When Transcriptomics and Metabolomics Work Hand in Hand: A Case Study Characterizing Plant CDF Transcription Factors
Abstract
:1. Introduction
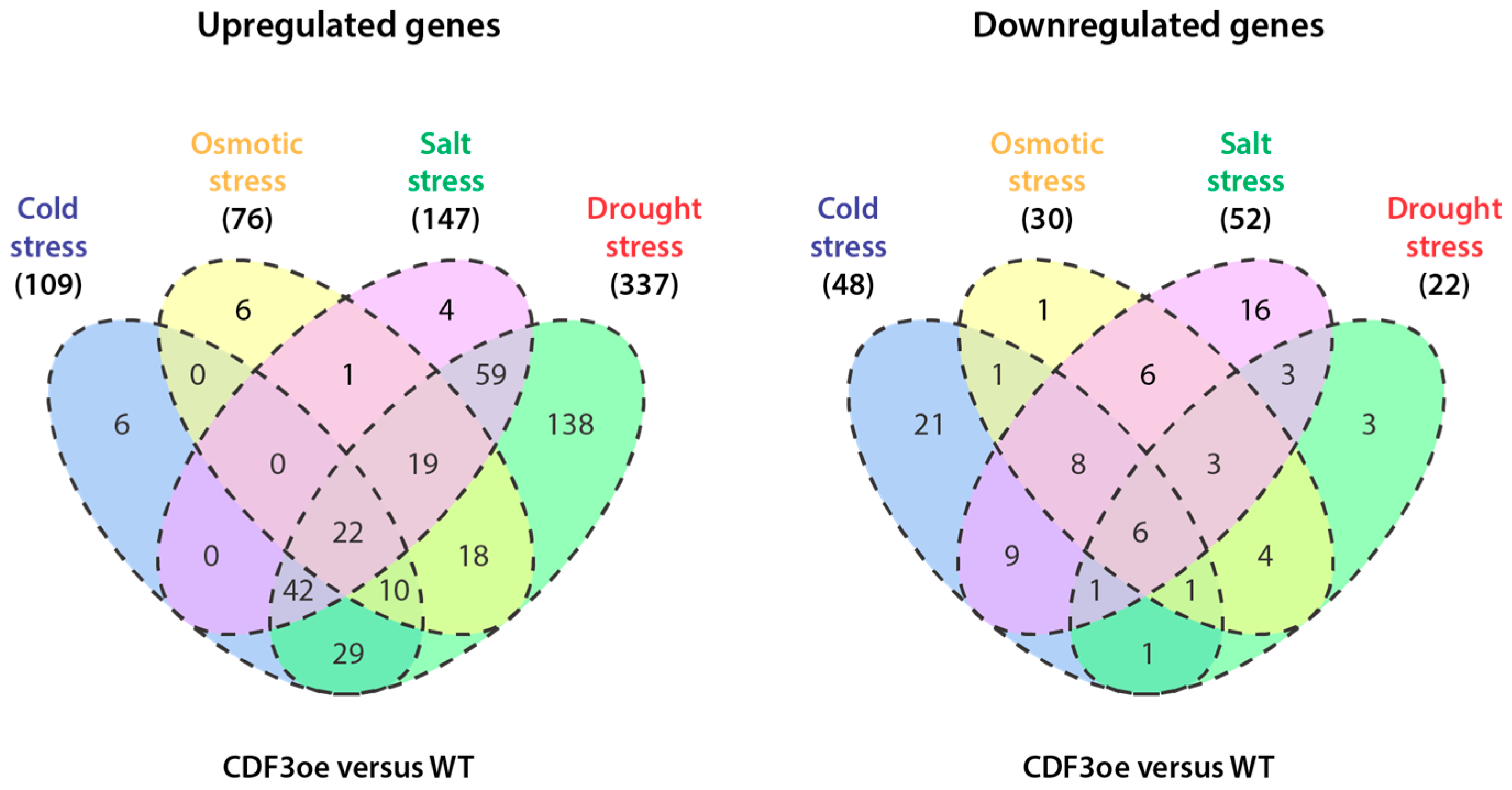
2. Genome-Wide Expression Studies
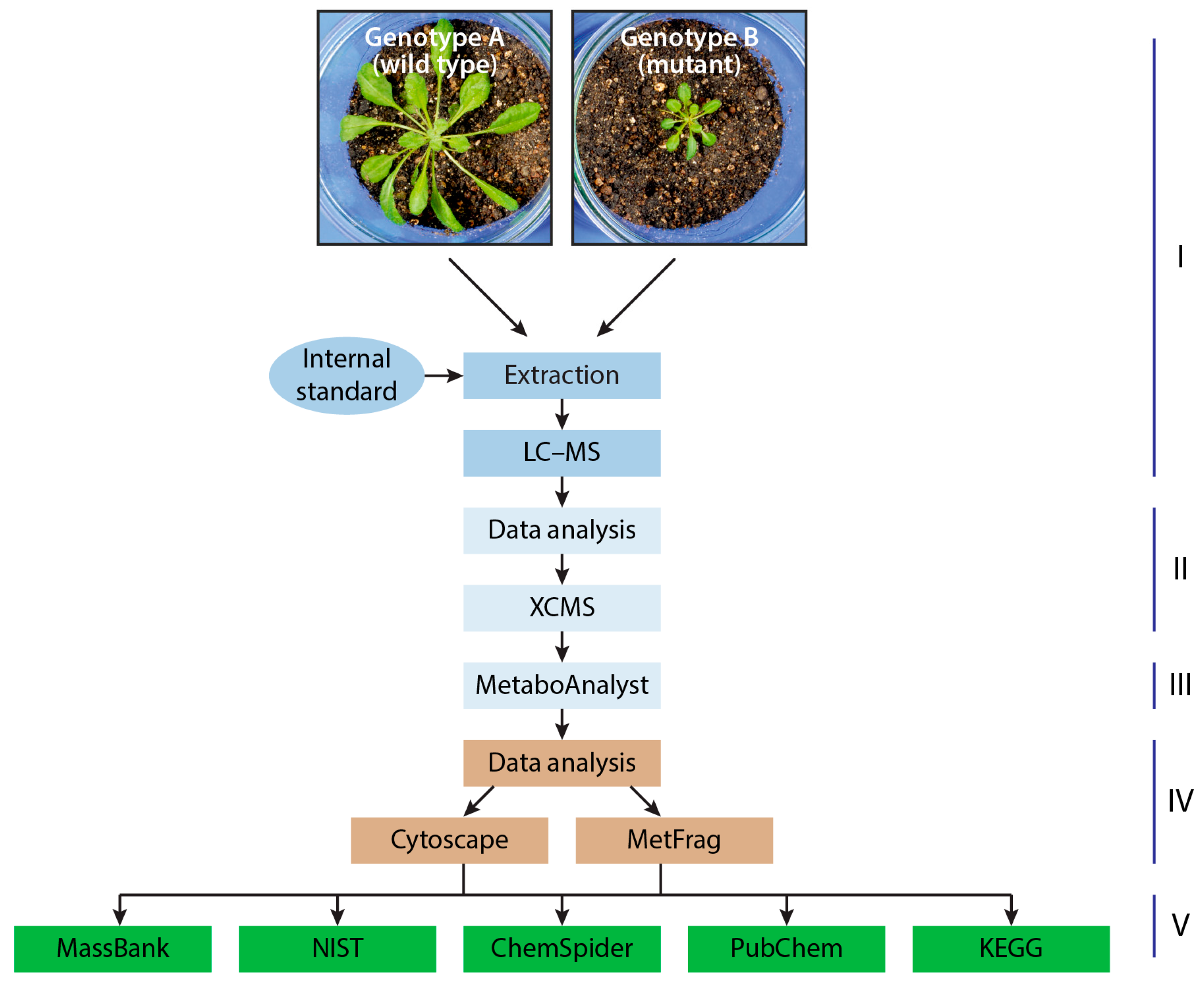
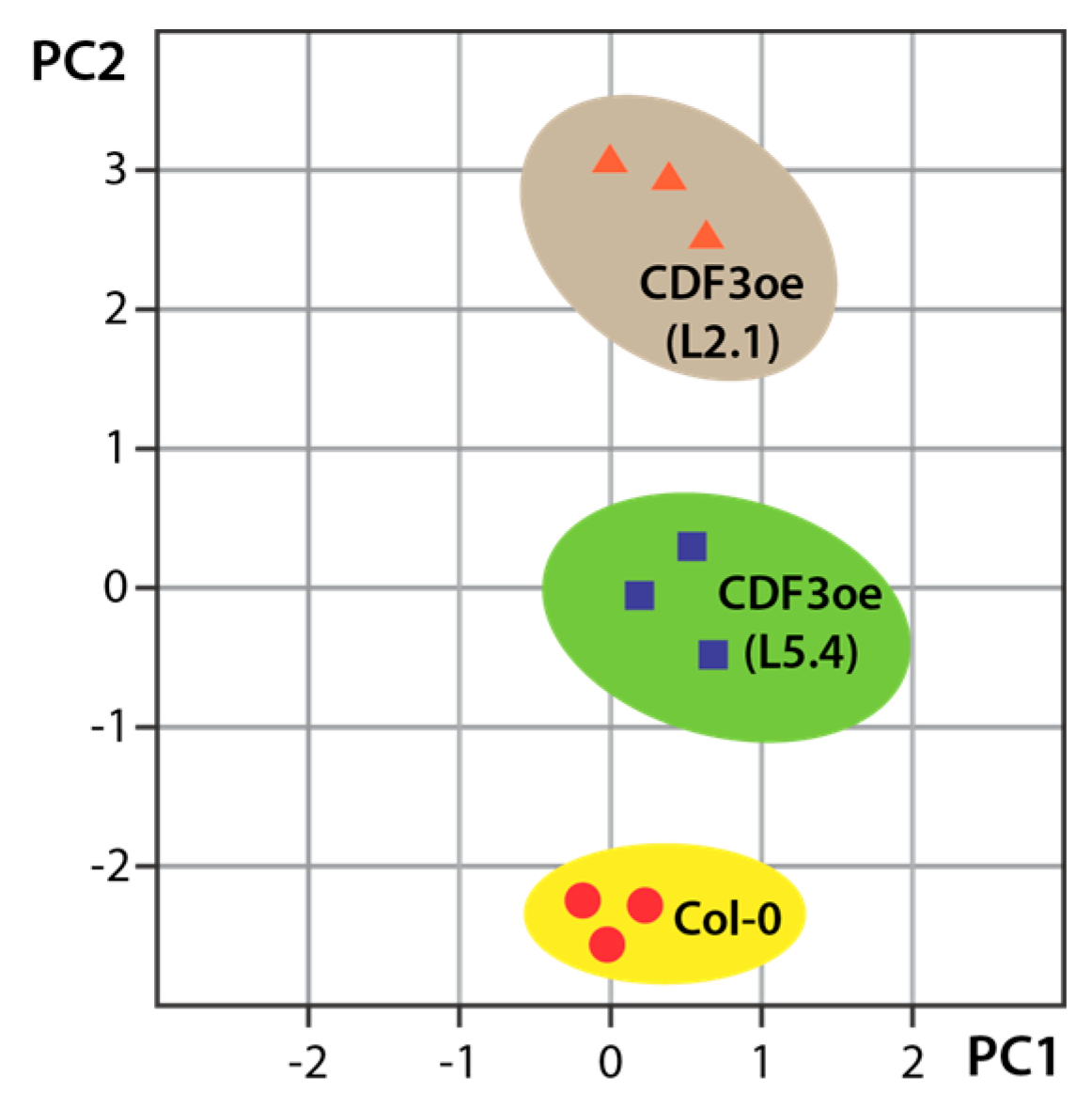
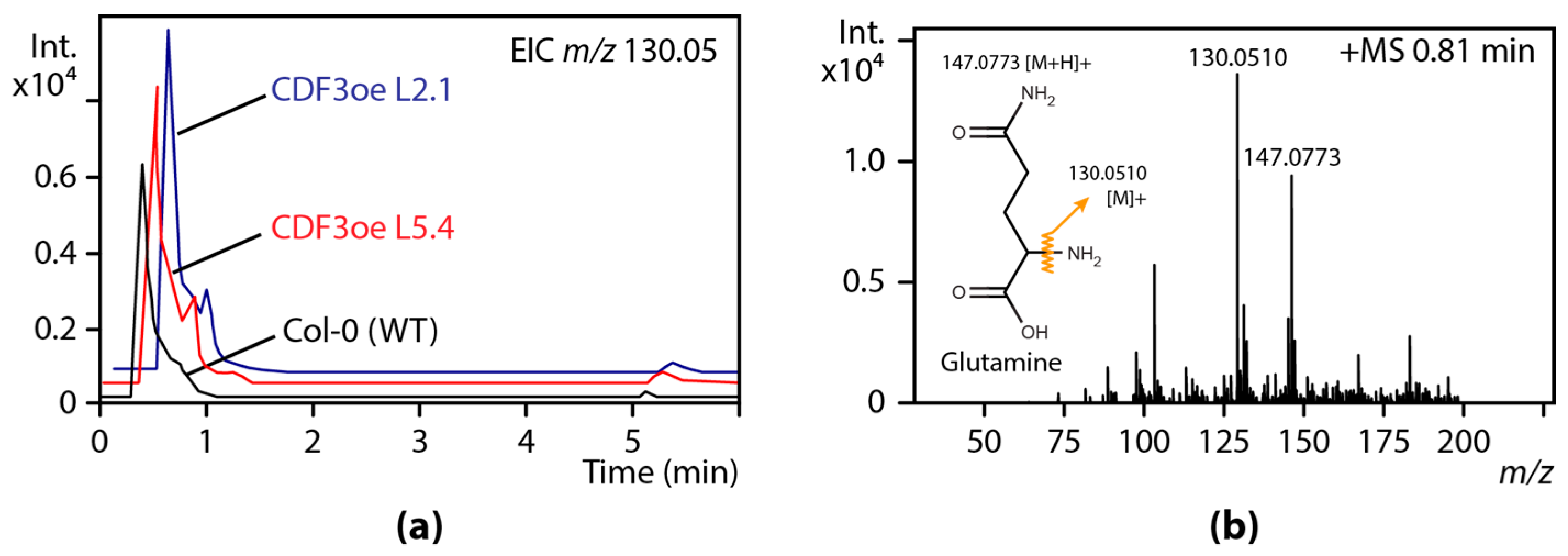
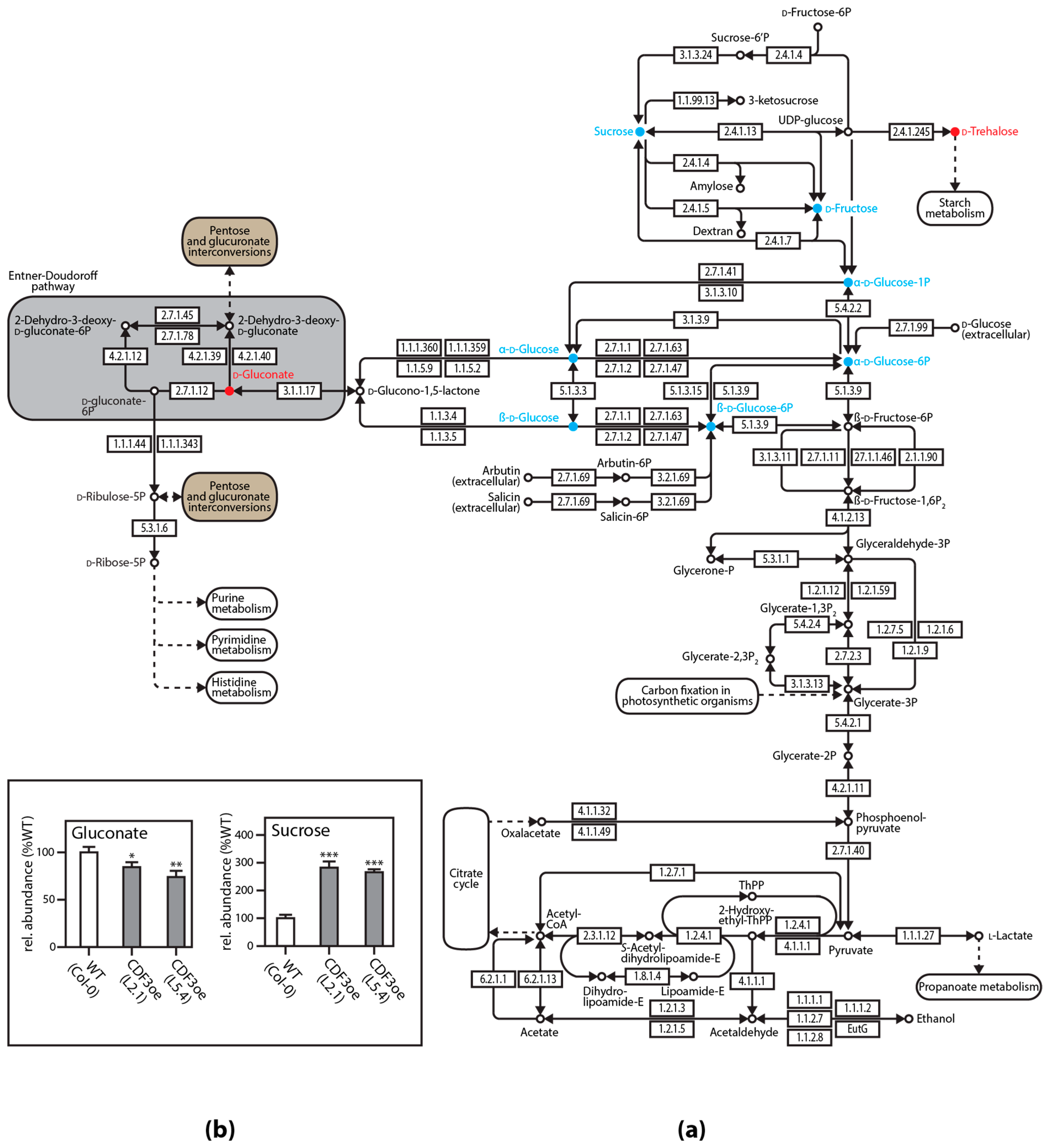
3. Untargeted and Targeted Metabolomics Studies
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pruneda-Paz, J.L.; Breton, G.; Nagel, D.H.; Kang, S.E.; Bonaldi, K.; Doherty, C.J.; Ravelo, S.; Galli, M.; Ecker, J.R.; Kay, S.A. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 2014, 8, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.; Callis, J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 2007, 99, 787–822. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-oxylipin crosstalk: Relationship of antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Baumann, S.; Kalkhof, S.; Hackermuller, J.; Otto, W.; Tomm, J.M.; Wissenbach, D.K.; Rolle-Kampczyk, U.; von Bergen, M. Requirements and Perspectives for Integrating Metabolomics with other Omics Data. Curr. Metabolomics 2013, 1, 15–27. [Google Scholar]
- Jozefczuk, S.; Klie, S.; Catchpole, G.; Szymanski, J.; Cuadros-Inostroza, A.; Steinhauser, D.; Selbig, J.; Willmitzer, L. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 2010, 6, 364. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Vogel, C.; Wang, R.; Yao, X.; Marcotte, E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007, 25, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Nebauer, S.G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.V.; et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Domínguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Renau-Morata, B.; Molina, R.V.; Carrillo, L.; Cebolla-Cornejo, J.; Sánchez-Perales, M.; Pollmann, S.; Domínguez-Figueroa, J.; Corrales, A.R.; Flexas, J.; Vicente-Carbajosa, J.; et al. Ectopic Expression of CDF3 Genes in Tomato Enhances Biomass Production and Yield under Salinity Stress Conditions. Front. Plant Sci. 2017, 8, 660. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. ChIP-seq: Welcome to the new frontier. Nat. Methods 2007, 4, 613. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Godoy, M.; Franco-Zorrilla, J.M.; Pérez-Pérez, J.; Oliveros, J.C.; Lorenzo, Ó.; Solano, R. Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J. 2011, 66, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Morant, M.; Ekstrøm, C.; Ulvskov, P.; Kristensen, C.; Rudemo, M.; Olsen, C.E.; Hansen, J.; Jørgensen, K.; Jørgensen, B.; Møller, B.L.; et al. Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2. Mol. Plant 2010, 3, 192–211. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Degenkolbe, T.; Cuadros-Inostroza, A.; Klie, S.; Sulpice, R.; Leisse, A.; Steinhauser, D.; Fernie, A.R.; Willmitzer, L.; Hannah, M.A. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. The Plant Journal 2011, 67, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Noguero, M.; Atif, R.M.; Ochatt, S.; Thompson, R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Ruhl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Goralogia, G.S.; Liu, T.K.; Zhao, L.; Panipinto, P.M.; Groover, E.D.; Bains, Y.S.; Imaizumi, T. CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. 2017, 92, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Zhang, J.; Li, H.; Yang, C.; Zhang, C.; Zhang, X.; Khurram, Z.; Zhang, Y.; Wang, T.; Fei, Z.; et al. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 2010, 61, 3563–3575. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Guo, S.Q.; Yang, X.; Bao, Y.M.; Tang, H.J.; Sun, H.; Huang, J.; Zhang, H.S. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 2010, 61, 2807–2818. [Google Scholar] [CrossRef] [PubMed]
- Brazma, A.; Hingamp, P.; Quackenbush, J.; Sherlock, G.; Spellman, P.; Stoeckert, C.; Aach, J.; Ansorge, W.; Ball, C.A.; Causton, H.C.; et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001, 29, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, H.; Sarkans, U.; Kolesnikov, N.; Abeygunawardena, N.; Burdett, T.; Dylag, M.; Emam, I.; Farne, A.; Hastings, E.; Holloway, E.; et al. ArrayExpress update—An archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011, 39, D1002–D1004. [Google Scholar] [CrossRef] [PubMed]
- Functional Genomics Data Society. Available online: http://www.fged.org (accessed on 21 February 2018).
- Encyclopedia of DNA Elements. Available online: https://www.encodeproject.org (accessed on 21 February 2018).
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, M.D.; Verborgh, R.; Bonino da Silva Santos, L.O.; Clark, T.; Swertz, M.A.; Kelpin, F.D.L.; Gray, A.J.G.; Schultes, E.A.; van Mulligen, E.M.; Ciccarese, P.; et al. Interoperability and FAIRness through a novel combination of Web technologies. PeerJ Comput. Sci. 2017, 3, e110. [Google Scholar] [CrossRef]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, E.; Provart, N.J. The Botany Array Resource: E-Northerns, Expression Angling, and promoter analyses. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.A.; Zhang, P.; Rhee, S.Y. AraCyc: A Biochemical Pathway Database for Arabidopsis. Plant Physiol. 2003, 132, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Clifford, S.C.; Arndt, S.K.; Corlett, J.E.; Joshi, S.; Sankhla, N.; Popp, M.; Jones, H.G. The role of solute accumulation, osmotic adjustment and changes in cell wall elasticity in drought tolerance in Ziziphus mauritiana (Lamk.). J. Exp. Bot. 1998, 49, 967–977. [Google Scholar] [CrossRef]
- Épron, D.; Dreyer, E. Starch and soluble carbohydrates in leaves of water-stressed oak saplings. Ann. For. Sci. 1996, 53, 263–268. [Google Scholar] [CrossRef]
- Morales, C.G.; Pino, M.T.; del Pozo, A. Phenological and physiological responses to drought stress and subsequent rehydration cycles in two raspberry cultivars. Sci. Hort. 2013, 162, 234–241. [Google Scholar] [CrossRef]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, N.; Saghatelian, A.; Patti, G.J. Defining the metabolome: Size, flux, and regulation. Mol. Cell 2015, 58, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Banton, S.; Tran, V.T.; Konomi, J.V.; Li, S.; Jones, D.P.; Vos, M.B. Amino Acid Metabolism is Altered in Adolescents with Nonalcoholic Fatty Liver Disease-An Untargeted, High Resolution Metabolomics Study. J. Pediatr. 2016, 172, 14–19 e5. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Sumner, L.W.; Rhee, S.Y.; Ward, J.; Dickerson, J.; Lange, B.M.; Lane, G.; Roessner, U.; Last, R.; Nikolau, B. Minimum reporting standards for plant biology context information in metabolomic studies. Metabolomics 2007, 3, 195–201. [Google Scholar] [CrossRef]
- Fiehn, O.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Lee, D.Y.; Lu, Y.; Moon, S.; Nikolau, B. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J. 2008, 53, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, H.; Hardy, N.; Beckmann, M.; Draper, J.; Smith, A.R.; Taylor, J.; Fiehn, O.; Goodacre, R.; Bino, R.J.; Hall, R.; et al. A proposed framework for the description of plant metabolomics experiments and their results. Nat. Biotechnol. 2004, 22, 1601. [Google Scholar] [CrossRef] [PubMed]
- Sansone, S.A.; Fan, T.; Goodacre, R.; Griffin, J.L.; Hardy, N.W.; Kaddurah-Daouk, R.; Kristal, B.S.; Lindon, J.; Mendes, P.; Morrison, N.; et al. The Metabolomics Standards Initiative. Nat. Biotechnol. 2007, 25, 846. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.J.; Badger, M.R.; Harvey Millar, A. The MetabolomeExpress Project: Enabling web-based processing, analysis and transparent dissemination of GC/MS metabolomics datasets. BMC Bioinform. 2010, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.J.; Zhang, P.; Whitehead, L.; Kaines, S.; Tcherkez, G.; Badger, M.R. PhenoMeter: A Metabolome Database Search Tool Using Statistical Similarity Matching of Metabolic Phenotypes for High-Confidence Detection of Functional Links. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Ferry-Dumazet, H.; Gil, L.; Deborde, C.; Moing, A.; Bernillon, S.; Rolin, D.; Nikolski, M.; de Daruvar, A.; Jacob, D. MeRy-B: A web knowledgebase for the storage, visualization, analysis and annotation of plant NMR metabolomic profiles. BMC Plant Biol. 2011, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P.; et al. MetaboLights—An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
- PhenoMeNal. Available online: http://phenomenal-h2020.eu/ (accessed on 21 February 2018).
- MetaboFlow. Available online: http://www.metaboflow.org/ (accessed on 21 February 2018).
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüuller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007, 2, 2366–2382. [Google Scholar] [CrossRef] [PubMed]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chemi. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Riemann, M.; Müller, A.; Duchting, P.; Weiler, E.W.; Nick, P. Cholodny-Went revisited: A role for jasmonate in gravitropism of rice coleoptiles. Planta 2005, 222, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Krug, D.; Zurek, G.; Schneider, B.; Garcia, R.; Muller, R. Efficient mining of myxobacterial metabolite profiles enabled by liquid chromatography-electrospray ionisation-time-of-flight mass spectrometry and compound-based principal component analysis. Anal. Chim. Acta 2008, 624, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Rosin, F.M.; Watanabe, N.; Cacas, J.L.; Kato, N.; Arroyo, J.M.; Fang, Y.; May, B.; Vaughn, M.; Simorowski, J.; Ramu, U.; et al. Genome-wide transposon tagging reveals location-dependent effects on transcription and chromatin organization in Arabidopsis. Plant J. 2008, 55, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Ojanperä, S.; Pelander, A.; Pelzing, M.; Krebs, I.; Vuori, E.; Ojanperä, I. Isotopic pattern and accurate mass determination in urine drug screening by liquid chromatography/time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Loba, V.; Pérez-Alonso, M.M.; Pollmann, S. Monitoring of Crosstalk Between Jasmonate and Auxin in the Framework of Plant Stress Responses of Roots. Methods Mol. Biol. 2017, 1569, 175–185. [Google Scholar]
- Carrasco-Loba, V.P.S. Highly Sensitive Salicylic Acid Quantification in Milligram Amounts of Plant Tissue. Methods Mol. Biol. 2017, 1497, 221–229. [Google Scholar]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Bain, J.R.; Stevens, R.D.; Wenner, B.R.; Ilkayeva, O.; Muoio, D.M.; Newgard, C.B. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes 2009, 58, 2429–2443. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Palsson, B.; Zengler, K. The challenges of integrating multi-omic data sets. Nat. Chem. Biol. 2010, 6, 787. [Google Scholar] [CrossRef] [PubMed]
- Droste, P.; Nöh, K.; Wiechert, W. Omix—A Visualization Tool for Metabolic Networks with Highest Usability and Customizability in Focus. Chem. Ing. Tech. 2013, 85, 849–862. [Google Scholar] [CrossRef]
- QIAGEN. Ingenuity Pathway Analysis. Available online: https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ (accessed on 21 February 2018).
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Neuweger, H.; Persicke, M.; Albaum, S.P.; Bekel, T.; Dondrup, M.; Huser, A.T.; Winnebald, J.; Schneider, J.; Kalinowski, J.; Goesmann, A. Visualizing post genomics data-sets on customized pathway maps by ProMeTra-aeration-dependent gene expression and metabolism of Corynebacterium glutamicum as an example. BMC Syst. Biol. 2009, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- García-Alcalde, F.; García-López, F.; Dopazo, J.; Conesa, A. Paintomics: A web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics 2011, 27, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Grapov, D.; Wanichthanarak, K.; Fiehn, O. MetaMapR: Pathway independent metabolomic network analysis incorporating unknowns. Bioinformatics 2015, 31, 2757–2760. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline mechanisms of stress survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Kovács, Z.; Simon-Sarkadi, L.; Vashegyi, I.; Kocsy, G. Different accumulation of free amino acids during short- and long-term osmotic stress in wheat. Sci. World J. 2012, 2012, 216521. [Google Scholar] [CrossRef] [PubMed]
- Ayre, B.G. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 2011, 4, 377–394. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Alonso, M.-M.; Carrasco-Loba, V.; Medina, J.; Vicente-Carbajosa, J.; Pollmann, S. When Transcriptomics and Metabolomics Work Hand in Hand: A Case Study Characterizing Plant CDF Transcription Factors. High-Throughput 2018, 7, 7. https://doi.org/10.3390/ht7010007
Pérez-Alonso M-M, Carrasco-Loba V, Medina J, Vicente-Carbajosa J, Pollmann S. When Transcriptomics and Metabolomics Work Hand in Hand: A Case Study Characterizing Plant CDF Transcription Factors. High-Throughput. 2018; 7(1):7. https://doi.org/10.3390/ht7010007
Chicago/Turabian StylePérez-Alonso, Marta-Marina, Víctor Carrasco-Loba, Joaquín Medina, Jesús Vicente-Carbajosa, and Stephan Pollmann. 2018. "When Transcriptomics and Metabolomics Work Hand in Hand: A Case Study Characterizing Plant CDF Transcription Factors" High-Throughput 7, no. 1: 7. https://doi.org/10.3390/ht7010007
APA StylePérez-Alonso, M.-M., Carrasco-Loba, V., Medina, J., Vicente-Carbajosa, J., & Pollmann, S. (2018). When Transcriptomics and Metabolomics Work Hand in Hand: A Case Study Characterizing Plant CDF Transcription Factors. High-Throughput, 7(1), 7. https://doi.org/10.3390/ht7010007






