Prognostic Significance of JMJD3 Expression in Pleural Mesotheliomas
Abstract
:1. Introduction
2. Results
2.1. Clinicopathological Features
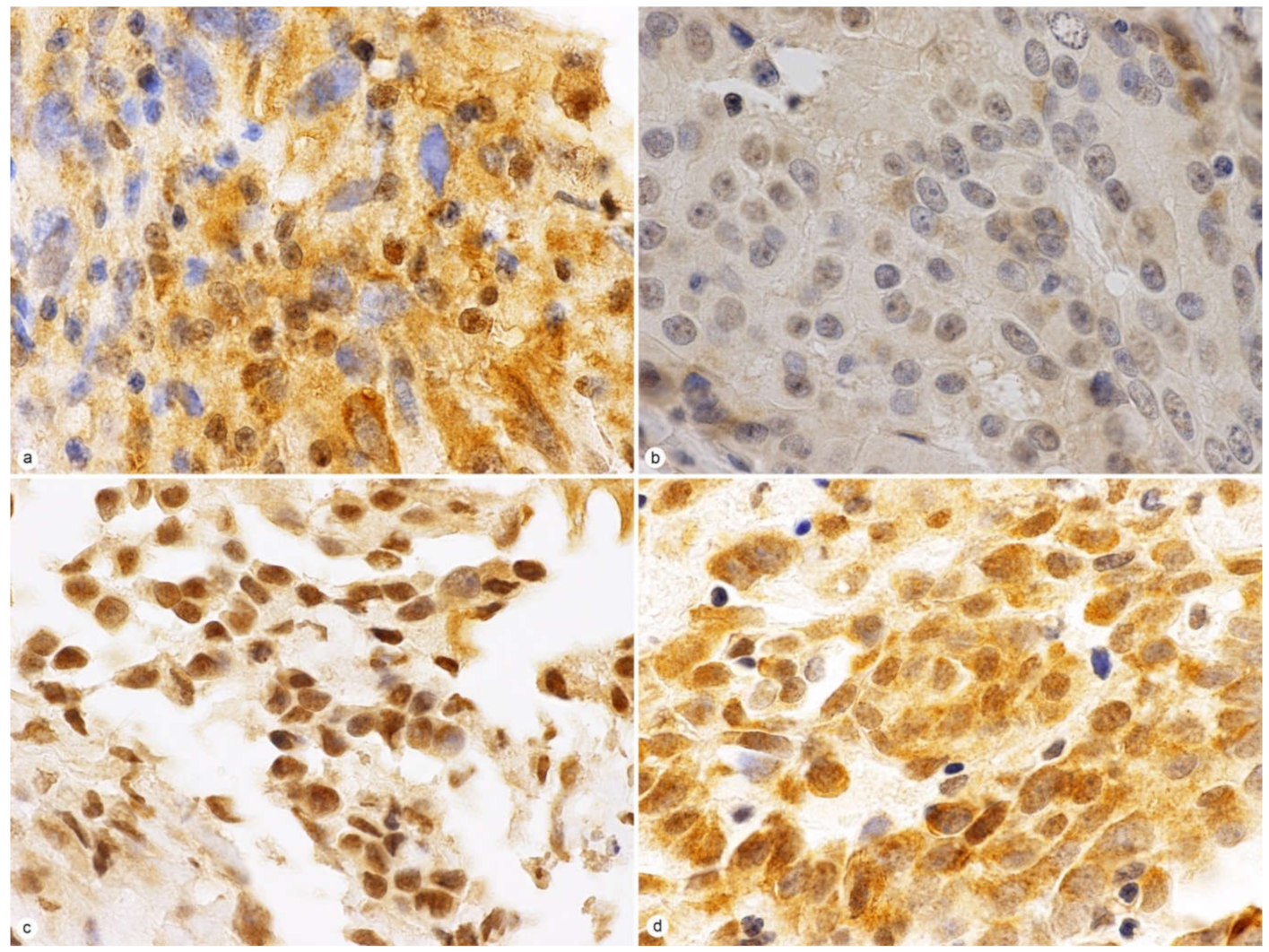
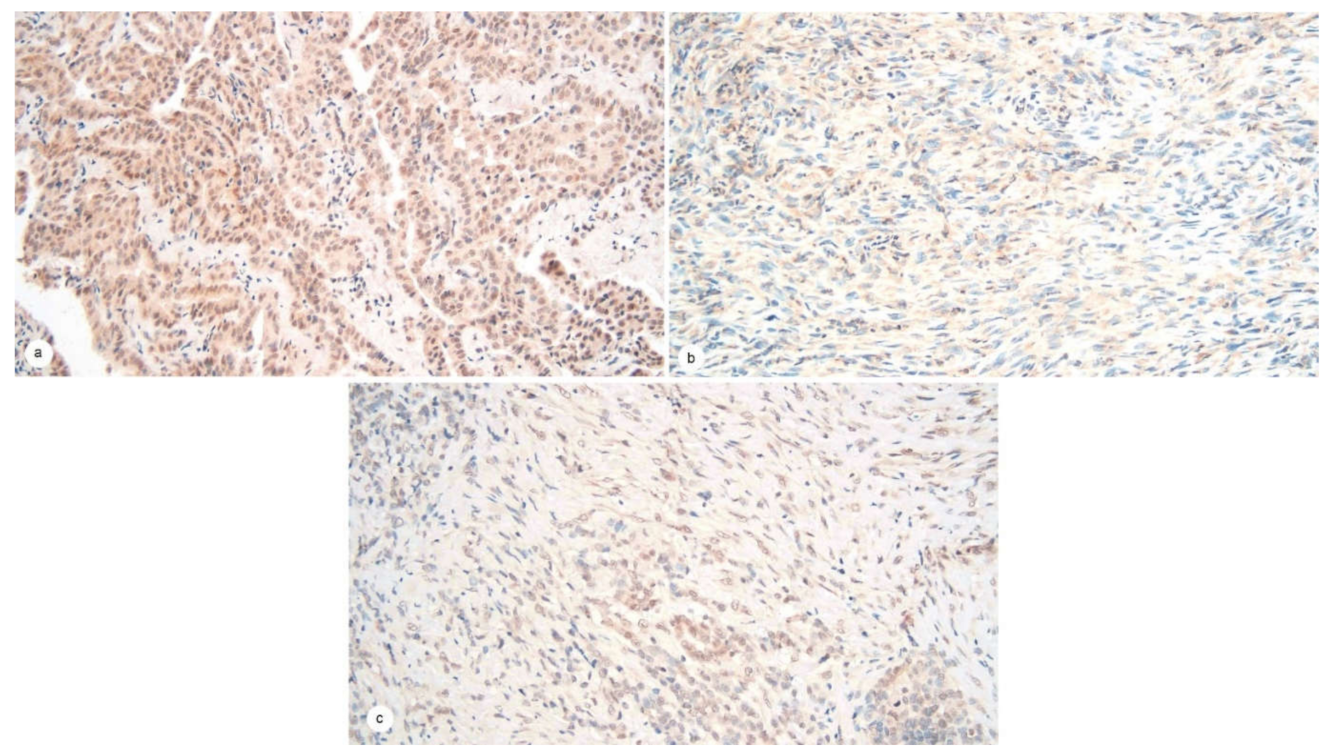
2.2. JMJD3 Expression on IHC
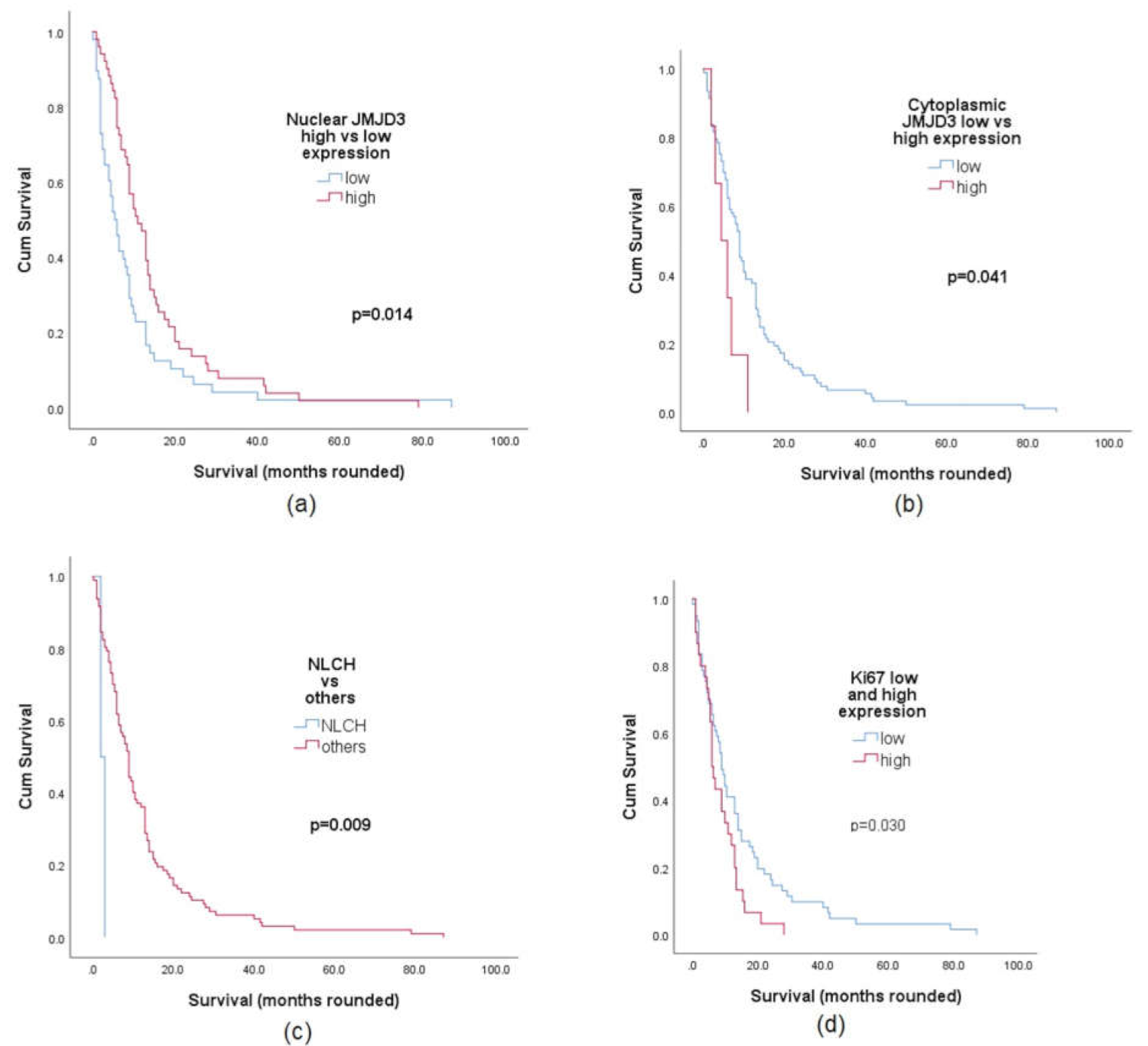
2.3. JMJD3 Association with Survival
2.4. Univariate and Multivariate Analysis of JMJD3 Expression
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. Tissue Microarrays and JMJD3 Immunohistochemical Analysis
4.3. JMJD3 Scoring
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Mesothelioma Fact Sheet. Available online: Gco.iarc.fr/today/data/factsheets/cancers/18-Mesothelioma-fact-sheet.pdf (accessed on 18 June 2021).
- Australian Institute of Health and Welfare. Mesothelioma in Australia 2019; AIHW: Canberra, Australia, 2020. [Google Scholar]
- Meyerhoff, R.R.; Yang, C.F.; Speicher, P.J.; Gulack, B.C.; Hartwig, M.G.; D’Amico, T.A.; Harpole, D.H.; Berry, M.F. Impact of mesothelioma histologic subtype on outcomes in the Surveillance, Epidemiology, and End Results database. J. Surg. Res. 2015, 196, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapel, D.B.; Schulte, J.J.; Husain, A.N.; Krausz, T. Application of immunohistochemistry in diagnosis and management of malignant mesothelioma. Transl. Lung Cancer Res. 2020, 9, S3–S27. [Google Scholar] [CrossRef] [PubMed]
- Galateau-Salle, F.; Churg, A.; Roggli, V.; Travis, W.D.; World Health Organization Committee for Tumors of the Pleura. The 2015 World Health Organization Classification of Tumors of the Pleura: Advances since the 2004 Classification. J. Thorac. Oncol. 2016, 11, 142–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husain, A.N.; Colby, T.V.; Ordonez, N.G.; Allen, T.C.; Attanoos, R.L.; Beasley, M.B.; Butnor, K.J.; Chirieac, L.R.; Churg, A.M.; Dacic, S.; et al. Guidelines for Pathologic Diagnosis of Malignant Mesothelioma 2017 Update of the Consensus Statement From the International Mesothelioma Interest Group. Arch. Pathol. Lab. Med. 2018, 142, 89–108. [Google Scholar] [CrossRef] [Green Version]
- Marchevsky, A.M.; LeStang, N.; Hiroshima, K.; Pelosi, G.; Attanoos, R.; Churg, A.; Chirieac, L.; Dacic, S.; Husain, A.; Khoor, A.; et al. The differential diagnosis between pleural sarcomatoid mesothelioma and spindle cell/pleomorphic (sarcomatoid) carcinomas of the lung: Evidence-based guidelines from the International Mesothelioma Panel and the MESOPATH National Reference Center. Hum. Pathol. 2017, 67, 160–168. [Google Scholar] [CrossRef]
- Eccher, A.; Girolami, I.; Lucenteforte, E.; Troncone, G.; Scarpa, A.; Pantanowitz, L. Diagnostic mesothelioma biomarkers in effusion cytology. Cancer Cytopathol. 2021. [Google Scholar] [CrossRef]
- Agger, K.; Cloos, P.A.; Christensen, J.; Pasini, D.; Rose, S.; Rappsilber, J.; Issaeva, I.; Canaani, E.; Salcini, A.E.; Helin, K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007, 449, 731–734. [Google Scholar] [CrossRef]
- Perrigue, P.M.; Najbauer, J.; Barciszewski, J. Histone demethylase JMJD3 at the intersection of cellular senescence and cancer. Biochim. Biophys. Acta 2016, 1865, 237–244. [Google Scholar] [CrossRef]
- Hong, Z.; Li, H.; Li, L.; Wang, W.; Xu, T. Different expression patterns of histone H3K27 demethylases in renal cell carcinoma and bladder cancer. Cancer Biomark 2017, 18, 125–131. [Google Scholar] [CrossRef]
- Ma, J.; Wang, N.; Zhang, Y.; Wang, C.; Ge, T.; Jin, H.; Deng, X.; Huo, X.; Gu, D.; Ge, Z.; et al. KDM6B Elicits Cell Apoptosis by Promoting Nuclear Translocation of FOXO1 in Non-Small Cell Lung Cancer. Cell Physiol. Biochem. 2015, 37, 201–213. [Google Scholar] [CrossRef]
- Ramadoss, S.; Chen, X.; Wang, C.Y. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J. Biol. Chem. 2012, 287, 44508–44517. [Google Scholar] [CrossRef] [Green Version]
- Tokunaga, R.; Sakamoto, Y.; Nakagawa, S.; Miyake, K.; Izumi, D.; Kosumi, K.; Taki, K.; Higashi, T.; Imamura, Y.; Ishimoto, T.; et al. The Prognostic Significance of Histone Lysine Demethylase JMJD3/KDM6B in Colorectal Cancer. Ann. Surg. Oncol. 2016, 23, 678–685. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, L.; Stupack, D.G.; Bai, N.; Xun, J.; Ren, G.; Han, J.; Li, L.; Luo, Y.; Xiang, R.; et al. JMJD3 promotes survival of diffuse large B-cell lymphoma subtypes via distinct mechanisms. Oncotarget 2016, 7, 29387–29399. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Zhang, D.; Xu, Z.; Wen, X.; Zhang, Y. JMJD3 promotes the epithelial-mesenchymal transition and migration of glioma cells via the CXCL12/CXCR4 axis. Oncol. Lett. 2019, 18, 5930–5940. [Google Scholar] [CrossRef]
- Tang, B.; Qi, G.; Tang, F.; Yuan, S.; Wang, Z.; Liang, X.; Li, B.; Yu, S.; Liu, J.; Huang, Q.; et al. Aberrant JMJD3 Expression Upregulates Slug to Promote Migration, Invasion, and Stem Cell-Like Behaviors in Hepatocellular Carcinoma. Cancer Res. 2016, 76, 6520–6532. [Google Scholar] [CrossRef] [Green Version]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000132510-KDM6B/tissue (accessed on 11 March 2021).
- De Santa, F.; Narang, V.; Yap, Z.H.; Tusi, B.K.; Burgold, T.; Austenaa, L.; Bucci, G.; Caganova, M.; Notarbartolo, S.; Casola, S.; et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009, 28, 3341–3352. [Google Scholar] [CrossRef]
- Kamikawa, Y.F.; Donohoe, M.E. The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated. Epigenetics 2014, 9, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Agger, K.; Cloos, P.A.; Rudkjaer, L.; Williams, K.; Andersen, G.; Christensen, J.; Helin, K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009, 23, 1171–1176. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Xiao, X.; Guo, F. Roles of H3K27me3 Demethylase JMJD3 in Inflammation and Cancers. J. Rare Dis. Res. Treat. 2019, 4, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Jiang, L.; He, Q.; Wei, W.; Wang, Y.; Zhang, X.; Liu, J.; Chen, K.; Chen, J.; Xie, D. The Prognostic Significance of JMJD3 in Primary Sarcomatoid Carcinoma of The Lung, a Rare Subtype of Lung Cancer. OncoTargets Ther. 2019, 12, 9385–9393. [Google Scholar] [CrossRef] [Green Version]
- Li, S.H.; Lu, H.I.; Chen, Y.H.; Lo, C.M.; Huang, W.T.; Tien, W.Y.; Lan, Y.C.; Tsai, H.T.; Chen, C.H. JMJD3 expression is an independent prognosticator in patients with esophageal squamous cell carcinoma. Surgery 2019, 165, 946–952. [Google Scholar] [CrossRef]
- Xu, Z.; Xia, Y.; Xiao, Z.; Jia, Y.; Li, L.; Jin, Y.; Zhao, Q.; Wan, L.; Yi, T.; Yu, Y.; et al. Comprehensive profiling of JMJD3 in gastric cancer and its influence on patient survival. Sci. Rep. 2019, 9, 868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.A.; Mohn, S.E.; Weinmann, A.S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 2010, 40, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Ohguchi, H.; Harada, T.; Sagawa, M.; Kikuchi, S.; Tai, Y.T.; Richardson, P.G.; Hideshima, T.; Anderson, K.C. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia 2017, 31, 2661–2669. [Google Scholar] [CrossRef] [Green Version]
- Salminen, A.; Kaarniranta, K.; Hiltunen, M.; Kauppinen, A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J. Mol. Med. 2014, 92, 1035–1043. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Y.; Gao, Y.; Geng, P.; Lu, Y.; Liu, X.; Yao, R.; Hou, P.; Liu, D.; Lu, J.; et al. JMJD3 promotes SAHF formation in senescent WI38 cells by triggering an interplay between demethylation and phosphorylation of RB protein. Cell Death Differ. 2015, 22, 1630–1640. [Google Scholar] [CrossRef]
- Sola, S.; Xavier, J.M.; Santos, D.M.; Aranha, M.M.; Morgado, A.L.; Jepsen, K.; Rodrigues, C.M. p53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation. PLoS ONE 2011, 6, e18421. [Google Scholar] [CrossRef] [Green Version]
- Cregan, S.; Breslin, M.; Roche, G.; Wennstedt, S.; MacDonagh, L.; Albadri, C.; Gao, Y.; O’Byrne, K.J.; Cuffe, S.; Finn, S.P.; et al. Kdm6a and Kdm6b: Altered expression in malignant pleural mesothelioma. Int. J. Oncol. 2017, 50, 1044–1052. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.; Tateishi, K.; Kudo, Y.; Sato, T.; Yamamoto, S.; Miyabayashi, K.; Matsusaka, K.; Asaoka, Y.; Ijichi, H.; Hirata, Y.; et al. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPalpha. Carcinogenesis 2014, 35, 2404–2414. [Google Scholar] [CrossRef]
- Canino, C.; Mori, F.; Cambria, A.; Diamantini, A.; Germoni, S.; Alessandrini, G.; Borsellino, G.; Galati, R.; Battistini, L.; Blandino, R.; et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene 2012, 31, 3148–3163. [Google Scholar] [CrossRef] [Green Version]
- Lin, B.T.-Y.; Colby, T.; Gown, A.M.; Hammar, S.P.; Mertens, R.B.; Churg, A.; Battifora, H. Malignant vascular tumors of the serous membranes mimicking mesothelioma: A report of 14 cases. Am. J. Surg. Pathol. 1996, 20, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-C.; Chow, J.-M.; Wang, K.-M.; Fang, C.-L.; Chu, J.-S.; Chen, C.-L. Primary pleural angiosarcoma as a mimicker of mesothelioma: A case report. Diagn. Pathol. 2011, 6, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Del Frate, C.; Mortele, K.; Zanardi, R.; Hunsaker, A.R.; Nikpoor, N.; Cibas, E.S.; Silverman, S.G. Pseudomesotheliomatous angiosarcoma of the chest wall and pleura. J. Thorac. Imaging 2003, 18, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Pulford, E.; Huilgol, K.; Moffat, D.; Henderson, D.W.; Klebe, S. Malignant Mesothelioma, BAP1 Immunohistochemistry, and VEGFA: Does BAP1 Have Potential for Early Diagnosis and Assessment of Prognosis? Dis. Markers 2017, 2017, 1310478. [Google Scholar] [CrossRef]
- Kao, S.C.; Lee, K.; Armstrong, N.J.; Clarke, S.; Vardy, J.; van Zandwijk, N.; Reid, G.; Burn, J.; McCaughan, B.C.; Henderson, D.W.; et al. Validation of tissue microarray technology in malignant pleural mesothelioma. Pathology 2011, 43, 128–132. [Google Scholar] [CrossRef]
| Clinicopathological Parameters | n (%) |
|---|---|
| Age at diagnosis [median years(range)] | 75 years (47–90) |
| Sex | |
| Male | 80 (81) |
| Female | 19 (19) |
| Histological subtype | |
| Epithelioid | 50 (51) |
| Sarcomatoid | 27 (27) |
| Biphasic | 22 (22) |
| Ki-67 expression | |
| low | 61 (67) |
| high | 30 (33) |
| GATA3 IHC expression | |
| present | 13 (13) |
| absent | 84 (87) |
| JMJD3 nuclear expression | |
| low | 48 (48) |
| high | 51 (52) |
| JMJD3 cytoplasmic expression | |
| low | 93 (94) |
| high | 6 (6) |
| Combined score groups | |
| NHCL | 47 (48) |
| NHCH | 4 (4) |
| NLCL | 46 (46) |
| NLCH | 2 (2) |
| Subtype | N | Nuclear | p Value | Cytoplasmic | p Value | ||
|---|---|---|---|---|---|---|---|
| High n (%) | Low n (%) | High n (%) | Low n (%) | ||||
| Total | 99 | 51 | 48 | 6 | 93 | ||
| Epithelioid | 50 | 42 (84) | 8 (16) | <0.001 | 4 (8) | 46 (92) | 0.399 |
| Sarcomatoid | 27 | 3 (11) | 24 (89) | 2 (7) | 25 (93) | ||
| Biphasic | 22 | 6 (27) | 16 (73) | 0 | 22 (100) | ||
| UVA | MVA | |||||||
|---|---|---|---|---|---|---|---|---|
| Sig. | HR | 95.0% CI for Exp(B) | Sig. | HR | 95.0% CI for Exp(B) | |||
| Lower | Upper | Lower | Upper | |||||
| Ki67 | 0.036 | 1.631 | 1.033 | 2.576 | 0.009 | 1.959 | 1.187 | 3.233 |
| Nuclear JMJ | 0.018 | 1.622 | 1.088 | 2.42 | 0.369 | 1.389 | 0.678 | 2.844 |
| Cyto JMJ | 0.052 | 2.316 | 0.992 | 5.405 | 0.045 | 2.463 | 1.019 | 5.958 |
| Gata3 | 0.55 | 1.207 | 0.651 | 2.237 | 0.563 | 0.811 | 0.398 | 1.651 |
| Subtype: | ||||||||
| Ref Epithelioid | 1 | 1 | ||||||
| Sarcomatoid | 0.002 | 2.157 | 1.338 | 3.478 | 0.083 | 2.014 | 0.914 | 4.437 |
| Biphasic | 0.311 | 1.305 | 0.78 | 2.183 | 0.399 | 1.404 | 0.638 | 3.088 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rask-Nielsen, L.; Prabhakaran, S.; Hocking, A.J.; Hussey, M.; Klebe, S. Prognostic Significance of JMJD3 Expression in Pleural Mesotheliomas. J. Mol. Pathol. 2021, 2, 223-232. https://doi.org/10.3390/jmp2030019
Rask-Nielsen L, Prabhakaran S, Hocking AJ, Hussey M, Klebe S. Prognostic Significance of JMJD3 Expression in Pleural Mesotheliomas. Journal of Molecular Pathology. 2021; 2(3):223-232. https://doi.org/10.3390/jmp2030019
Chicago/Turabian StyleRask-Nielsen, Lauren, Sarita Prabhakaran, Ashleigh J. Hocking, Matthew Hussey, and Sonja Klebe. 2021. "Prognostic Significance of JMJD3 Expression in Pleural Mesotheliomas" Journal of Molecular Pathology 2, no. 3: 223-232. https://doi.org/10.3390/jmp2030019
APA StyleRask-Nielsen, L., Prabhakaran, S., Hocking, A. J., Hussey, M., & Klebe, S. (2021). Prognostic Significance of JMJD3 Expression in Pleural Mesotheliomas. Journal of Molecular Pathology, 2(3), 223-232. https://doi.org/10.3390/jmp2030019






