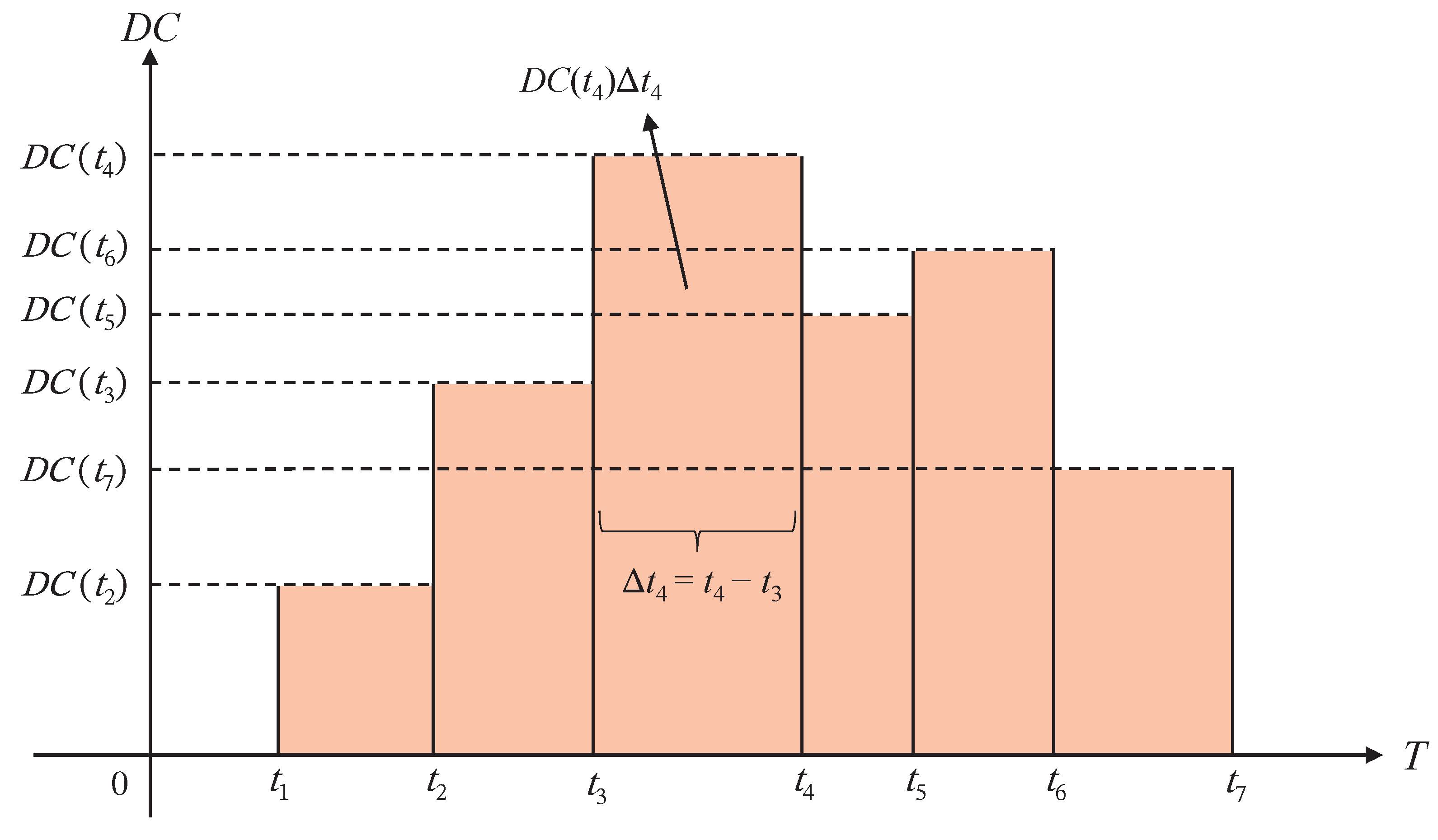3.1. Simulation Study
We conduct several simulated examples to evaluate the screening performance of LDCS and LCDCS, and compare the results with those of some existing methods, including partial residual sure independence screening (PRSIS) [
3], time-varying coefficient models’ sure independence screening (TVCM-SIS) [
19], distance correlation sure independence screening (DC-SIS) [
7], conditional distance correlation sure independence screening (CDC-SIS) [
9], and covariate information number sure independence screening (CIS) [
10]. PRSIS and TVCM-SIS are longitudinal data feature screening procedures where PRSIS is based on partially linear models and TVCM-SIS is based on time-varying coefficient models. DC-SIS, CDC-SIS, and CIS are all model-free methods for cross-sectional data.
We repeat each simulation example 500 times. In all examples, the number of predictors is , and the sample size is , yielding a submodel size of . The predictor and the random error are generated independently from , where and are different in each example. The time points are sampled from a standard uniform distribution and do not change in a simulation. We use the following four criteria to assess the screening performance:
: The minimum submodel size containing all important predictors. The median and robust standard deviation (RSD = IQR/1.34) of are reported, where IQR denotes interquartile range.
: The proportion of the important predictor selected in the submodel.
: The proportion of all important predictors selected in the submodel.
: The average number of important predictors selected in the submodel.
3.1.1. Example 1: Partially Linear Models
We generate the response from the following two models:
Model (1.a):
Model (1.b):
Model (1.a) considers categorical predictor variables, while Model (1.b) incorporates interaction effects. These two models are based on Model (1.a) and Model (1.b) in Li et al. [
7], but the difference is that we introduced longitudinal measurements and nonlinear time variables. We set
,
and
in Model (1.a), and
,
, and
in Model (1.b). Furthermore, we set the repeated measurements
and the within-subject correlation structure as first-order autoregressive (AR(1)) structure. The within-subject correlations are set to
and
for each model, respectively.
The detailed results are reported in
Table 1. For the four different situations, our proposed method LDCS achieves the smallest MMS (including median and RSD), along with the largest
and
. When a strong nonlinear time effect is present, the model-free feature screening procedures DC-SIS and CIS perform less effectively. Furthermore, we observe that TVCM-SIS and PRSIS are not as effective in identifying interaction effects. While CIS relatively successfully identifies interaction effects, it tends not to retain important predictors with and without interaction effects simultaneously. Example 1 demonstrates that our proposed method LDCS is unaffected by strong time effects and performs better in handling situations with interaction effects in partially linear models.
3.1.2. Example 2: Time-Varying Coefficient Models
In this example, we generate the response from the following two models:
Model (2.a):
Model (2.b):
Model (2.a) is similar to Example I in Chu et al. [
19], but we set a smaller sample size and measurements. Model (2.b) considers a count response with Poisson distribution, which is based on Example II in Chu et al. [
20], but we use time-varying coefficient model, rather than the generalized varying coefficient mixed-effect model. In Model (2.a), we set
,
,
,
and
. In Model (2.b), we set
,
,
,
, and
. Furthermore, we set the within-subject correlation structure as a compound symmetry (CS) structure. The within-subject correlations are set to
and
for each model, respectively.
Table 2 presents the detailed simulation results for Example 2. It can be seen that the results of Example 2 are similar to those of Example 1, i.e., our proposed method LDCS achieves the smallest MMS (including median and RSD), along with the largest
and
. Specifically, for Model (2.a), although TVCM-SIS is based on the assumption of varying coefficient models, its performance is not ideal when the categorical predictors have a stronger impact on the response. For Model (2.b), the screening performance of TVCM-SIS, PRSIS, DC-SIS, and CIS is all unsatisfactory, indicating that these methods are not well suited for varying coefficient models with count responses. These examples further demonstrate that, for some special varying coefficient models, our proposed method LDCS may achieve better screening performance.
3.1.3. Example 3: Partially Linear Single-Index Models
We assess the screening performance of LCDCS when the predictors are correlated with each other. The response is generated from Model (3):
Model (3):
We set , , , the repeated measurements and the within-subject correlation structure as AR(1) structure with within-subject correlation as . The correlation structure between predictors is set as CS structure, with two scenarios considered: and .
We first use LDCS and DC-SIS to screen the most relevant predictor as a conditional predictor for LCDCS and CDC-SIS, respectively.
Table 3 presents the detailed simulation results for Example 3. It can be seen that our proposed method, LCDCS, also achieves the smallest MMS (including median and RSD), along with the largest
and
. Since the number of repeated measurements is only 3, the improvement in screening performance of LCDCS over CDC-SIS is not particularly obvious. Additionally, the screening performance of TVCM-SIS and PRSIS is very poor, indicating that these methods, which rely on specific model assumptions, may not perform well in some complex model structures. CIS is also not well suited when the predictors are correlated. This example demonstrates that our proposed method, LCDCS, is more suitable for longitudinal data with correlated predictors in some special model structures.
3.2. Application to Gut Microbiota Data
We analyze real-world data to demonstrate the empirical performance of our methods in this section. This is a gut microbiota data from Bangladeshi children, as reported by Subramanian et al. [
21]. The longitudinal cohort study aimed at analyzing the effects of therapeutic foods on children. For details, they monitored the growth and development of
infants over a two-year period after birth, ultimately collecting a total of
fecal samples. Each sample contained 79,597 operational taxonomic units sharing at least
nucleotide sequence identity (97%-identity OTUs). Following Subramanian et al. [
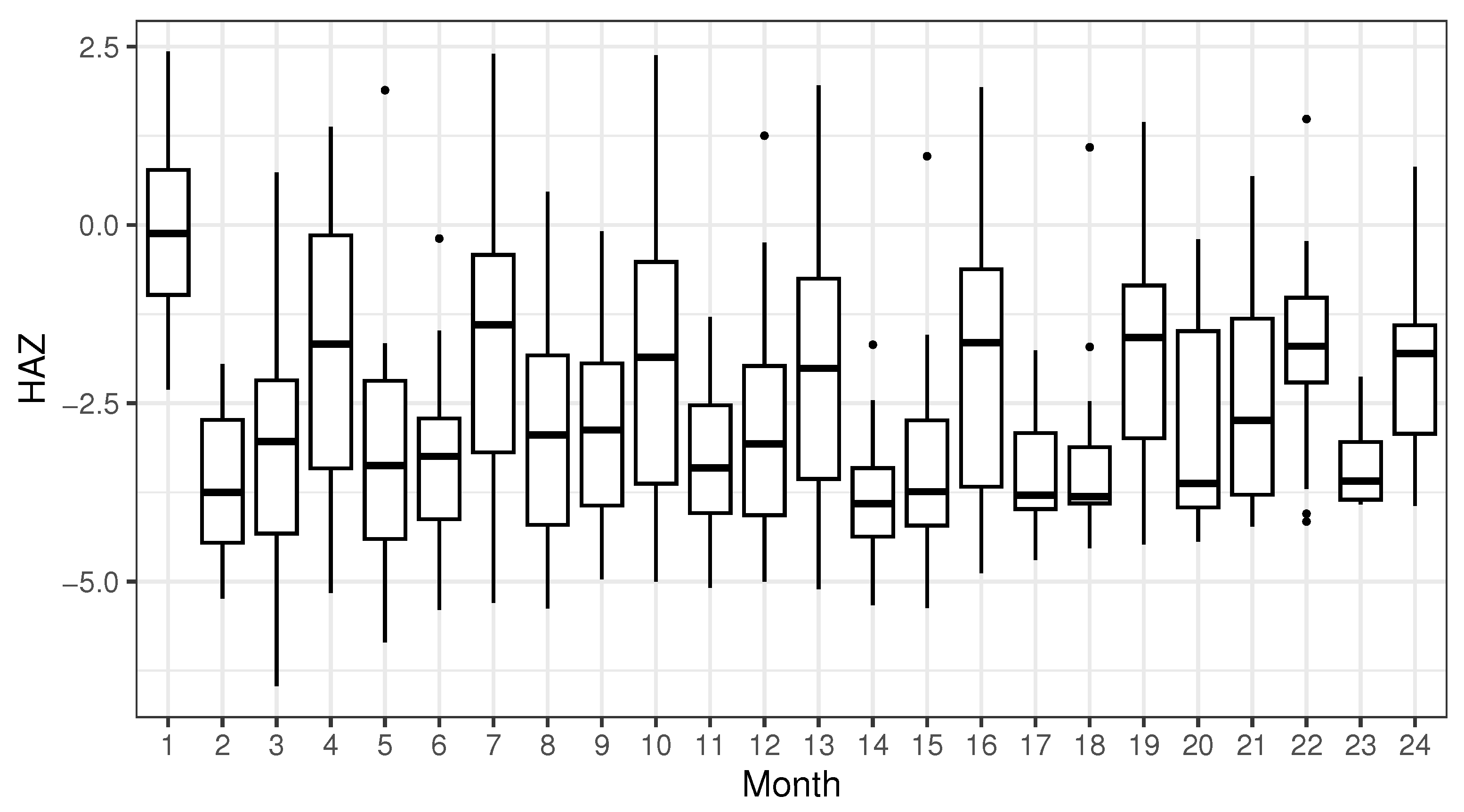
21], we obtain 1222 97%-identity OTUs that have at least two fecal samples with an average relative abundance greater than 0.1%. In this article, we consider height-for-age Z-scores (HAZ) as the response, which reflects how a child’s height compares to the average height of the same age and gender group, indicating whether growth and development are within the normal range. The HAZ of each child was measured between 6 and 22 times, as illustrated in
Figure 2. As a further preprocessing step, we retain only the months in which at least 24 children had their HAZ measured. Thus, we eventually work with a dataset comprising 1222 97%-identity OTUs (the predictors) and HAZ (the response), measured on
children over 13 months, for a total of 433 measurements. On this dataset, we apply our LDCS and LCDCS, along with TVCM-SIS, PRSIS, DC-SIS, CDC-SIS, and CIS for feature screening, and we compare the results with those of Subramanian et al. [
21].
First, we compare the top
97%-identity OTUs selected by each method, as summarized in
Table 4. Our methods, LDCS and LCDCS, demonstrate a moderate overlap in the number of selected 97%-identity OTUs with PRSIS, DC-SIS, and CDC-SIS, while the selection results of TVCM-SIS and CIS differ significantly from those of the other methods. Next, we compare the top
97%-identity OTUs, as Subramanian et al. [
21] identified 220 gut microbiota that were significantly different between severe acute malnutrition (SAM) and healthy children. It is obvious that LDCS and LCDCS have the maximum number of 97%-identity OTUs, which align with those of Subramanian et al. [
21] among all screening procedures.
Furthermore, we evaluate the screening performance through regression analysis. Following Chu et al. [
19], we establish time-varying coefficient models with different numbers of 97%-identity OTUs and use the total heritability commonly used in genetic analysis to assess the goodness of fit. The total heritability of all 97%-identity OTUs is calculated through
Here, fetal means whether the child is a singleton birth and RSS is the residual sum of squares defined as
where
denotes the actual value, and
denotes the predicted value from the time-varying coefficient models.
We calculate the total heritability of the top
to 49 97%-identity OTUs and remove more irrelevant 97%-identity OTUs using forward regression to achieve better regression fitting results. Our proposed method, LCDCS, has the highest total heritability at 64.76%, followed by LDCS with 60.24%. The final number of 97%-identity OTUs selected and the total heritability for each method are shown in
Table 5.
We also plot the curves of total heritability for the different screening procedures, as shown in
Figure 3. We note that, overall, when the number of 97%-identity OTUs selected is between 4 and 23, the total heritability of LCDCS is higher than that of other screening procedures. Additionally, when the number of 97%-identity OTUs selected is between 24 and 33, the LDCS method shows a higher total heritability. Therefore, when the number of selected 97%-identity OTUs is moderate, we recommend using LCDCS, while for a larger number of selected 97%-identity OTUs, we recommend using LDCS.
We also calculate the heritability of each 97%-identity OTU, which depends on the sequence of selection in the forward regression. The heritability of the
kth 97%-identity OTU is calculated through
Table 6 presents the IDs and taxonomic annotations of the top 10 97%-identity OTUs based on the heritability, as selected via LCDCS. The taxonomic annotations are given by Subramanian et al. [
21], representing the genus of the gut microbiota. Among the 10 gut microbiota, 4 gut microbiota belong to the Bifidobacterium genus. Bifidobacterium is a common group of probiotics found primarily in the human gut, particularly in infants, and it has been shown to be closely associated with children’s nutrient absorption and growth development. Furthermore, there are 5 97%-identity OTUs that differ from the results of Subramanian et al. [
21], which may represent a new discovery. We also perform a sensitivity analysis to evaluate how the parameter
influences the empirical results, with detailed information provided in
Appendix B.