Current Topics on the Integration of Artificial Intelligence in the Histopathological and Molecular Diagnosis of Uveal Melanoma
Abstract
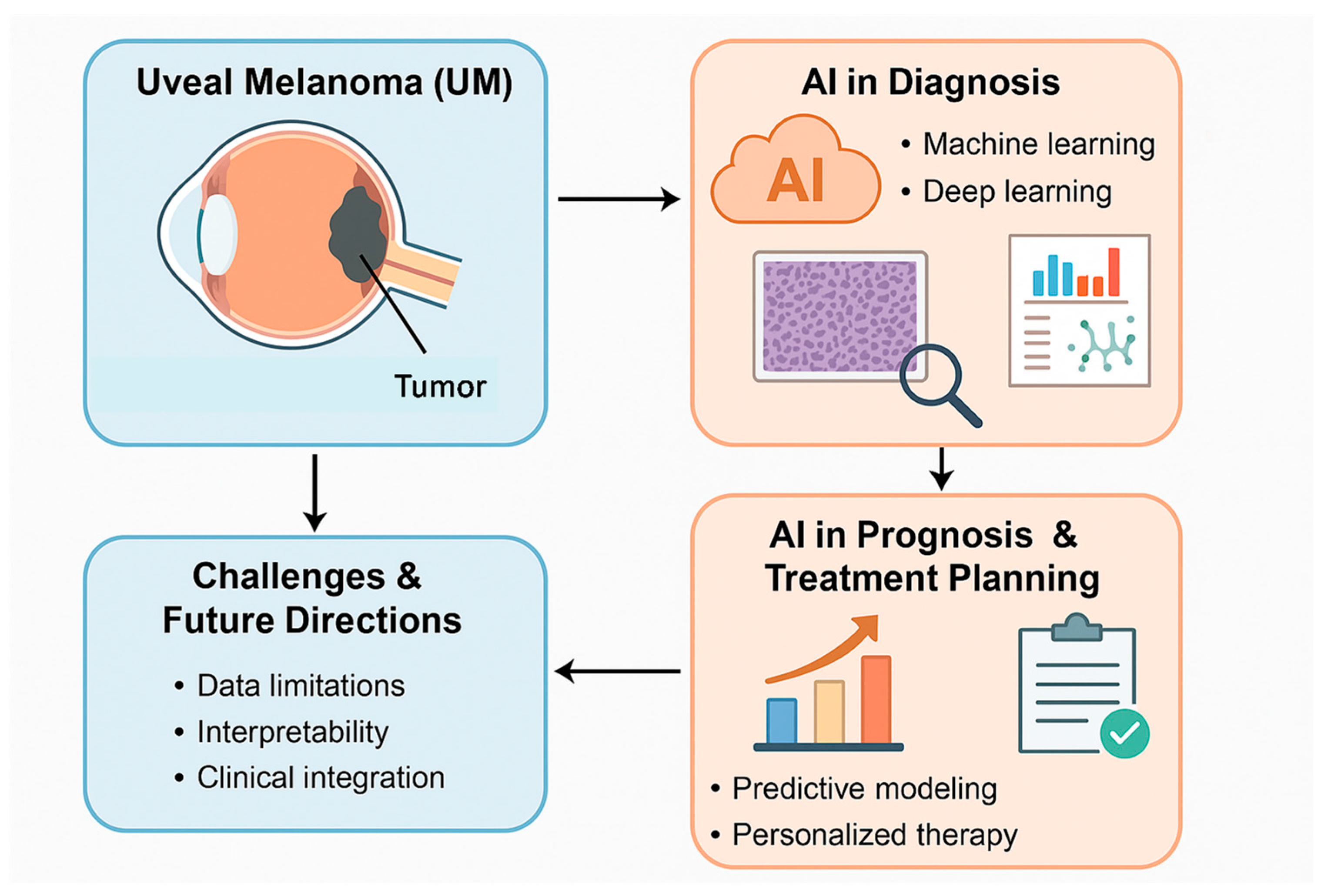
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
- (i)
- Involve patients diagnosed with UM;
- (ii)
- Use AI for histopathological or molecular diagnosis of UM;
- (iii)
- Use AI in predicting patient outcomes associated with UM.
3. Results
4. Discussion
4.1. AI in Histopathological Diagnosis
4.2. AI in Molecular and Genomic Analysis
4.3. AI in Prognostic Prediction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bai, H.; Bosch, J.J.; Heindl, L.M. Current management of uveal melanoma: A review. Clin. Exp. Ophthalmol. 2023, 51, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Dadzie, A.K.; Iddir, S.P.; Abtahi, M.; Ebrahimi, B.; Le, D.; Ganesh, S.; Son, T.; Heiferman, M.J.; Yao, X. Colour fusion effect on deep learning classification of uveal melanoma. Eye 2024, 38, 2781–2787. [Google Scholar] [CrossRef]
- Chandrabhatla, A.S.; Horgan, T.M.; Cotton, C.C.; Ambati, N.K.; Shildkrot, Y.E. Clinical Applications of Machine Learning in the Management of Intraocular Cancers: A Narrative Review. Invest. Ophthalmol. Vis. Sci. 2023, 64, 29. [Google Scholar] [CrossRef]
- Sabazade, S.; Lumia Michalski, M.A.; Bartoszek, J.; Fili, M.; Holmström, M.; Stålhammar, G. Development and Validation of a Deep Learning Algorithm for Differentiation of Choroidal Nevi from Small Melanoma in Fundus Photographs. Ophthalmol. Sci. 2024, 5, 100613. [Google Scholar] [CrossRef]
- Wan, Q.; Ren, X.; Wei, R.; Yue, S.; Wang, L.; Yin, H.; Tang, J.; Zhang, M.; Ma, K.; Deng, Y.P. Deep learning classification of uveal melanoma based on histopathological images and identification of a novel indicator for prognosis of patients. Biol. Proced. Online 2023, 25, 15. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.; Zhang, K.; Hui, S.; Feng, Y.; Luo, J.; Li, Y.; Wei, W. Validation of the Relationship Between Iris Color and Uveal Melanoma Using Artificial Intelligence With Multiple Paths in a Large Chinese Population. Front. Cell Dev. Biol. 2021, 9, 713209. [Google Scholar] [CrossRef] [PubMed]
- Iddir, S.P.; Love, J.; Ma, J.S.; Bryan, J.M.; Ganesh, S.; Heiferman, M.J.; Yi, D. Predicting Malignant Transformation of Choroidal Nevi Using Machine Learning. Res. Sq. 2023; rs.3. [Google Scholar] [CrossRef]
- Kulbay, M.; Marcotte, E.; Remtulla, R.; Lau, T.H.A.; Paez-Escamilla, M.; Wu, K.Y.; Burnier, M.N. Uveal Melanoma: Comprehensive Review of Its Pathophysiology, Diagnosis, Treatment, and Future Perspectives. Biomedicines 2024, 12, 1758. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, L.; Runkel, C.B.; Künzel, S.; Kabiri, P.; Rübsam, A.; Bonaventura, T.; Marquardt, P.; Haas, V.; Biniaminov, N.; Biniaminov, S.; et al. Using Deep Learning to Distinguish Highly Malignant Uveal Melanoma from Benign Choroidal Nevi. J. Clin. Med. 2024, 13, 4141. [Google Scholar] [CrossRef]
- Murali, R.; Wiesner, T.; Scolyer, R.A. Tumours associated with BAP1 mutations. Pathology 2013, 45, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhou, W.; Qi, X.; Zhang, G.; Girnita, L.; Seregard, S.; Grossniklaus, H.E.; Yao, Z.; Zhou, X.; Stålhammar, G. Prediction of BAP1 Expression in Uveal Melanoma Using Densely-Connected Deep Classification Networks. Cancers 2019, 11, 1579. [Google Scholar] [CrossRef]
- Zhang, H.; Kalirai, H.; Acha-Sagredo, A.; Yang, X.; Zheng, Y.; Coupland, S.E. Piloting a Deep Learning Model for Predicting Nuclear BAP1 Immunohistochemical Expression of Uveal Melanoma from Hematoxylin-and-Eosin Sections. Transl. Vis. Sci. Technol. 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.A.; Chen, H.; Gomez, C.; Correa, Z.M.; Unberath, M. Direct Gene Expression Profile Prediction for Uveal Melanoma from Digital Cytopathology Images via Deep Learning and Salient Image Region Identification. Ophthalmol. Sci. 2022, 3, 100240. [Google Scholar] [CrossRef] [PubMed]
- Akram, F.; de Bruyn, D.P.; van den Bosch, Q.C.C.; Trandafir, T.E.; van den Bosch, T.P.P.; Verdijk, R.M.; de Klein, A.; Kiliç, E.; Stubbs, A.P.; Brosens, E.; et al. Prediction of molecular subclasses of uveal melanoma by deep learning using routine haematoxylin-eosin-stained tissue slides. Histopathology 2024, 85, 909–919. [Google Scholar] [CrossRef]
- Liu, T.Y.A.; Zhu, H.; Chen, H.; Arevalo, J.F.; Hui, F.K.; Yi, P.H.; Wei, J.; Unberath, M.; Correa, Z.M. Gene Expression Profile Prediction in Uveal Melanoma Using Deep Learning: A Pilot Study for the Development of an Alternative Survival Prediction Tool. Ophthalmol. Retina. 2020, 4, 1213–1215. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef]
- Reggiani, F.; El Rashed, Z.; Petito, M.; Pfeffer, M.; Morabito, A.; Tanda, E.T.; Spagnolo, F.; Croce, M.; Pfeffer, U.; Amaro, A. Machine Learning Methods for Gene Selection in Uveal Melanoma. Int. J. Mol. Sci. 2024, 25, 1796. [Google Scholar] [CrossRef]
- Xie, J.; Wu, Z.; Xu, X.; Liang, G.; Xu, J. Screening and identification of key genes and pathways in metastatic uveal melanoma based on gene expression using bioinformatic analysis. Medicine 2020, 99, e22974. [Google Scholar] [CrossRef]
- Hou, P.; Bao, S.; Fan, D.; Yan, C.; Su, J.; Qu, J.; Zhou, M. Machine learning-based integrative analysis of methylome and transcriptome identifies novel prognostic DNA methylation signature in uveal melanoma. Brief. Bioinform. 2021, 22, bbaa371. [Google Scholar] [CrossRef]
- Bassi, A.; Krance, S.H.; Pucchio, A.; Pur, D.R.; Miranda, R.N.; Felfeli, T. The Application of Artificial Intelligence in the Analysis of Biomarkers for Diagnosis and Management of Uveitis and Uveal Melanoma: A Systematic Review. Clin. Ophthalmol. 2022, 16, 2895–2908. [Google Scholar] [CrossRef]
- Lever, M.; Bogner, S.; Giousmas, M.; Mairinger, F.D.; Baba, H.A.; Richly, H.; Gromke, T.; Schuler, M.; Bechrakis, N.E.; Kalkavan, H. Prognostic value of clinical and radiomic parameters in patients with liver metastases from uveal melanoma. Pigment. Cell Melanoma Res. 2024, 37, 831–838. [Google Scholar] [CrossRef]
- Chen, Y.N.; Wang, Y.N.; Chen, M.X.; Zhang, K.; Chen, R.T.; Fang, R.; Wang, H.; Zhang, H.H.; Huang, Y.N.; Feng, Y.; et al. Machine learning models for outcome prediction of Chinese uveal melanoma patients: A 15-year follow-up study. Cancer Commun. 2022, 42, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Donizy, P.; Krzyzinski, M.; Markiewicz, A.; Karpinski, P.; Kotowski, K.; Kowalik, A.; Orlowska-Heitzman, J.; Romanowska-Dixon, B.; Biecek, P.; Hoang, M.P. Machine learning models demonstrate that clinicopathologic variables are comparable to gene expression prognostic signature in predicting survival in uveal melanoma. Eur. J. Cancer. 2022, 174, 251–260. [Google Scholar] [CrossRef]
- Luo, J.; Chen, Y.; Yang, Y.; Zhang, K.; Liu, Y.; Zhao, H.; Dong, L.; Xu, J.; Li, Y.; Wei, W. Prognosis Prediction of Uveal Melanoma After Plaque Brachytherapy Based on Ultrasound With Machine Learning. Front. Med. 2022, 8, 777142. [Google Scholar] [CrossRef]
- Wu, S.N.; Qin, D.Y.; Zhu, L.; Guo, S.J.; Li, X.; Huang, C.H.; Hu, J.; Liu, Z. Uveal melanoma distant metastasis prediction system: A retrospective observational study based on machine learning. Cancer Sci. 2024, 115, 3107–3126. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Eleuteri, A.; Fisher, A.C.; Coupland, S.E.; Taktak, A.F. Artificial neural networks estimating survival probability after treatment of choroidal melanoma. Ophthalmology 2008, 115, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Tailor, P.D.; Kopinski, P.K.; D’Souza, H.S.; Leske, D.A.; Olsen, T.W.; Shields, C.L.; Shields, J.A.; Dalvin, L.A. Predicting Choroidal Nevus Transformation to Melanoma Using Machine Learning. Ophthalmol. Sci. 2024, 5, 100584. [Google Scholar] [CrossRef]
- Broggi, G.; Mazzucchelli, M.; Salzano, S.; Barbagallo, G.M.V.; Certo, F.; Zanelli, M.; Palicelli, A.; Zizzo, M.; Koufopoulos, N.; Magro, G.; et al. The emerging role of artificial intelligence in neuropathology: Where are we and where do we want to go? Pathol. Res. Pract. 2024, 263, 155671. [Google Scholar] [CrossRef] [PubMed]
- Broggi, G.; Maniaci, A.; Lentini, M.; Palicelli, A.; Zanelli, M.; Zizzo, M.; Koufopoulos, N.; Salzano, S.; Mazzucchelli, M.; Caltabiano, R. Artificial Intelligence in Head and Neck Cancer Diagnosis: A Comprehensive Review with Emphasis on Radiomics, Histopathological, and Molecular Applications. Cancers 2024, 16, 3623. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- McLean, M.J.; Foster, W.D.; Zimmerman, L.E. Prognostic factors in small malignant melanomas of choroid and ciliary body. Arch. Ophthalmol. 1977, 95, 48–58. [Google Scholar] [CrossRef]
- Folberg, R.; Hendrix, M.J.; Maniotis, A.J. Vasculogenic mimicry and tumor angiogenesis. Am. J. Pathol. 2000, 156, 361–381. [Google Scholar] [CrossRef] [PubMed]
- Folberg, R.; Rummelt, V.; Parys-Van Ginderdeuren, R.; Hwang, T.; Woolson, R.F.; Pe’er, J.; Gruman, L.M. The prognostic value of tumor blood vessel morphology in primary uveal melanoma. Ophthalmology 1993, 100, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Salzano, S.; Vecchio, G.M.; Failla, M.; Russo, A.; Avitabile, T.; Longo, A.; Caltabiano, R.; Broggi, G. Metastases from uveal melanoma may lack S100 expression: A clinico-pathologic and immunohistochemical study with emphasis on potential causes and diagnostic implications. Ann. Diagn. Pathol. 2025, 76, 152464. [Google Scholar] [CrossRef] [PubMed]
- Koseoglu, N.D.; Corrêa, Z.M.; Liu, T.Y.A. Artificial intelligence for ocular oncology. Curr. Opin. Ophthalmol. 2023, 34, 437–440. [Google Scholar] [CrossRef]
- Tahmasebzadeh, A.; Sadeghi, M.; Naseripour, M.; Mirshahi, R.; Ghaderi, R. Artificial intelligence and different image modalities in uveal melanoma diagnosis and prognosis: A narrative review. Photodiagnosis Photodyn. Ther. 2025, 52, 104528. [Google Scholar] [CrossRef]
- Longhitano, L.; Giallongo, S.; Orlando, L.; Broggi, G.; Longo, A.; Russo, A.; Caltabiano, R.; Giallongo, C.; Barbagallo, I.; Di Rosa, M.; et al. Lactate Rewrites the Metabolic Reprogramming of Uveal Melanoma Cells and Induces Quiescence Phenotype. Int. J. Mol. Sci. 2022, 24, 24. [Google Scholar] [CrossRef]
| Study | Advantages | Limitations |
|---|---|---|
| Dadzie AK et al. (2024) [2] | AI provides consistent results, eliminating intraobserver and interobserver variability commonly seen among human specialists; DL, especially CNNs, has demonstrated significant capability in distinguishing UM from choroidal nevi, achieving high F1-scores, accuracy, and area under the curve (AUC) metrics in classification tasks | The performance of AI models heavily relies on the quality and diversity of training datasets. Limited or non-representative datasets can negatively affect results; lesions with unusual features or low-quality images (e.g., those affected by hemorrhage or retinal detachment) may not be accurately classified, limiting AI’s applicability |
| Chandrabhatla AS et al. (2023) [3] | The majority of ML research has demonstrated strong performance despite challenges like small datasets; tools developed with ML may assist in predicting metastatic spread for UM, which is a key area for improving patient outcomes | Limited sample sizes can lead to variability in algorithm performance and restrict generalizability |
| Sabazade S et al. (2024) [4] | The AI algorithm demonstrated superior sensitivity compared to resident and consultant ophthalmologists and matched the performance of ocular oncologists in differentiating small choroidal melanomas from nevi; the algorithm provides identical assessments for the same images, eliminating intraobserver and interobserver variability that can occur with human specialists | The algorithm’s accuracy depends heavily on the quality and precision of the ocular oncologists’ initial diagnoses and the representativeness of the dataset; a relatively small sample size was used for training and validation, which could limit the algorithm’s performance, particularly with atypical lesions not represented in the dataset |
| Wan Q et al. (2023) [5] | The algorithm achieved high accuracy (90%) in predicting vital status from histopathological images; AI identifies subtypes (Cluster1 and Cluster2) with distinct molecular profiles, mutations, and immune microenvironments, aiding in personalized therapeutic approaches | AI identified mutations and proposed biomarkers; although these results are promising, further validation of AI’s predictive value in independent clinical studies is essential for generalizing conclusions |
| Zhang H et al. (2021) [6] | AI was used in analyzing iris color for the potential diagnosis of uveal melanoma | There is a need for more comprehensive approaches, incorporating additional clinical and molecular data to improve diagnostic accuracy |
| Iddir SP et al. (2023) [7] | AI models demonstrated high accuracy in detecting risk factors for the malignant transformation of choroidal lesions using single ultra-widefield (UWF) or B-scan ultrasound (US) images; ML can streamline diagnosis by accurately analyzing risk factors such as subretinal fluid, orange pigment, and tumor thickness, which traditionally rely on subjective evaluation | The study involved a limited number of patients from a single institution, reducing the generalizability of the findings; small features like orange pigment and drusen are difficult to identify accurately, suggesting the need for alternative techniques such as image cropping |
| Kulbay M et al. (2024) [8] | AI models have shown high accuracy in distinguishing between benign choroidal nevi and malignant melanoma; combining histopathological data with clinical features enhances prediction accuracy | There is a need for larger datasets, better integration of imaging data, and the exploration of deep learning techniques |
| Hoffmann L et al. (2024) [9] | AI reduces dependency on expensive multimodal imaging, making it suitable for resource-limited settings; advanced techniques like attention mechanisms and larger datasets can enhance performance | Models were trained on small datasets with no external validation, affecting generalizability; misclassifications often occur in lesions with moderate or borderline features; real-world efficacy has not been confirmed through prospective clinical studies |
| Study | Advantages | Limitations |
|---|---|---|
| Sun et al. (2019) [11] | Good accuracy in predicting BAP1 expression based on histopathology images through DL models | Histopathology slides from two centers were used, which may not adequately capture the full spectrum of nuclear BAP1 expression in UM or account for interlaboratory variability. A significant limitation for model training was the small sample size and the uneven distribution of the four categories (positive, negative, excluded, and blurred). Additionally, a loss of BAP1 expression can occur in a notable subset of cases without a corresponding BAP1 mutation, and the reverse is also possible, potentially creating a misleading impression of normal protein levels |
| Zhang et al. (2020) [12] | Good accuracy in predicting nuclear BAP1 expression through DL models on H&E-stained histopathology slides | Patches from the same slide are similar in tissue morphology, pigment, etc., so this represents “limited knowledge” from the perspective of a DL model; there is a need for larger case series, with multicenter validation |
| Liu et al. (2022) [13] | Considerable sensitivity, accuracy, and specificity for predicting GEP in UM from digital cytopathology images | Small or thin UM tumors may not be suitable for FNABs, which are necessary to produce cytopathology slides. The DL algorithm was trained and validated using cellular aspirates stained with H&E. To ensure compatibility with specimens prepared using Papanicolaou staining, the algorithm requires further adaptation. Additionally, the study population lacked substantial diversity, highlighting the need for prospective validation to confirm the model’s generalizability |
| Akram et al. (2024) [14] | Predicting UM molecular subclasses by CNN using routine H&E-stained slides with promising results | There was a limited dataset size, which requires further training and validation on external cohorts |
| Liu et al. (2020) [15] | Predicting GEP in UM using DL models based on H&E-stained slides from FNAB | FNABs can be technically challenging and susceptible to sampling errors. There was a small patient sample size. There is a need to train an algorithm with a larger database and actual survival data |
| Reggiani et al. (2024) [17] | Developing ML models to identify molecular classes of UM based on different gene alterations | Technical variations in sequencing experiments can obscure or imitate biological differences, potentially leading to model overfitting. This may hinder the extraction of meaningful biological features and limit the model’s ability to perform accurate classification when applied to a different dataset. Furthermore, it remains uncertain whether molecular classification approaches would retain their efficiency when applied to larger datasets, necessitating further investigation |
| Xie et al. (2020) [18] | Employing bioinformatic analysis to identify key genes in metastatic UM | All data were obtained from the Gene Expression Omnibus (GEO) database rather than directly from tissues of UM patients. All conclusions were based on bioinformatic analysis; therefore, caution must be exercised in interpreting the results. The dataset consisted of 39 primary tumor samples and 2 metastatic samples, so there may be a possible bias due to the imbalance between groups |
| Hou et al. (2021) [19] | Identifying DNA methylation signatures in UM, which may be of prognostic relevance, stratifying patients into two risk groups, with significantly different prognosis in terms of survival. | The discovery of the DNA methylation signature was based on in silico analysis. Independent validation of the ML model was challenged by the lack of an available patient cohort with clinical information |
| Study | Advantages | Limitations |
|---|---|---|
| Bassi et al. (2022) [20] | Large amount of information regarding significant biomarkers | There was no mention of the reliability of the biomarker sample collection process. There was a large degree of heterogeneity |
| Lever et al. (2024) [21] | Using ML algorithms to stratify prognosis of UM patients with liver metastasis, identifying clinical, laboratory, and radiomic parameters to predict overall survival | This was a retrospective analysis, with the need for external validation. Combining parameters does not increase the risk stratification accuracy. Training and test groups were not ideally balanced with respect to gender and global outcomes, leading to a smaller proportion of high-risk patients in the test group and, more importantly, a different median risk score |
| Chen et al. (2022) [22] | Identifying clinico-pathological features which may be predictive of death and metastasis in UM; high sensitivity and specificity | This retrospective study was conducted only on a Chinese population |
| Donizy et al. (2022) [23] | Three ML models which identified predictive features for OS and PFS in UM | Possible bias: only patients who underwent ocular enucleation were included in the study |
| Luo et al. (2022) [24] | Prediction of risk of death and metastasis in patients with UM who have undergone plaque brachytherapy | The possibility of metastatic disease at diagnosis may affect the model’s predictive value for metastasis. Due to the retrospective nature of the study, the follow-up interval after surgery was not fixed. |
| Wu et al. (2024) [25] | Developing ML models to identify risk factors for metastatic UM; highlighting differing metastatic potential between ciliary body and choroid melanoma | This was a retrospective analysis, with potential information and selection bias difficulties in establishing causal relationships. There was a lack of external validation with data from different hospitals. The performance of ML models may vary across patients from different regions and hospitals |
| Damato et al. (2008) [26] | Using neural network to estimate survival probability after treatment of UM, generating curves which were similar to Kaplan–Meier | Some combinations of risk factors were rare, so survival could be modeled only imprecisely. There is a need for external validation. Data were obtained from patients treated by enucleation or local resection. There is a need for further studies to determine whether survival predictions would be accurate in patients undergoing non-surgical treatment |
| Tailor et al. (2024) [27] | First study developing ML models to predict malignant transformation of choroidal nevi into melanoma; large multicenter cohort, use of multiple imaging modalities to comprehensively phenotype nevi, and external validation demonstrating model robustness | The retrospective design of the studies, which rely on existing records, introduces inherent limitations, including a lack of tissue confirmation of the disease in most cases. Both datasets predominantly represent a White population, limiting the generalizability of findings to more diverse populations. The reliance on clinician annotations and tabular data, rather than the direct extraction of raw imaging features, may reduce the potential for uncovering novel predictive markers. Furthermore, the imbalanced nature of the classification problem, due to the relatively rare occurrence of transformation events, poses challenges for model training and evaluation. Without further validation and evidence of clinical utility, there is a risk of overstating the practical value of these approaches in guiding clinical integration |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salzano, S.; Broggi, G.; Russo, A.; Avitabile, T.; Longo, A.; Caltabiano, R.; Mazzucchelli, M. Current Topics on the Integration of Artificial Intelligence in the Histopathological and Molecular Diagnosis of Uveal Melanoma. J. Mol. Pathol. 2025, 6, 7. https://doi.org/10.3390/jmp6020007
Salzano S, Broggi G, Russo A, Avitabile T, Longo A, Caltabiano R, Mazzucchelli M. Current Topics on the Integration of Artificial Intelligence in the Histopathological and Molecular Diagnosis of Uveal Melanoma. Journal of Molecular Pathology. 2025; 6(2):7. https://doi.org/10.3390/jmp6020007
Chicago/Turabian StyleSalzano, Serena, Giuseppe Broggi, Andrea Russo, Teresio Avitabile, Antonio Longo, Rosario Caltabiano, and Manuel Mazzucchelli. 2025. "Current Topics on the Integration of Artificial Intelligence in the Histopathological and Molecular Diagnosis of Uveal Melanoma" Journal of Molecular Pathology 6, no. 2: 7. https://doi.org/10.3390/jmp6020007
APA StyleSalzano, S., Broggi, G., Russo, A., Avitabile, T., Longo, A., Caltabiano, R., & Mazzucchelli, M. (2025). Current Topics on the Integration of Artificial Intelligence in the Histopathological and Molecular Diagnosis of Uveal Melanoma. Journal of Molecular Pathology, 6(2), 7. https://doi.org/10.3390/jmp6020007









