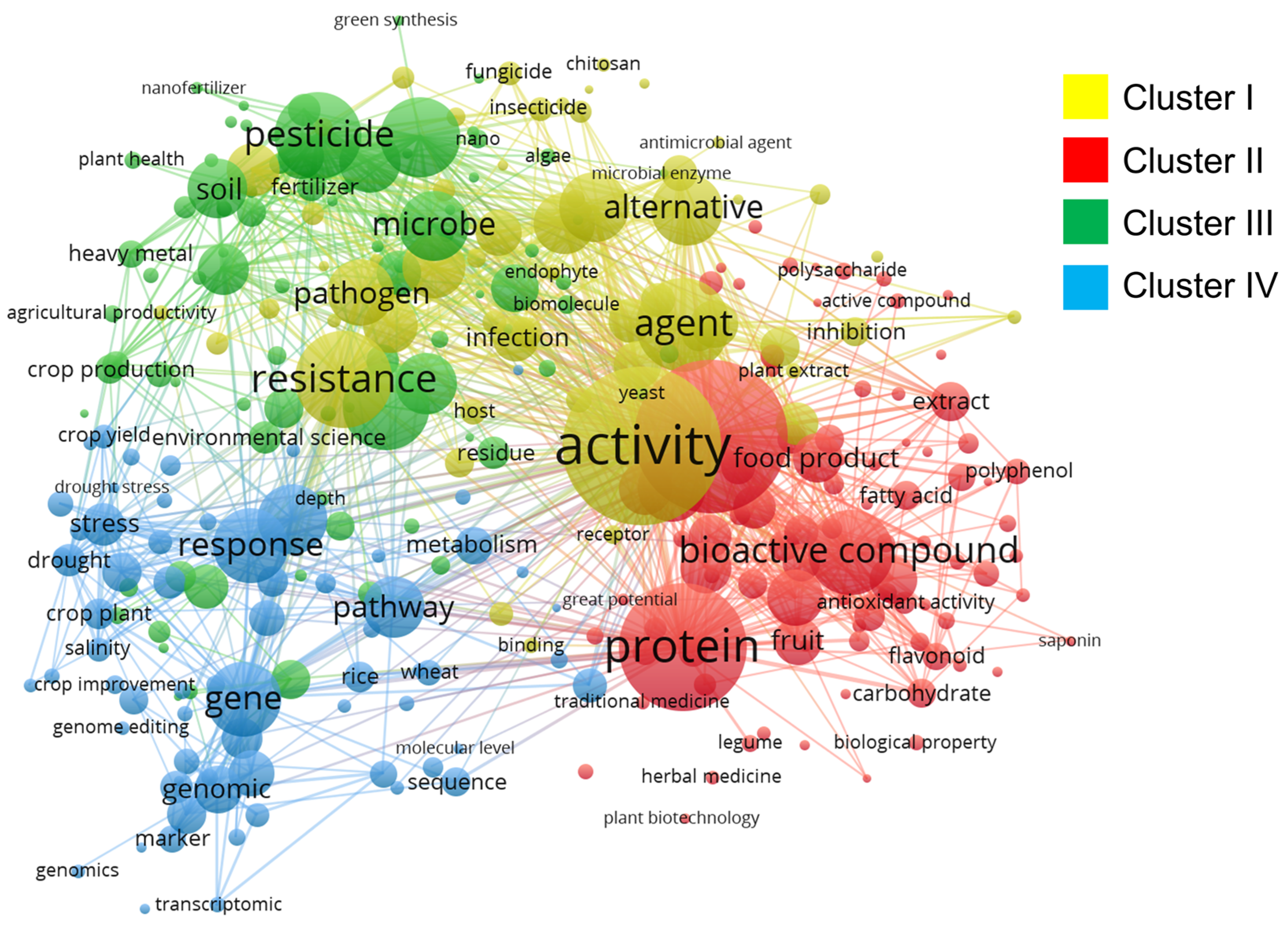
3. Trends in Molecular Docking Research for Agri-Food Security
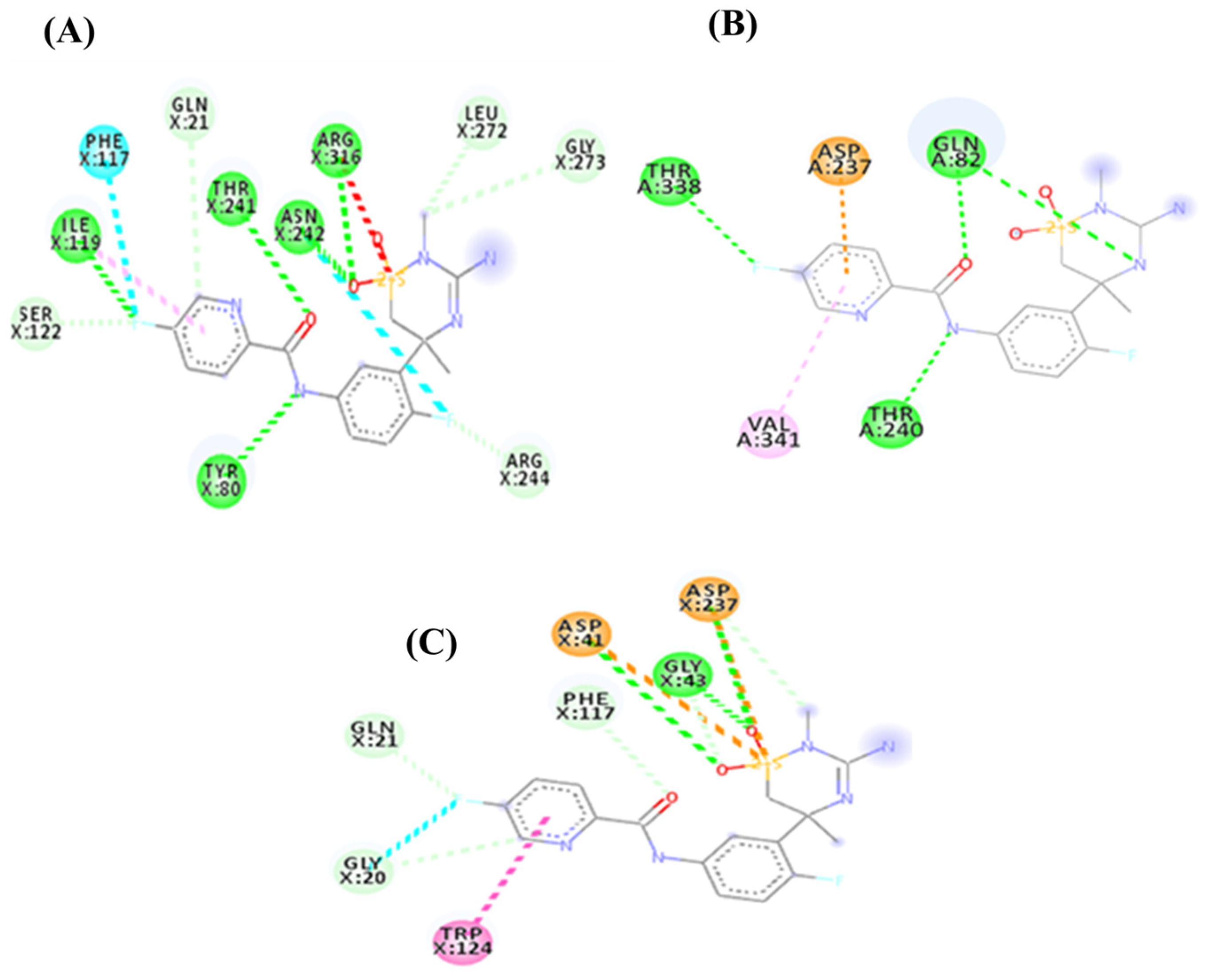
4. Fundamentals of the Technique and Its Application in Agri-Food Security
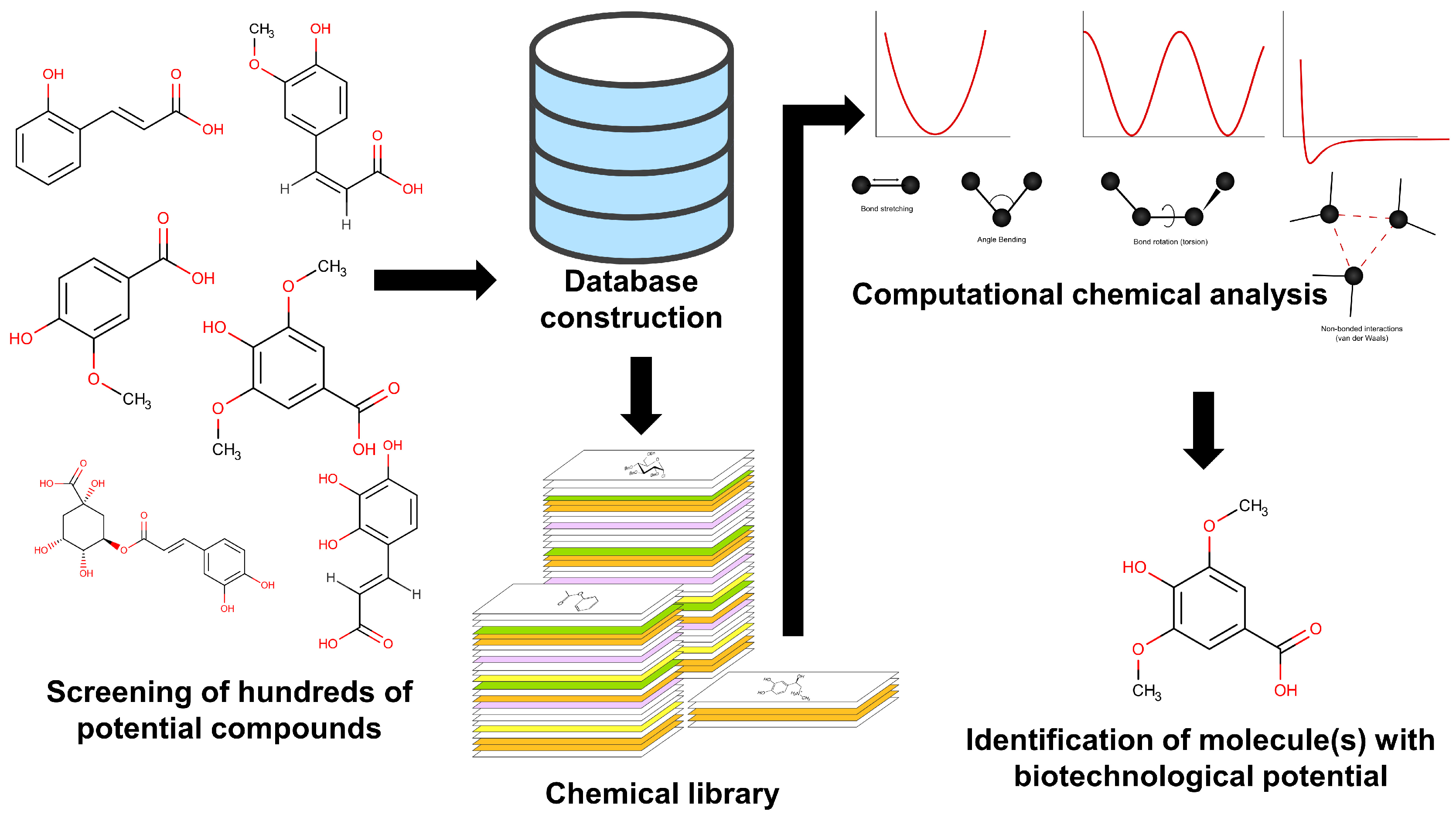
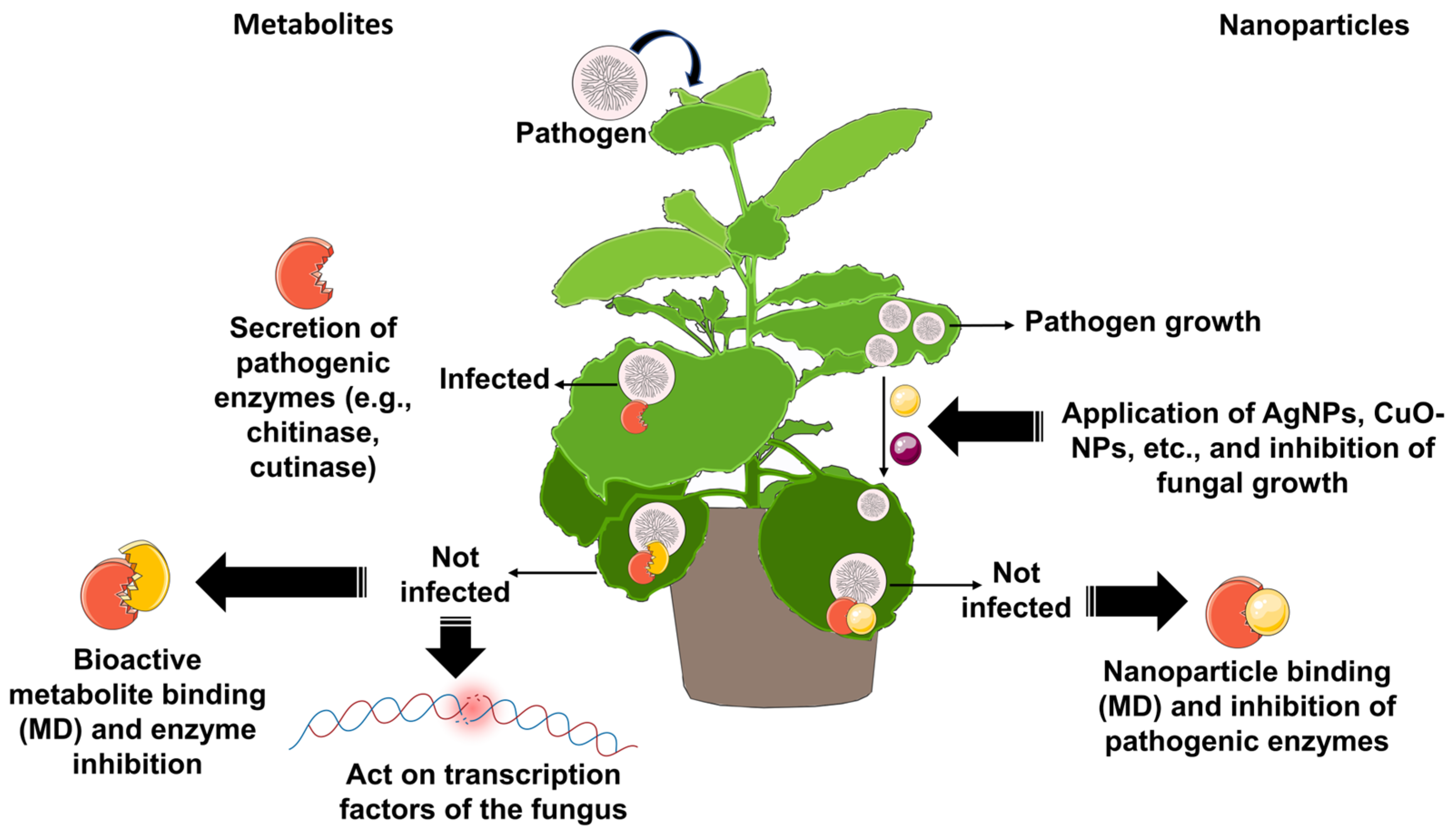
5. Molecular Docking of Natural Origin Molecules and Nanoparticles for Sustainable Crop Protection
6. Applications of Molecular Docking in Major Agri-Food Crops
6.1. Rice
6.2. Wheat
6.3. Sorghum
6.4. Maize
7. Technical Challenges and Computational Limitations in Molecular Docking
7.1. Ligand Conformation and Flexibility
7.2. Protein Flexibility
7.3. Quality of Input Data
7.4. Scoring Functions and Optimization
7.5. Computational Resources
7.6. Statistical Assessment Limitations
8. Biological Challenges and Limitations
8.1. Genetic Diversity in Crops
8.2. Environmental Context
9. Future Directions and Perspectives
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic Acid |
| AI | Artificial Intelligence |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| GC-MS | Gas Chromatography-Mass Spectrometry |
| MD | Molecular Dynamics |
| ML | Machine Learning |
| PDB | Protein Data Bank |
| PR proteins | Pathogenesis-Related Proteins |
| qPCR | Quantitative Polymerase Chain Reaction |
| RNA | Ribonucleic Acid |
| SSR | Simple Sequence Repeat (cuando lo uses en contexto filogenético) |
| T1R1/T1R3 | Taste receptor type 1, subunit 1/Taste receptor type 1, subunit 3 |
| UPLC-Q-TOF-MS | Ultra-Performance Liquid Chromatography coupled to Quadrupole Time-of-Flight Mass Spectrometry |
References
- Clapp, J.; Moseley, W.G.; Burlingame, B.; Termine, P. The case for a six-dimensional food security framework. Food Policy 2022, 106, 102164. [Google Scholar] [CrossRef]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 2023, 21, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Molotoks, A.; Smith, P.; Dawson, T.P. Impacts of land use, population, and climate change on global food security. Food Energy Secur. 2021, 10, e261. [Google Scholar] [CrossRef]
- Sangaraju, R.; Kumar, R.; Huynh, T.; Sinha, S.N. Pesticide Effects on Human Health and Pest Management; IntechOpen: London, UK, 2024. [Google Scholar]
- Muhammed, M.T.; Aki-Yalcin, E. Molecular docking: Principles, advances, and its applications in drug discovery. Lett. Drug Des. Discov. 2024, 21, 480–495. [Google Scholar] [CrossRef]
- Paggi, J.M.; Pandit, A.; Dror, R.O. The art and science of molecular docking. Annu. Rev. Biochem. 2024, 93, 389–410. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.J.; Corte, D.D. Using molecular docking and molecular dynamics to investigate protein-ligand interactions. Mod. Phys. Lett. B 2021, 35, 2130002. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, S.; He, R.; Liang, G. Application of molecular simulation methods in food science: Status and prospects. J. Agric. Food Chem. 2023, 71, 2684–2703. [Google Scholar] [CrossRef] [PubMed]
- Pranckutė, R. Web of Science (WoS) and Scopus: The titans of bibliographic information in today’s academic world. Publications 2021, 9, 12. [Google Scholar] [CrossRef]
- Arruda, H.; Silva, E.R.; Lessa, M.; Proença, D., Jr.; Bartholo, R. VOSviewer and bibliometrix. J. Med. Libr. Assoc. JMLA 2022, 110, 392. [Google Scholar] [CrossRef]
- McAllister, J.T.; Lennertz, L.; Atencio Mojica, Z. Mapping a discipline: A guide to using VOSviewer for bibliometric and visual analysis. Sci. Technol. Libr. 2022, 41, 319–348. [Google Scholar] [CrossRef]
- Santos, M.; Cajaiba, R.L.; Bastos, R.; Gonzalez, D.; Petrescu Bakış, A.-L.; Ferreira, D.; Leote, P.; Barreto da Silva, W.; Cabral, J.A.; Gonçalves, B. Why do agroforestry systems enhance biodiversity? Evidence from habitat amount hypothesis predictions. Front. Ecol. Evol. 2022, 9, 630151. [Google Scholar] [CrossRef]
- Mourao, P.R.; Martinho, V.D. Forest entrepreneurship: A bibliometric analysis and a discussion about the co-authorship networks of an emerging scientific field. J. Clean. Prod. 2020, 256, 120413. [Google Scholar] [CrossRef]
- Van Eck, N.; Waltman, L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scim 2010, 84, 523–538. [Google Scholar] [CrossRef]
- Sood, S.K.; Kumar, N.; Saini, M. Scientometric analysis of literature on distributed vehicular networks: VOSViewer visualization techniques. Artif. Intell. Rev. 2021, 54, 6309–6341. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Sidhu, K.S.; Bhangu, S.K.; Pathak, R.K.; Yadav, I.S.; Chhuneja, P. Identification of natural lead compounds for leaf rust of Wheat: A molecular docking and simulation study. J. Proteins Proteom. 2020, 11, 283–295. [Google Scholar] [CrossRef]
- El-Zemity, S.R.; Esmaiel, K.E.; Badawy, M.E. Design, synthesis, pharmacophore modeling, and molecular docking of some novel chloroacetamide derivatives as herbicidal agents. Chem. Biol. Technol. Agric. 2024, 11, 124. [Google Scholar] [CrossRef]
- Choudhir, G.; Shamsi, A.; Shahid, M.; Ahmed, A.; Hassan, M.I.; Islam, A. Targeting Rice Blast Disease: Evaluating Binding Affinity of Trichoderma Metabolite to Pyricularia oryzae GSK-1 through through molecular modeling approaches. Next Res. 2025, 2, 100363. [Google Scholar] [CrossRef]
- Mascarenhas, A.M.S.; de Almeida, R.B.M.; de Araujo Neto, M.F.; Mendes, G.O.; da Cruz, J.N.; Dos Santos, C.B.R.; Botura, M.B.; Leite, F.H.A. Pharmacophore-based virtual screening and molecular docking to identify promising dual inhibitors of human acetylcholinesterase and butyrylcholinesterase. J. Biomol. Struct. Dyn. 2021, 39, 6021–6030. [Google Scholar] [CrossRef]
- Zhang, B.; Li, H.; Yu, K.; Jin, Z. Molecular docking-based computational platform for high-throughput virtual screening. CCF Trans. High Perform. Comput. 2022, 4, 63–74. [Google Scholar] [CrossRef]
- Molla, M.H.R.; Aljahdali, M.O.; Sumon, M.A.A.; Asseri, A.H.; Altayb, H.N.; Islam, M.S.; Alsaiari, A.A.; Opo, F.D.M.; Jahan, N.; Ahammad, F. Integrative ligand-based pharmacophore modeling, virtual screening, and molecular docking simulation approaches identified potential lead compounds against pancreatic cancer by targeting FAK1. Pharmaceuticals 2023, 16, 120. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Prieto-Martínez, F.D.; Arciniega, M.; Medina-Franco, J.L. Molecular docking: Current advances and challenges. TIP. Rev. Espec. en Cienc. Quím.-Biol. 2018, 21. [Google Scholar] [CrossRef]
- Joshi, T.; Joshi, T.; Sharma, P.; Chandra, S.; Pande, V. Molecular docking and molecular dynamics simulation approach to screen natural compounds for inhibition of Xanthomonas oryzae pv. Oryzae by targeting peptide deformylase. J. Biomol. Struct. Dyn. 2021, 39, 823–840. [Google Scholar] [CrossRef]
- Hou, Y.; Bai, Y.; Lu, C.; Wang, Q.; Wang, Z.; Gao, J.; Xu, H. Applying molecular docking to pesticides. Pest Manag. Sci. 2023, 79, 4140–4152. [Google Scholar] [CrossRef]
- Stankovic, S.; Kostic, M.; Kostic, I.; Krnjajic, S. Practical approaches to pest control: The use of natural compounds. In Pests, Weeds and Diseases in Agricultural Crop and Animal Husbandry Production; IntechOpen: London, UK, 2020. [Google Scholar]
- Koul, O. Plant Biodiversity as a Resource for Natural Products for Insect Pest Management; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 85–105. [Google Scholar]
- Souto, A.L.; Sylvestre, M.; Tölke, E.D.; Tavares, J.F.; Barbosa-Filho, J.M.; Cebrián-Torrejón, G. Plant-derived pesticides as an alternative to pest management and sustainable agricultural production: Prospects, applications and challenges. Molecules 2021, 26, 4835. [Google Scholar] [CrossRef]
- Hashimi, M.H.; Hashimi, R.; Ryan, Q. Toxic effects of pesticides on humans, plants, animals, pollinators and beneficial organisms. Asian Plant Res. J. 2020, 5, 37–47. [Google Scholar] [CrossRef]
- Berestetskiy, A. Modern approaches for the development of new herbicides based on natural compounds. Plants 2023, 12, 234. [Google Scholar] [CrossRef]
- Charpentier, T.; Viault, G.; Le Ray, A.-M.; Bataillé-Simoneau, N.; Helesbeux, J.-J.; Blon, N.; Bastide, F.; Marchi, M.; Aligon, S.; Bruguière, A. Natural Products Targeting the Fungal Unfolded Protein Response as an Alternative Crop Protection Strategy. J. Agric. Food Chem. 2023, 71, 13706–13716. [Google Scholar] [CrossRef]
- Rani, P.; Rajak, B.K.; Mahato, G.K.; Rathore, R.S.; Chandra, G.; Singh, D.V. Strategic lead compound design and development utilizing computer-aided drug discovery (CADD) to address herbicide-resistant Phalaris minor in wheat fields. Pest Manag. Sci. 2024, 81, 2469–2479. [Google Scholar] [CrossRef]
- Dutta, A.; Mandal, A.; Kundu, A.; Malik, M.; Chaudhary, A.; Khan, M.R.; Shanmugam, V.; Rao, U.; Saha, S.; Patanjali, N. Deciphering the behavioral response of Meloidogyne incognita and Fusarium oxysporum toward mustard essential oil. Front. Plant Sci. 2021, 12, 714730. [Google Scholar] [CrossRef] [PubMed]
- Taruna, A.; Dewi, R.R.; Hadiwijoyo, E.; Yulianah, I.; Syib’li, M.A.; Abadi, A.L. Molecular Docking and In vitro Study Revealed the Inhibition Mechanism of Cutinase of Fusarium oxsyporum f. sp lycopersici by Natural Compounds of Local Turmeric in Indonesia. AGRIVITA J. Agric. Sci. 2023, 45, 554–569. [Google Scholar] [CrossRef]
- Malik, A.; Afaq, S.; El Gamal, B.; Abd Ellatif, M.; Hassan, W.N.; Dera, A.; Noor, R.; Tarique, M. Molecular docking and pharmacokinetic evaluation of natural compounds as targeted inhibitors against Crz1 protein in Rhizoctonia solani. Bioinformation 2019, 15, 277. [Google Scholar] [CrossRef]
- Hamdy, E.; El-Gendi, H.; Al-Askar, A.; El-Far, A.; Kowalczewski, P.; Behiry, S.; Abdelkhalek, A. Copper oxide nanoparticles-mediated Heliotropium bacciferum leaf extract: Antifungal activity and molecular docking assays against strawberry pathogens. Open Chem. 2024, 22, 20240028. [Google Scholar] [CrossRef]
- Islam, A.S.; Bhuiyan, R.; Nihad, S.A.I.; Akter, R.; Khan, M.A.I.; Akter, S.; Islam, M.R.; Khokon, M.A.R.; Latif, M.A. Green synthesis and characterization of silver nanoparticles and its efficacy against Rhizoctonia solani, a fungus causing sheath blight disease in rice. PLoS ONE 2024, 19, e0304817. [Google Scholar] [CrossRef] [PubMed]
- Mare, A.D.; Ciurea, C.N.; Man, A.; Mareș, M.; Toma, F.; Berța, L.; Tanase, C. In vitro antifungal activity of silver nanoparticles biosynthesized with beech bark extract. Plants 2021, 10, 2153. [Google Scholar] [CrossRef]
- Irshad, M.A.; Hussain, A.; Nasim, I.; Nawaz, R.; Al-Mutairi, A.A.; Azeem, S.; Rizwan, M.; Al-Hussain, S.A.; Irfan, A.; Zaki, M.E. Exploring the antifungal activities of green nanoparticles for sustainable agriculture: A research update. Chem. Biol. Technol. Agric. 2024, 11, 133. [Google Scholar] [CrossRef]
- Ren, Z.; Chhetri, A.; Guan, Z.; Suo, Y.; Yokoyama, K.; Lee, S.-Y. Structural basis for inhibition and regulation of a chitin synthase from Candida albicans. Nat. Struct. Mol. Biol. 2022, 29, 653–664. [Google Scholar] [CrossRef]
- Chen, W.; Cao, P.; Liu, Y.; Yu, A.; Wang, D.; Chen, L.; Sundarraj, R.; Yuchi, Z.; Gong, Y.; Merzendorfer, H. Structural basis for directional chitin biosynthesis. Nature 2022, 610, 402–408. [Google Scholar] [CrossRef]
- Ravindran, K.; Sivaramakrishnan, S.; Hussain, M.; Dash, C.K.; Bamisile, B.S.; Qasim, M.; Liande, W. Investigation and molecular docking studies of Bassianolide from Lecanicillium lecanii against Plutella xylostella (Lepidoptera: Plutellidae). Comp. Biochem. Physiol. Part. C Toxicol. Pharmacol. 2018, 206, 65–72. [Google Scholar] [CrossRef]
- Hashem, A.S.; Ramadan, M.M.; Abdel-Hady, A.A.; Sut, S.; Maggi, F.; Dall’Acqua, S. Pimpinella anisum essential oil nanoemulsion toxicity against Tribolium castaneum? Shedding light on its interactions with aspartate aminotransferase and alanine aminotransferase by molecular docking. Molecules 2020, 25, 4841. [Google Scholar] [CrossRef]
- Méndez-Álvarez, D.; Herrera-Mayorga, V.; Juárez-Saldivar, A.; Paz-González, A.D.; Ortiz-Pérez, E.; Bandyopadhyay, D.; Pérez-Sánchez, H.; Rivera, G. Ligand-based virtual screening, molecular docking, and molecular dynamics of eugenol analogs as potential acetylcholinesterase inhibitors with biological activity against Spodoptera frugiperda. Mol. Divers. 2022, 26, 2025–2037. [Google Scholar] [CrossRef]
- Darrag, H.M.; Almuhanna, H.T.; Hakami, E.H. Secondary metabolites in basil, bio-insecticide, inhibition effect, and in silico molecular docking against proteolytic enzymes of the red palm weevil (Rhynchophorus ferrugineus). Plants 2022, 11, 1087. [Google Scholar] [CrossRef]
- Herrera-Mayorga, V.; Guerrero-Sánchez, J.A.; Méndez-Álvarez, D.; Paredes-Sánchez, F.A.; Rodríguez-Duran, L.V.; Niño-García, N.; Paz-González, A.D.; Rivera, G. Insecticidal activity of organic extracts of Solidago graminifolia and its main metabolites (quercetin and chlorogenic acid) against Spodoptera frugiperda: An in vitro and in silico approach. Molecules 2022, 27, 3325. [Google Scholar] [CrossRef]
- Mangat, H.K.; Rani, M.; Pathak, R.K.; Yadav, I.S.; Utreja, D.; Chhuneja, P.K.; Chhuneja, P. Virtual screening, molecular dynamics and binding energy-MM-PBSA studies of natural compounds to identify potential EcR inhibitors against Bemisia tabaci Gennadius. PLoS ONE 2022, 17, e0261545. [Google Scholar] [CrossRef] [PubMed]
- Houzi, G.; El Abdali, Y.; Beniaich, G.; Chebaibi, M.; Taibi, M.; Elbouzidi, A.; Kaioua, S.; Asehraou, A.; Addi, M.; Chaabane, K. Antifungal, Insecticidal, and Repellent Activities of Rosmarinus officinalis Essential Oil and Molecular Docking of Its Constituents against Acetylcholinesterase and β-Tubulin. Scientifica 2024, 2024, 5558041. [Google Scholar] [CrossRef]
- Zhu, F.; He, S.; Ni, C.; Wu, Y.; Wu, H.; Wen, L. Study on the structure-activity relationship of rice immunopeptides based on molecular docking. Food Chem. X 2024, 21, 101158. [Google Scholar] [CrossRef] [PubMed]
- Meher, J.; Sarkar, A.; Sarma, B.K. Binding of stress-responsive OsWRKY proteins through WRKYGQK heptapeptide residue with the promoter region of two rice blast disease resistance genes Pi2 and Pi54 is important for development of blast resistance. 3 Biotech 2023, 13, 294. [Google Scholar] [CrossRef]
- Chen, Y.H.; Dai, K.; Zhang, H.; Wu, Y.H.; Wang, C.T.; Liu, X.Q.; Liu, X.Q. Spectroscopic and molecular docking study on the interaction between salicylic acid and the induced disease-resistant protein OsAAA1 of rice. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2017, 173, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Liu, M.; Yu, Y.; Yang, Y.; Fang, A.; Tian, B.; Wang, J.; Bi, C. Amino acid mutation of succinate dehydrogenase complex induced resistance to benzovindiflupyr in Magnaporthe oryzae. Pestic. Biochem. Physiol. 2024, 203, 106027. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, K.; Fujisaki, K.; Shimizu, M.; Takeda, T.; Nemoto, K.; Saitoh, H.; Hirabuchi, A.; Hiraka, Y.; Miyaji, N.; Białas, A. The blast pathogen effector AVR-Pik binds and stabilizes rice heavy metal-associated (HMA) proteins to co-opt their function in immunity. PLoS Path. 2024, 20, e1012647. [Google Scholar] [CrossRef]
- Yan, J.; Li, L.; Bao, J.; Wang, J.; Liu, X.; Lin, F.; Zhu, X. A glance at structural biology in advancing rice blast fungus research. Virulence 2024, 15, 2403566. [Google Scholar] [CrossRef]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525. [Google Scholar] [CrossRef]
- Zhao, S.; Li, M.; Ren, X.; Wang, C.; Sun, X.; Sun, M.; Yu, X.; Wang, X. Enhancement of broad-spectrum disease resistance in wheat through key genes involved in systemic acquired resistance. Front. Plant Sci. 2024, 15, 1355178. [Google Scholar] [CrossRef]
- Liu, W.; Liu, R.; Qin, Q.; Wang, H.; Zhang, X.; Meng, G. Molecular docking and molecular dynamics simulation of wheat gluten-derived antioxidant peptides acting through the Keap1-Nrf2 pathway. J. Sci. Food Agric. 2024, 104, 8150–8161. [Google Scholar] [CrossRef]
- Li, G.; Zhang, L.; Li, Y.; Li, X.; Gao, X.; Dai, T.; Miao, J.; Liu, X. Analysis of resistance risk and mechanism of the 14α-demethylation inhibitor ipconazole in Fusarium pseudograminearum. Pestic. Biochem. Physiol. 2024, 199, 105786. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, T.; An, X.; Ma, H.; Wang, M. Design, synthesis, antifungal activity and molecular docking of novel pyrazole-4-carboxamides containing tertiary alcohol and difluoromethyl moiety as potential succinate dehydrogenase inhibitors. Pest Manag. Sci. 2024, 80, 2032–2041. [Google Scholar] [CrossRef]
- Helal, N.M.; Saudy, H.S.; Hamada, M.M.; El-Yazied, A.A.; El-Gawad, H.G.A.; Mukherjee, S.; Al-Qahtani, S.M.; Al-Harbi, N.A.; El-Sayed, S.M.; Ibrahim, M.F. Potentiality of Melatonin for Reinforcing Salinity Tolerance in Sorghum Seedlings via Boosting Photosynthetic Pigments, Ionic and Osmotic Homeostasis and Reducing the Carbonyl/Oxidative Stress Markers. J. Soil Sci. Plant Nutr. 2024, 24, 4243–4260. [Google Scholar] [CrossRef]
- Jadhav, A.R.; War, A.R.; Nikam, A.N.; Adhav, A.S.; Gupta, V.S.; Sharma, H.C.; Giri, A.P.; Tamhane, V.A. Capsicum annuum proteinase inhibitor ingestion negatively impacts the growth of sorghum pest Chilo partellus and promotes differential protease expression. Biochem. Biophys. Rep. 2016, 8, 302–309. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mudasir, M.; Baig, M.M.A.; Sultan, Y.; Baig, A. Structural and functional characterization of pyrabactin resistance 1-like (PYL) proteins and molecular docking analysis provides insight into stress tolerance in Sorghum bicolor. Genet. Resour. Crop Evol. 2024, 72, 4771–4787. [Google Scholar] [CrossRef]
- Mustapha, T.; B, S.; Zubair, T.; Patil, R.B.; Bhongade, B.A.; Sangshetti, J.N.; Mali, A.; Babalola, B.J.; Moin, A.T.; Islam, T. In vitro and in silico investigation of effects of antimicrobial peptides from Solanaceae plants against rice sheath blight pathogen Rhizoctinia solani. PLoS ONE 2024, 19, e0302440. [Google Scholar] [CrossRef] [PubMed]
- Dehury, B.; Patra, M.C.; Maharana, J.; Sahu, J.; Sen, P.; Modi, M.K.; Choudhury, M.D.; Barooah, M. Structure-based computational study of two disease resistance gene homologues (Hm1 and Hm2) in maize (Zea mays L.) with implications in plant-pathogen interactions. PLoS ONE 2014, 9, e97852. [Google Scholar] [CrossRef] [PubMed]
- Shabir, S.; Tehmina, A.; Waheed, A.; Saddam, H.; Rafiq, H.; Li, G. Biological evaluation, GC-MS profiling, and molecular docking studies of some essential oils against postharvest pathogens of maize. Arab. J. Chem. 2023, 16, 105339. [Google Scholar] [CrossRef]
- Debnath, S.; Elgorban, A.M.; Bahkali, A.H.; Eswaramoorthy, R.; Verma, M.; Tiwari, P.; Wang, S.; Wong, L.S.; Syed, A. Exploring the efficacy of 1-amino-cyclopropane-1-carboxylic acid (ACCA) as a natural compound in strengthening maize resistance against biotic and abiotic stressors: An empirical computational study. Front. Microbiol. 2023, 14, 1232086. [Google Scholar] [CrossRef]
- Tessema, F.B.; Belachew, A.M.; Gonfa, Y.H.; Asfaw, T.B.; Admassie, Z.G.; Bachheti, A.; Bachheti, R.K.; Tadesse, M.G. Efficacy of fumigant compounds from essential oil of feverfew (Chrysanthemum parthenium L.) against maize weevil (Sitophilus zeamais Mots.): Fumigant toxicity test and in-silico study. Bull. Chem. Soc. Ethiop. 2024, 38, 457–472. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, W.; Yang, M.; Niu, J.; Huang, W.; Chen, Z.; Chen, Y.; Wang, D.; Zhang, J.; Wu, S. Structure-guided discovery of novel AflG inhibitors for aflatoxin contamination control in aspergillus flavus. Front. Microbiol. 2024, 15, 1425790. [Google Scholar] [CrossRef]
- Selim, S.; Al-Sanea, M.M.; Alhejely, A.; Moawad, H.; Masmali, I.; Hendawy, O.M. Degradative Potential of Laccase and Manganese Peroxidase to Mycotoxins on Infected Maize Grains by Fungi with Docking Interaction Studies. BioResources 2024, 19, 9773. [Google Scholar] [CrossRef]
- Menchaca, T.M.; Juárez-Portilla, C.; Zepeda, R.C. Past, Present, and Future of Molecular Docking; IntechOpen: London, UK, 2020. [Google Scholar]
- Shamim, S.; Munawar, R.; Rashid, Y.; Qadar, S.M.Z.; Bushra, R.; Begum, I.; Imran, M.; Quds, T. Molecular Docking: An Insight from Drug Discovery to Drug Repurposing Approach; IntechOpen: London, UK, 2024. [Google Scholar]
- Pons, C.; Grosdidier, S.; Solernou, A.; Pérez-Cano, L.; Fernández-Recio, J. Present and future challenges and limitations in protein-protein docking. Proteins: Struct. Funct. Bioinform. 2010, 78, 95–108. [Google Scholar] [CrossRef]
- Agu, P.; Afiukwa, C.; Orji, O.; Ezeh, E.; Ofoke, I.; Ogbu, C.; Ugwuja, E.; Aja, P. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 60, 273–343. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fu, A.; Zhang, L. An overview of scoring functions used for protein-ligand interactions in molecular docking. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 320–328. [Google Scholar] [CrossRef]
- Fan, J.; Fu, A.; Zhang, L. Progress in molecular docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Hajiahmadi, Z.; Abedi, A.; Wei, H.; Sun, W.; Ruan, H.; Zhuge, Q.; Movahedi, A. Identification, evolution, expression, and docking studies of fatty acid desaturase genes in wheat (Triticum aestivum L.). BMC Genom. 2020, 21, 778. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ghosh, A.; Mukherjee, A. Nanoencapsulation-based edible coating of essential oils as a novel green strategy against fungal spoilage, mycotoxin contamination, and quality deterioration of stored fruits: An overview. Front. Microbiol. 2021, 12, 768414. [Google Scholar] [CrossRef]
- Omar, H.S.; Al Mutery, A.; Osman, N.H.; Reyad, N.E.-H.A.; Abou-Zeid, M.A. Genetic diversity, antifungal evaluation and molecular docking studies of Cu-chitosan nanoparticles as prospective stem rust inhibitor candidates among some Egyptian wheat genotypes. PLoS ONE 2021, 16, e0257959. [Google Scholar] [CrossRef] [PubMed]
- Broughton, W.J.; Hernández, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.)-model food legumes. Plant Soil 2003, 252, 55–128. [Google Scholar] [CrossRef]
- Basavaraja, T.; Pratap, A.; Dubey, V.; Gurumurthy, S.; Gupta, S.; Singh, N. Molecular and Conventional Breeding Strategies for Improving Biotic Stress Resistance in Common Bean. In Accelerated Plant Breeding, Volume 3: Food Legumes; Springer International Publishing: Cham, Switzerland, 2020; pp. 389–421. [Google Scholar]
- Aamir, M.; Singh, V.K.; Dubey, M.K.; Meena, M.; Kashyap, S.P.; Katari, S.K.; Upadhyay, R.S.; Umamaheswari, A.; Singh, S. In silico prediction, characterization, molecular docking, and dynamic studies on fungal SDRs as novel targets for searching potential fungicides against Fusarium wilt in tomato. Front. Pharmacol. 2018, 9, 1038. [Google Scholar] [CrossRef]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef]
- Pratap, A.; Kumar, S.; Polowick, P.L.; Blair, M.W.; Baum, M. Accelerating genetic gains in pulses. Front. Plant Sci. 2022, 13, 879377. [Google Scholar] [CrossRef]
- Langridge, P.; Reynolds, M.P. Genomic tools to assist breeding for drought tolerance. Curr. Opin. Biotechnol. 2015, 32, 130–135. [Google Scholar] [CrossRef]
- Muhammad, A.; Kong, X.; Zheng, S.; Bai, N.; Li, L.; Khan, M.H.U.; Fiaz, S.; Zhang, Z. Exploring plant-microbe interactions in adapting to abiotic stress under climate change: A review. Front. Plant Sci. 2024, 15, 1482739. [Google Scholar] [CrossRef] [PubMed]
- Alazmi, M.; Alshammari, N.; Alanazi, N.A.; Sulieman, A.M.E. In silico characterization, docking, and simulations to understand host-pathogen interactions in an effort to enhance crop production in date palms. J. Mol. Model. 2021, 27, 339. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gao, T.; Guo, H.; Zhao, L.; Lv, S.; Lv, J.; Yao, R.; Yu, Y.; Ma, F. Application of molecular dynamics simulation for exploring the roles of plant biomolecules in promoting environmental health. Sci. Total Environ. 2023, 869, 161871. [Google Scholar] [CrossRef]
- Ruff, K.M.; Pappu, R.V. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Nussinov, R.; Zhang, M.; Liu, Y.; Jang, H. AlphaFold, artificial intelligence (AI), and allostery. J. Phys. Chem. 2022, 126, 6372–6383. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Ma, L.; Zhou, J. Applications of CRISPR/Cas genome editing in economically important fruit crops: Recent advances and future directions. Mol. Hortic. 2023, 3, 1. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, P.; Yu, X.; Xu, J.; Liu, G. Physiological and molecular mechanisms of rice tolerance to salt and drought stress: Advances and future directions. Int. J. Mol. Sci. 2024, 25, 9404. [Google Scholar] [CrossRef]
- Zhang, M.; Guo, M.; Chen, N.; Tang, Z.; Xiang, J.; Yang, L.; Wang, G.; Yang, B.; Li, H. An Integration of UPLC-Q-TOF-MS, GC-MS, Electronic Nose, Electronic Tongue, and Molecular Docking for the Study of the Chemical Properties and Flavor Profiles of Moringa oleifera Leaves. Chemosensors 2024, 12, 199. [Google Scholar] [CrossRef]
- Li, W.; Chen, W.; Wu, D.; Zhang, Z.; Yang, Y. Taste peptides derived from Stropharia rugosoannulata fermentation mycelium and molecular docking to the taste receptor T1R1/T1R3. Front. Nutr. 2022, 9, 960218. [Google Scholar] [CrossRef]
- Saha, A.; Arantes, P.R.; Palermo, G. Dynamics and mechanisms of CRISPR-Cas9 through the lens of computational methods. Curr. Opin. Struct. Biol. 2022, 75, 102400. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-m.; Park, Y.; Berani, U.; Bang, E.; Vankerschaver, J.; Van Messem, A.; De Neve, W.; Shim, H. In silico optimization of RNA-protein interactions for CRISPR-Cas13-based antimicrobials. Biol. Direct 2022, 17, 27. [Google Scholar] [CrossRef]
- Nuñez-Muñoz, L.; Vargas-Hernández, B.; Hinojosa-Moya, J.; Ruiz-Medrano, R.; Xoconostle-Cázares, B. Plant drought tolerance provided through genome editing of the trehalase gene. Plant Signal. Behav. 2021, 16, 1877005. [Google Scholar] [CrossRef]
- Sivula, T.; Yetukuri, L.; Kalliokoski, T.; Kasnanen, H.; Poso, A.; Pohner, I. Machine learning-boosted docking enables the efficient structure-based virtual screening of giga-scale enumerated chemical libraries. J. Chem. Inf. Model. 2023, 63, 5773–5783. [Google Scholar] [CrossRef]
- Gorgulla, C.; Nigam, A.; Koop, M.; Selim Çınaroğlu, S.; Secker, C.; Haddadnia, M.; Kumar, A.; Malets, Y.; Hasson, A.; Li, M. VirtualFlow 2.0-the next generation drug discovery platform enabling adaptive screens of 69 billion molecules. bioRxiv 2023. [Google Scholar] [CrossRef]
- Lai, H.; Wang, L.; Qian, R.; Huang, J.; Zhou, P.; Ye, G.; Wu, F.; Wu, F.; Zeng, X.; Liu, W. Interformer: An interaction-aware model for protein-ligand docking and affinity prediction. Nat. Commun. 2024, 15, 10223. [Google Scholar] [CrossRef]
- Marin, E.; Kovaleva, M.; Kadukova, M.; Mustafin, K.; Khorn, P.; Rogachev, A.; Mishin, A.; Guskov, A.; Borshchevskiy, V. Regression-based active learning for accessible acceleration of ultra-large library docking. J. Chem. Inf. Model. 2023, 64, 2612–2623. [Google Scholar] [CrossRef]
- Pal, P.; Singh, A.K.; Srivastava, R.K.; Rathore, S.S.; Sahoo, U.K.; Subudhi, S.; Sarangi, P.K.; Prus, P. Circular bioeconomy in action: Transforming food wastes into renewable food resources. Foods 2024, 13, 3007. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Chen, G.; Guo, S.; Lin, Z.; Zeng, Y.; Ren, J.; Wang, Q.; Yang, W.; Liang, Y.; Li, J. Anti-Bacterial and Anti-Biofilm activities of essential oil from citrus reticulata Blanco Cv. Tankan Peel against Listeria monocytogenes. Foods 2024, 13, 3841. [Google Scholar] [PubMed]
- Ahmad, T.; Esposito, F.; Cirillo, T. Valorization of agro-food by-products: Advancing sustainability and sustainable development goals 2030 through functional compounds recovery. Food Biosci. 2024, 62, 105194. [Google Scholar] [CrossRef]
- Craven, N.C.; Singh, R.; Quach, C.D.; Gilmer, J.B.; Crawford, B.; Marin-Rimoldi, E.; Smith, R.; DeFever, R.; Dyukov, M.S.; Fothergill, J.W. Achieving Reproducibility and Replicability of Molecular Dynamics and Monte Carlo Simulations Using the Molecular Simulation Design Framework (MoSDeF). J. Chem. Eng. Data 2025, 70, 2178–2199. [Google Scholar] [CrossRef] [PubMed]
| Species | Metabolite | Tool | Findings | Validation | Reference |
|---|---|---|---|---|---|
| Puccinia triticina (wheat leaf rust) | Cynaroside, Prodelphinidin | AutoDock Vina and GROMACS | Cynaroside and Prodelphinidin showed high binding affinity with MAPK1, suggesting their potential as natural fungicides | Molecular Dynamics Simulation (20 ns); RMSD & PCA confirm complex stability | [18,19] |
| Chenopodium album, Anagallis arvensis, Lolium temulentum, Echinochloa crus-galli | Chloroacetamide derivatives | Molecular Operating Environment (MOE) | Chloroacetamide derivatives showed high affinity for VLCFAS, inhibiting fatty acid synthesis in weeds | In planta: EC50 by foliar bioassay (SPAD); Docking + Pharmacophore modeling | [19] |
| Oryza sativa (rice) | Trichotetronine, Bisorbibutenolide | Molecular Docking and MD | Metabolites from Trichoderma species inhibited the fungal enzyme GSK-1 of Pyricularia oryzae | Molecular Dynamics Simulations (100 ns) | [20] |
| Source | Compound | Pathogen | Tool | Finding | Validation | Reference |
|---|---|---|---|---|---|---|
| Lecanicillium lecanii | Bassianolide | Plutella xylostella | Molecular docking | Bassianolide showed high toxicity against P. xylostella larvae with a mortality rate > 80%. A potent natural insecticide. | In vitro bioassay (3 doses); full chemical characterization (LC-MS, NMR, FTIR); docking study | [44] |
| Piper longum L., Ocimum gratissimum L., Phaseolus vulgaris L., Curcuma longa L., Inula helenium Asso | Bisdemethoxycurcumin, rosmarinic acid, chlorogenic acid, piperanine, dihydropiperlonguminine, piperdardine, dihydrocurcumin, longumosides B | Xanthomonas oryzae pv. oryzae | AutoDock Vina, GROMACS | Compounds with high binding affinity to peptide deformylase, inhibiting the growth of X. oryzae, the cause of bacterial leaf blight in rice. | MD simulation (80 ns), RMSD, MM-PBSA, H-bonds, PCA, ADMET | [26] |
| Fragaria vesca L., Solanum tuberosum L., Vitis vinifera L. | Phenolic acids (caffeic, chlorogenic, ferulic acids) | Botrytis cinerea | AutoDock Vina | Phenolic acids showed high binding affinity with laccase enzyme of B. cinerea, suggesting their use as natural inhibitors. | MD simulation | [31] |
| Pimpinella anisum L. | (E)-anethole, Limonene, α-Himachalene, Linalool, trans-Verbenol | Tribolium castaneum | Molecular Operating Environment (MOE) | The main compounds of the essential oil and its nanoemulsion showed high binding affinity with ALT and AST enzymes, suggesting a potent insecticidal effect. | GC-MS, in vivo toxicity assay (LC50, feeding indices), in vitro enzyme assays (AST, ALT), docking with modeled proteins | [45] |
| Syzygium aromaticum (L.) Merr. and L.M.Perry | Eugenol and its analogs | Spodoptera frugiperda | AutoDock Vina, GROMACS, MM-PBSA | Three new eugenol analogs showed high insecticidal activity against S. frugiperda larvae (LC50 of 0.042 mg/mL by diet and 0.027 mg/mL by topical application). | Homology modeling, MD (50 ns), MM-PBSA, Probit LC50 (in vivo), per-residue binding analysis | [46] |
| Artemisia vulgaris L. | Scopoletin | Amaranthus retroflexus | Virtual Screening and Molecular docking analysis | Scopoletin showed selective inhibitory activity against weed growth such as A. retroflexus. | In vivo: plant growth suppression assay (extract-level only) | [32] |
| Ocimum basilicum L. | Chicoric acid, ursolic acid, salvigenin, nepetoidin B, rosmarinic acid | Rhynchophorus ferrugineus | AutoDock, ADMET, UPLC-MS/MS | Secondary metabolites inhibited proteolytic enzymes of the red palm weevil. Chicoric acid showed the most insecticidal activity with an LC50 of 1132 µg/mL. | Docking + ADMET + in vitro enzymatic inhibition + in vivo larval mortality | [47] |
| Solidago graminifolia (L.) Nutt. | Quercetin, chlorogenic Acid | Spodoptera frugiperda | AutoDock Vina, SwissModel, UPLC-MS/MS | Quercetin showed an LC50 of 0.157 mg/mL with interactions at the AChE active site. Chlorogenic acid showed no insecticidal activity but antagonized the effect of quercetin. | Docking + homology modeling + UPLC-MS + larval mortality assay (LC50) + interaction profiling | [48] |
| Compounds from ZINC Database | ZINC08952607, ZINC04264850 | Bemisia tabaci | Virtual Screening, Molecular Dynamics, MM-PBSA | Both compounds showed high binding affinity with the ecdysteroid receptor of S. frugiperda, suggesting their use as potential insecticides. | Docking + MD (50 ns) + MM-PBSA + physico-chemical profiling; no in vivo validation | [49] |
| Rosmarinus officinalis Spenn. | 1,8-cineole, α-pinene, camphor | Botrytis cinerea, Fusarium oxysporum, Alternaria alternata, Callosobruchus maculatus | GC-MS, Glide (Schrödinger v11.5) | Borneol showed the highest activity against insect acetylcholinesterase, and α-caryophyllene against the β-tubulin of B. cinerea, with Glide Scores of −7.254 and −7.025 kcal/mol, respectively. | Docking + in vitro antifungal (disk diffusion) + insect bioassay (mortality, oviposition, repellency) | [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iñiguez-Luna, M.I.; Cadena-Zamudio, J.D.; Ramírez-Mosqueda, M.A.; Aguirre-Noyola, J.L.; Cadena-Zamudio, D.A.; Cadena-Iñiguez, J.; Armenta-Medina, A. Molecular Docking as a Key Driver of Biocontrol for Agri-Food Security. BioTech 2025, 14, 80. https://doi.org/10.3390/biotech14040080
Iñiguez-Luna MI, Cadena-Zamudio JD, Ramírez-Mosqueda MA, Aguirre-Noyola JL, Cadena-Zamudio DA, Cadena-Iñiguez J, Armenta-Medina A. Molecular Docking as a Key Driver of Biocontrol for Agri-Food Security. BioTech. 2025; 14(4):80. https://doi.org/10.3390/biotech14040080
Chicago/Turabian StyleIñiguez-Luna, María Isabel, Jorge David Cadena-Zamudio, Marco A. Ramírez-Mosqueda, José Luis Aguirre-Noyola, Daniel Alejandro Cadena-Zamudio, Jorge Cadena-Iñiguez, and Alma Armenta-Medina. 2025. "Molecular Docking as a Key Driver of Biocontrol for Agri-Food Security" BioTech 14, no. 4: 80. https://doi.org/10.3390/biotech14040080
APA StyleIñiguez-Luna, M. I., Cadena-Zamudio, J. D., Ramírez-Mosqueda, M. A., Aguirre-Noyola, J. L., Cadena-Zamudio, D. A., Cadena-Iñiguez, J., & Armenta-Medina, A. (2025). Molecular Docking as a Key Driver of Biocontrol for Agri-Food Security. BioTech, 14(4), 80. https://doi.org/10.3390/biotech14040080








