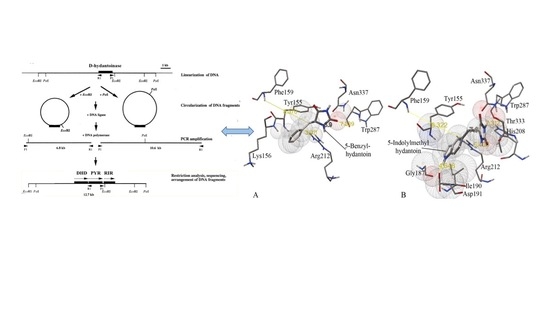
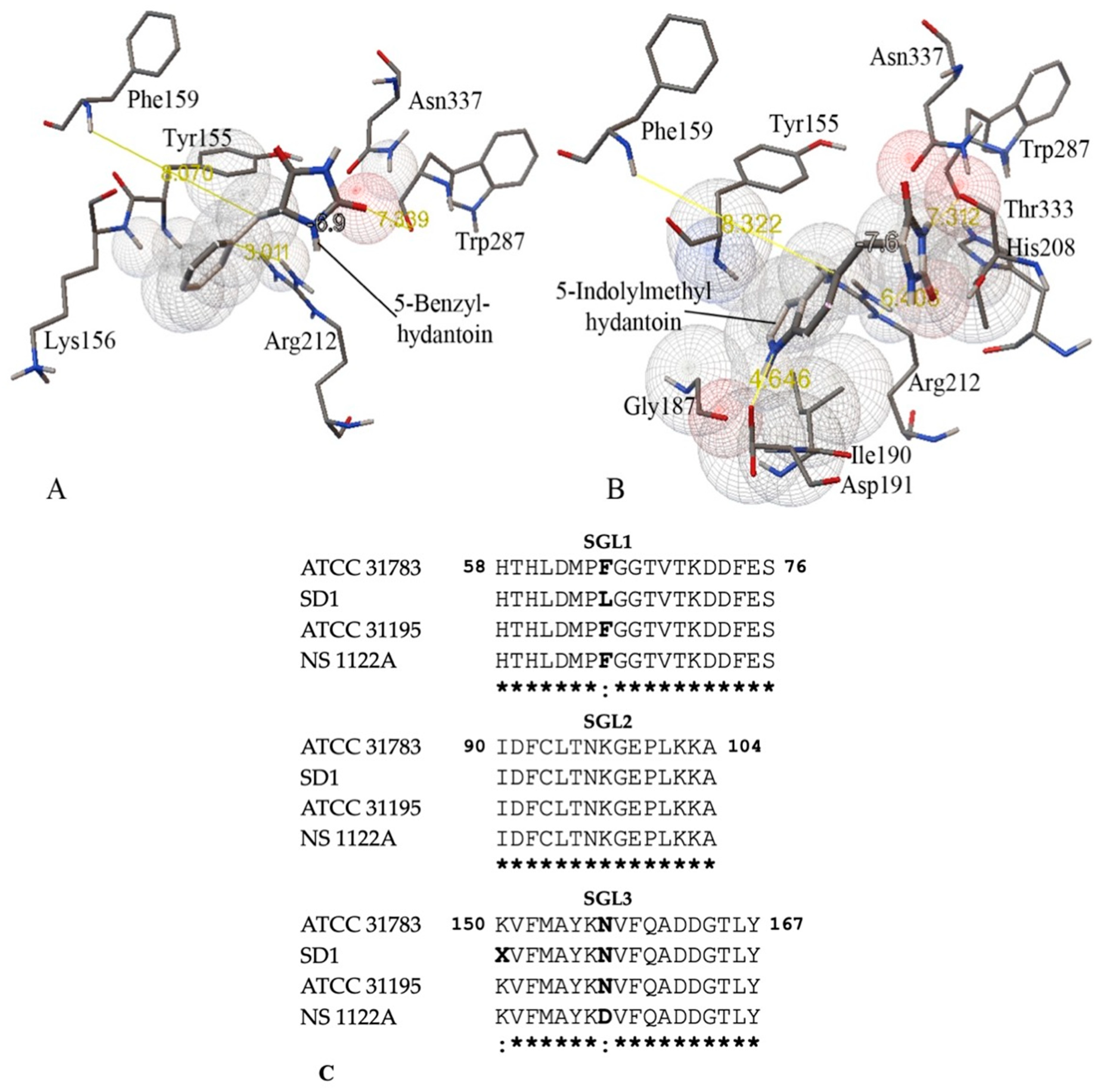
Rational Engineering of the Substrate Specificity of a Thermostable D-Hydantoinase (Dihydropyrimidinase)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Enzyme Assays
2.3. Homology Modeling
2.4. Site-Directed Mutagenesis
2.5. Long Inverse PCR
2.6. Molecular Docking
2.7. Statistics
3. Results
3.1. Characterization of G. stearothermophilus Hydantoinase
3.2. Homology Modeling of Thermostable D-Hydantoinase
3.3. Substrate Specificity and Kinetic Constants of Engineered Mutants of D-Hydantoinase
3.4. D-Hydantoinase Is the Enzyme of the Reductive Catabolism of Pyrimidines in G. stearothermophilus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
GenBank Accession Number
References
- Vogels, G.D.; van der Drift, C. Degradation of purines and pyrimidines by micro-organisms. Bacteriol. Rev. 1976, 40, 403–468. [Google Scholar] [CrossRef]
- Holm, L.; Sander, C. An evolutionary treasure: Unification of a broad set of amidohydrolases related to urease. Proteins 1997, 28, 72–82. [Google Scholar] [CrossRef]
- Barba, M.; Glansdorff, N.; Labedan, B. Evolution of cyclic amidohydrolases: A highly diversified superfamily. J. Mol. Evol. 2013, 77, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Cecere, F.; Galli, G.; Morisi, F. Substrate and steric specificity of hydropyrimidine hydrase. FEBS Lett. 1975, 57, 192–194. [Google Scholar] [CrossRef] [Green Version]
- Syldatk, C.; May, O.; Altenbuchner, J.; Mattes, R.; Siemann, M. Microbial hydantoinases - industrial enzymes from the origin of life? Appl. Microb. Biotechnol. 1999, 51, 293–309. [Google Scholar] [CrossRef] [PubMed]
- Engel, U.; Syldatk, C.; Rudat, J. Stereoselective hydrolysis of aryl-substituted dihydropyrimidines by hydantoinases. Appl. Microb. Biotechnol. 2012, 94, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, G.J.; Kim, H.S. Production of D-Amino Acid Using Whole Cells of Recombinant Escherichia coli with Separately and Coexpressed D-Hydantoinase and N-Carbamoylase. Biotechnol. Prog. 2000, 16, 564–570. [Google Scholar] [CrossRef]
- Kananavičiūté, R.; Čitavičius, D. Genetic engineering of Geobacillus spp. J. Microbiol. Methods 2015, 111, 31–39. [Google Scholar] [CrossRef]
- Shi, T.; Han, P.; You, C.; Zhan, Y.P.J. An in vitro synthetic biology platform for emerging industrial biomanufacturing: Bottom-uppathway design. Synth. Syst. Biotechnol. 2018, 3, 186–195. [Google Scholar] [CrossRef]
- Sakanyan, V.; Weigel, P.; Lecocq, M.; Marc, F.; Batisse, N. Hydantoinase and carbamoylase of thermophilic bacteria. Recent. Res. Dev. Microb. 1998, 2, 553–565. [Google Scholar]
- Mukohara, Y.; Ishikawa, T.; Watabe, K.; Nakamura, H. A thermostable hydantoinase of Bacillus stearothermophilus NS1122A: Cloning, sequencing, and high expression of the enzyme gene, and some properties of the expressed enzyme. Biosci. Biotechnol. Biochem. 1994, 58, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Lee, D.C.; Kim, H.S. Purification and characterization of thermostable D-hydantoinase from thermophilic Bacillus stearothermophilus SD1. Appl. Biochem. Biotechnol. 1997, 62, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, G.J.; Lee, D.C.; Kim, H.S. Purification and characterization of thermostable D-hydantoinase from Bacillus thermocatenulatus GH-2. Appl. Biochem. Biothecnol. 1999, 81, 53–65. [Google Scholar] [CrossRef]
- Cheon, Y.H.; Kim, H.S.; Han, K.H.; Abendroth, J.; Niefind, K.; Schomburg, D.; Wang, J.; Kim, Y. Crystal structure of D-hydantoinase from Bacillus stearothermophilus: Insight into the stereochemistry of enantioselectivity. Biochemistry 2002, 41, 9410–9417. [Google Scholar] [CrossRef] [PubMed]
- Cain, J.A.; Solis, N.; Cordwell, S.J. Beyond of gene expression: The impact of protein post-translational modifications in bacteria. J. Proteom. 2014, 97, 265–286. [Google Scholar] [CrossRef] [PubMed]
- Cheon, Y.H.; Park, H.S.; Kim, J.H.; Kim, Y.; Kim, H.S. Manipulation of the active site loops of D-hydantoinase, a (beta/alpha)8-barrel protein, for modulation of the substrate specificity. Biochemistry 2004, 43, 7413–7420. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Niu, L.X.; Shi, Y.W.; Yuan, J.M. The flexibility of the non-conservative region at the C terminus of D-hydantoinase from Pseudomonas putida YZ-26 is extremely limited. Appl. Biochem. Biotechnol. 2008, 144, 237–247. [Google Scholar] [CrossRef]
- Abendroth, J.; Niefind, K.; Schomburg, D. X-ray structure of a dihydropyrimidinase from Thermus sp. at 1.3 Å resolution. J. Mol. Biol. 2002, 320, 143–156. [Google Scholar] [CrossRef]
- Kim, G.J.; Lee, D.E.; Kim, H.S. Functional expression and characterization of the two cyclic amidohydrolase enzymes, allantoinase and a novel phenylhydantoinase, from Escherichia coli. J. Bacteriol. 2000, 182, 7021–7028. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Chang, Y.J.; Shin, D.M.; Han, J.; Seo, M.-H.; Fazelinia, H.; Maranas, C.D.; Kim, H.S. Designing the substrate specificity of D-hydantoinase using a rational approach. Enzym. Microb. Technol. 2009, 44, 170–175. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Kang, Z. In Silico Protein Design Promotes the Rapid Evolution of Industrial Enzymes. Biochemistry 2019, 58, 1451–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ware, E. The Chemistry of Hydantoins. Chem. Rev. 1950, 46, 403–470. [Google Scholar] [CrossRef] [PubMed]
- Guivarch, M.; Gillonier, C.; Brunie, J.-C. Obtention d’amino acides optiquement actifs à l’aide d’hydantoinases. Bull. Soc. Chim. 1980, 1–2, 91–95. [Google Scholar]
- Peterson, G.L. Determination of total protein. Meth. Enzym. 1983, 91, 95–119. [Google Scholar] [PubMed]
- Cornish-Bowden, E. Fundamentals of Enzyme Kinetics; Medicina: Moscow, Russia, 1979. (In Russian) [Google Scholar]
- Hambardzumyan, A.A. Statistical analyses of enzyme kinetics: Simple Michaelis-Menten and bi-bi ping-pong. Biol. J. Armen. 2017, 69, 6–12. [Google Scholar]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-Pdb Viewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Peternel, Š.; Grdadolnik, J.; Gaberc-Porekar, V.; Komel, R. Engineering inclusion bodies for nondenaturing extraction of functional proteins. Microb. Cell Fact. 2008, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Sakanyan, V.; Desmarez, L.; Legrain, C.; Charlier, D.; Mett, I.; Kochikyan, A.; Savchenko, A.; Boyen, A.; Falmagne, P.; Pierard, A.; et al. Gene cloning, sequence analysis, purification, and characterization of a thermostable aminoacylase from Bacillus Stearothermophilus. Appl. Environ. Microbiol. 1993, 59, 3878–3888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Niu, L.; Feng, X.; Yuan, J. Purification, enzymatic properties of a recombinant D-hydantoinase and its dissociation by zinc ion. World J. Microbiol. Biotechnol. 2006, 22, 675–680. [Google Scholar] [CrossRef]
- Eisenthal, R.; Danson, M.J.; Hough, D.W. Catalytic efficiency and kcat/Km: A useful comparator? Trends Biotechnol. 2007, 25, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Watabe, K.; Ishikawa, T.; Mukohara, Y.; Nakamura, H. Cloning and sequencing of the genes involved in the conversion of 5-sustituted hydantoins to the corresponding L-amino acids from the native plasmid of Pseudomonas sp. strain NS671. J. Bacteriol. 1992, 174, 962–969. [Google Scholar] [CrossRef] [Green Version]
- Weigel, P.; Marc, F.; Aganyants, H.A.; Sakanyan, V.A. Characterization of thermostable hydantoinases cloned from Geobacillus stearothermophilus. Rep. NAS RA 2013, 113, 92–98. [Google Scholar]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic applications of an inverse polymerase chain reaction. Genetics 1988, 120, 621–623. [Google Scholar]
- Triglia, T.; Peterson, M.G.; Kemp, D.J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988, 16, 8186. [Google Scholar] [CrossRef] [Green Version]
- Seibert, C.M.; Raushel, F.M. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef]
- Goikovic, Z.; Rislund, L.; Andersen, B.; Sandrini, M.P.; Cook, P.E.; Schnackerz, K.D.; Pisker, J. Dihydropyrimidine amidohydrolases and dihydroorotases share the same origin and several enzymatic properties. Nucleic Acids Res. 2003, 31, 1683–1692. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Hsu, C.C.; Chen, M.C.; Yang, Y.S. Effect of metal binding and posttranslational lysine carboxylation on the activity of recombinant hydantoinase. J. Biol. Inorg. Chem. 2009, 14, 111–121. [Google Scholar] [CrossRef]
- Cheng, J.H.; Huang, C.C.; Huang, Y.H.; Huang, C.Y. Structural Basis for pH-Dependent Oligomerization of Dihydropyrimidinase from Pseudomonas aeruginosa PAO1. Bioorg. Chem. Appl. 2018, 2018, 9564391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.Y.; Hsieh, H.C.; Huang, C.Y. Biochemical characterization of allantoinase from Escherichia coli BL21. Protein J. 2011, 30, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Sommer, T.; Bjerregaard-Andersen, K.; Uribe, L.; Etzerodt, M.; Diezemann, G.; Gauss, J.; Cascella, M.; Morth, J.P. A fundamental catalytic difference between zinc and manganese dependent enzymes revealed in a bacterial isatin hydrolase. Sci. Rep. 2018, 8, 13104. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.B.; Husted, S. The Biochemical Properties of Manganese in Plants. Plants 2019, 8, 381. [Google Scholar] [CrossRef] [Green Version]
- Lohkamp, B.; Andersen, B.; Piskur, J.; Dobritzsch, D. The crystal structures of dihydropyrimidinases reaffirm the close relationship between cyclic amidohydrolases and explain their substrate specificity. J. Biol. Chem. 2006, 281, 3762–3776. [Google Scholar] [CrossRef] [Green Version]
- Torrents, E. Ribonucleotide reductases: Essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 2014, 4, 52. [Google Scholar] [CrossRef] [Green Version]
- Mathews, C.K. DNA precursor metabolism and genomic stability. FASEB J. 2006, 20, 1300–1314. [Google Scholar] [CrossRef] [Green Version]


| Divalent Metal Ion | Specific Enzyme Activity, U/mg | |
|---|---|---|
| 0.2 mM | 2 mM | |
| Mn2+ | 8.37 | 4.49 |
| Fe2+ | 1.76 | 1.76 |
| Co2+ | 1.76 | ND |
| Ni2+ | 0.66 | ND |
| Mg2+ | 0.61 | 1.26 |
| Cu2+ | 0.55 | 1.56 |
| Zn2+ | 0.55 | ND |
| EDTA | 0.35 | 0.35 |
| Substrate | D-Carbamoyl Methionine, mM |
|---|---|
| D, L-5-(2-methylthioethyl) hydantoin | 48.9 ± 2.1 |
| D-5-2-methylthioethyl) hydantoin | 98.3 ± 2.5 |
| L-5-(2-methylthioethyl) hydantoin | 1.6 ± 0.5 |
| Mutation | Hydantoin | Dihydrouracil | D,L-5-methyl-lhydantoin | D,L-5-(2-methyl-thioethyl)-hydantoin | D,L-5-benzyl-hydantoin | D,L-5-indolylmethyl Hydantoin |
|---|---|---|---|---|---|---|
| Wild type | 3.5 ± 0.30 | 4.0 ± 0.31 | 1.4 ± 0.10 | 1.2 ± 0.09 | < 0.04 | < 0.04 |
| W287A | 0.05 ± 0.01 | 0.43 ± 0.03 | 0.17 ± 0.02 | 0.54 ± 0.04 | 1.75 ± 0.12 | 1.57 ± 0.12 |
| F159A | 0.06 ± 0.01 | 5.50 ± 0.40 | < 0.04 | 0.58 ± 0.04 | 1.30 ± 0.10 | 1.10 ± 0.08 |
| W287A/F159A | 0.32 ± 0.02 | 0.85 ± 0.07 | 0.53 ± 0.03 | 0.08 ± 0.01 | 1.75 ± 0.15 | 1.16 ± 0.09 |
| I190A | 0.56 ± 0.04 | 2.2 ± 0.10 | 0.22 ± 0.01 | 0.53 ± 0.04 | 0.05 ± 0.01 | < 0.04 |
| R212K | 1.19 ± 0.10 | 0.06 ± 0.01 | 0.22 ± 0.01 | 0.5 ± 0.04 | < 0.04 | < 0.04 |
| W287A/R212K | 1.1 ± 0.10 | < 0.04 | < 0.04 | 0.27 ± 0.02 | 0.5 ± 0.04 | < 0.04 |
| Enzyme | Km, mM | kcat/Km, (s·mM) −1 | kcat, s−1 | |||
|---|---|---|---|---|---|---|
| Dihydrouracil | D,L-5-indolylmethyl Hydantoin | Dihydrouracil | D,L-5-indolyl-methyl Hydantoin | Dihydrouracil | D,L-5-indolyl-methyl hydantoin | |
| Wild | 0.97 ± 0.22 | ND | 11.3 ± 2.20 | ND | 11.2 ± 0.70 | ND |
| W287A | 0.43 ± 0.13 | 1.66 ± 0.85 | 6.53 ± 1.83 | 4.13 ± 1.63 | 2.76 ± 0.17 | 6.83 ± 0.97 |
| F159A | 2.89 ± 1.27 | 0.60 ± 0.58 | 7.53 ± 2.50 | 6.89 ± 5.83 | 21.8 ± 2.60 | 4.16 ± 0.60 |
| W287A/F159A | 5.61 ± 2.32 | 4.30 ± 0.40 | 1.10 ± 0.30 | 2.90 ± 0.17 | 6.16 ± 1.00 | 12.5 ± 0.50 |
| I190A | 1.12 ± 0.61 | ND | 7.09 ± 3.20 | ND | 7.99 ± 0.90 | ND |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aganyants, H.; Weigel, P.; Hovhannisyan, Y.; Lecocq, M.; Koloyan, H.; Hambardzumyan, A.; Hovsepyan, A.; Hallet, J.-N.; Sakanyan, V. Rational Engineering of the Substrate Specificity of a Thermostable D-Hydantoinase (Dihydropyrimidinase). High-Throughput 2020, 9, 5. https://doi.org/10.3390/ht9010005
Aganyants H, Weigel P, Hovhannisyan Y, Lecocq M, Koloyan H, Hambardzumyan A, Hovsepyan A, Hallet J-N, Sakanyan V. Rational Engineering of the Substrate Specificity of a Thermostable D-Hydantoinase (Dihydropyrimidinase). High-Throughput. 2020; 9(1):5. https://doi.org/10.3390/ht9010005
Chicago/Turabian StyleAganyants, Hovsep, Pierre Weigel, Yeranuhi Hovhannisyan, Michèle Lecocq, Haykanush Koloyan, Artur Hambardzumyan, Anichka Hovsepyan, Jean-Noël Hallet, and Vehary Sakanyan. 2020. "Rational Engineering of the Substrate Specificity of a Thermostable D-Hydantoinase (Dihydropyrimidinase)" High-Throughput 9, no. 1: 5. https://doi.org/10.3390/ht9010005
APA StyleAganyants, H., Weigel, P., Hovhannisyan, Y., Lecocq, M., Koloyan, H., Hambardzumyan, A., Hovsepyan, A., Hallet, J.-N., & Sakanyan, V. (2020). Rational Engineering of the Substrate Specificity of a Thermostable D-Hydantoinase (Dihydropyrimidinase). High-Throughput, 9(1), 5. https://doi.org/10.3390/ht9010005