Abstract
Introduction: Yeast pathogens are emerging agents of nosocomial as well as community-acquired infections and their rapid and accurate identification is crucial for a better management of high-risk patients and for an adequate treatment. Methods: We performed a retrospective review of 156 yeast isolates collected during a 17 months’ period of regular clinical practice at the Microbiology Department of San Camillo Hospital in Treviso, Italy and analyzed by the traditional culture-based method combined with matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Results: Out of all the samples collected MALDI-TOF MS was able to characterize with a MT score ≥1.7 (accurate result at species level) 12 different yeast and yeast-like species from 140 samples: Candida albicans (63.7%), Candida glabrata (13.6%), Saccharomyces cerevisiae (6.5%), Candida parapsilosis (5.7%), Candida tropicalis (2.1%), Candida pararugosa (2.1%), Candida guilliermondii (2.1%), Candida kefyr (1.4%), Candida lusitaniae (0.7%), Candida palmioleophila (0.7%), Geotrichum silvicola (0.7%), Rhodotorula mucilaginosa (0.7%). Susceptibility testing toward seven common antifungal agents showed a characteristic MIC distribution of C. albicans isolates for echinocandins: particularly we noticed that 72% and 46% of C. albicans showed an MIC value close to clinical breakpoint as defined by EUCAST, respectively for anidulafungin and micafungin. Conclusion: Accurate identification of microorganisms and the study of their antifungal susceptibility allow to understand the epidemiology of a particular area, permitting the choice of the most appropriate early antifungal treatment.
Introduction
Yeast pathogens are emerging agents of nosocomial and community-acquired infections, local or disseminated, particularly in immunocompromised patients (intensive care unit, cancer, neutropenic patients) and their incidence has greatly increased in the last few years [1].
Until recently, only few pathogenic species of Candida were known. However, recent years have seen an increase in the number of species responsible for human infections [2]. C. albicans currently represents the most commonly isolated species, while other species that cause superficial and/or invasive infections diversify into C. glabrata, C. parapsilosis, C. tropicalis, C. kefyr and the emerging multidrug resistant Candida auris [3].
In addition, yeasts such as Saccharomyces are often considered the cause of serious infections in high-risk patients, while Rhodotorula mucilaginosais widely distributed in the environment [4]. Although it was once considered to be less virulent than Candida spp., Rhodotorula spp. is now considered a potential pathogen in patients treated with immunosuppressant drugs and bearers of central venous catheter [4]. A late diagnosis and incorrect treatment can significantly contribute to the high mortality associated with invasive fungal infections. Even if the ESCMID recommendations for the therapeutic management of candidemia do not differentiate the therapeutic approach according to the species, meaning that as first line therapy a presumptive large spectrum antifungal drug (in practice echinocandins) can be used whatever is the causative species, rapid and accurate identification (ID) methods for fungal species causing invasive mycoses are crucial for a better management of high-risk patients and for an adequate treatment [5]. Although the standardization of antifungal susceptibility testing associated a huge advance, some non-albicans species may be intrinsically resistant to first-line antifungal agents [6,7]. For these reasons, in addition to the traditional ID methods, it is extremely important to apply new technologies and diagnostic pipelines able to provide specific information on properties of the pathogenic microorganisms during infection treatment, to aid clinicians in the therapeutic management of infections rather than to provide ordinary pathogen ID. Conventional phenotypic methods may fail to capture the growing diversity of fungal pathogens while offering inconclusive results, especially for unusual yeasts, because phenotypic tests often require some hours or even days for only ID results [8,9].
In this regard, MS (mass spectrometry), particularly MALDI-TOF MS (matrix-assisted laser desorption ionization-time of flight mass spectrometry), has been suggested as an effective alternative for microbial characterization thanks to the high efficiency of proteome profiling, allowing rapid and accurate ID of pathogenic microorganisms, including yeasts and filamentous fungi of medical interest [10,11,12]. One of the advantages of this technique is represented by the possibility to simultaneously and rapidly analyze complex proteins or peptide mixtures without a prior separation, or to analyze directly whole cells, with minimal sample preparation and negligible consumable costs [13,14,15].
The aim of this study was to carry out an analysis of Candida spp. and yeasts isolated in a period of 17 months from clinical samples collected at the Microbiology Department of San Camillo Hospital in Treviso, using either traditional culture-based and MALDI-TOF MS-based techniques, and to investigate their susceptibility distribution to azoles, echinocandins, and amphotericin B, in order to give useful information for comparative epidemiological studies of antifungal resistance pattern in the Italian population.
Methods
This work is a retrospective review of laboratory results yielded in regular clinical practice during 17 months. A total of 156 clinical consecutive yeasts isolates from in- and out-patient samples (i.e., vaginal swab, urine, stools, nail fragments, skin swabs, bone fragments, oropharyngeal swabs) were collected at the Microbiology Unit of San Camillo Hospital of Treviso, Italy, from August 2013 to November 2014. Yeast isolates were processed using conventional ID methods based on biochemical characterization and also, they were identified by MALDI-TOF MS-based method at the Parasitology Unit of Bambino Gesù Hospital of Rome.
Phenotypic characterization by culture and biochemical assays
Sample cultures were performed on Sabouraud agar supplemented by chloramphenicol (BD Biosciences, Franklin Lakes, NJ, USA) for 48 hours at 37 °C in a dry incubator according to routine diagnostic conditions employed at the Microbiology Unit. Subsequently, colony morphology, texture, and color were examined on BBL CHROMagar Candida plates (BD Biosciences) for preliminary C. albicans, C. tropicalis and C. krusei ID [16]. ATCC reference strains such as C. albicans ATCC 10231 and C. krusei ATCC 6258 (Oxoid, Thermo Fisher Scientific, Basingstoke, UK) were used as controls in phenotypic characterization.
Anti-fungal susceptibility testing
Anti-fungal susceptibility tests were performed by using Sensititre® Yeast-One®YO10 cards, according to the manufacturer's instructions (TREK Diagnostic Systems, Cleveland, OH, USA). The test used broth microdilution method to provide qualitative (S=sensitive, I=intermediate or R=resistant) and quantitative results, related to the minimum inhibitory concentration (MIC), by a 96 wells’ plate containing antimicrobial agents in dehydrated form and in appropriate dilutions. The antifungal agents assayed were: amphotericin B (AMB), itraconazole (ITC), voriconazole (VRC), posaconazole (POS), fluconazole (FLU), caspofungin (CAS), micafungin (MFG), anidulafungin (AFG) [6,17].
Susceptibility profiles were optically evaluated with strain growth associated to blue chromogen (S) or rose-red (R) indicator change. All MIC values were evaluated with EUCAST clinical breakpoint tables for Candida spp. and Aspergillus spp. Statistical and epidemiological analysis of antifungal susceptibility was reevaluated according to the latest version of EUCAST guidelines (v.9.0) to make the data applicable to the current epidemiological reality [18].
Spectra generation and interpretation
After growth of fungal isolates three or four colonies from each sample were mixed thoroughly in 300 mL double-distilled water. Absolute ethanol (900 µL) (Sigma–Aldrich, Milan, Italy) was added, tube contents carefully mixed, centrifuged at 14,000 g for 2 min at room temperature (RT), the supernatant discarded and the pellet air dried. The pellet was mixed thoroughly with 50 μL of formic acid (70%) (Sigma–Aldrich), before addition of an equivalent volume of acetonitrile (Sigma–Aldrich). The mixture was centrifuged at 14,000 g for 2 min, and 1.5 µL of the supernatant from each sample (i.e., two spots for each) were placed onto a MSP 96 polished steel target (Bruker Daltonik GmbH, Bremen, Germany) and allowed to dry at RT. Each sample spot was overlaid with 1 µL of matrix, which consisted of a saturated solution of α-cyano-4 hydroxy-cinnamic acid (HCCA) in 50% acetonitrile- and 2.5% trifluoroacetic acid (Sigma-Aldrich) (final concentration 10 mg HCCA/mL) and air-dried at RT.
Measurements were performed with a Microflex LT mass spectrometer (Bruker Daltonik GmbH) using FlexControl software (version 3.0, Bruker Daltonik GmbH). Spectra were recorded in the linear positive mode at a laser frequency of 20 Hz (ion source 1 voltage, 20 kV; ion source 2 voltage, 18.4 kV; lens voltage, 9.1 kV) within a mass range from 2,000 to 20,137 Da. Each sample was tested in duplicate to assess spectra reproducibility. Two hundred laser shots per sample spot were acquired employing Flex Control software package. Spectra were internally calibrated by using an Escherichia coli ribosomal proteins BTS (Bacterial Test Standard, Bruker Daltonik GmbH) according to the manufacturer's instructions. All the produced spectra were visually inspected before statistical analysis into BioTyper software (version 3.0; Bruker Daltonik GmbH) that provided the pattern matching with default settings. Results of the pattern-matching process were expressed with MT scores ranging from 0 to 3.0 as proposed by the manufacturer. For each isolate, the highest score of a match against a spectrum in the customized database was used for identification. Scores below 1.7 were considered not to have generated a reliable identification, a score ≥1.7 was used for reliable species identification.
Statistical analysis
Data were analyzed using Pearson’s Chi-squared test and Mann-Whitney U test. Furthermore, the degree of concordance between results obtained by traditional methods and MALDI-TOF MS was calculated using Cohen’s kappa coefficient. Statistical significance was defined as a two-tailed p<0.05 for all analyses which were carried out using the Stata 12 software package (College Station, TX, USA).
Results
One-hundred and fifty-six yeast isolates were collected at the Microbiology Unit of San Camillo Hospital of Treviso from August 2013 to November 2014. The median number of yeast isolations was eight per month, but we observed an increment in the isolation number in November 2013 (N=13), March 2014 (N=18) and from May (N=24) to June 2014 (N=13) (Figure 1A), even if the Mann-Whitney U Test did not show any statistical association (p>0.05) between yeast isolation and their seasonal-driven occurrence.
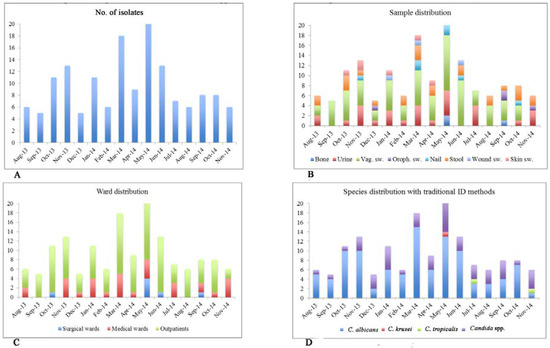
Figure 1.
Graphical distribution of Candida spp. isolates collected from August 2013 to November 2014 (N=156). The strains are grouped on the basis of collection date (Panel A), sample type (Panel B), ward distribution (Panel C), and species distribution by traditional ID methods (Panel D).
Out of the 156 fungal isolates, 75 were isolated from vaginal swabs (48%), 30 from urine (19%), 26 from feces (17%), and 25 from other sites (16%) (Figure 1B). One-hundred and seventeen of 156 samples derived from outpatients (75%), the remaining 39 belonged to hospitalized patients, specifically 32 were isolated from patients admitted to the medical wards (82%) and seven from patients admitted to surgical wards (18%) (Figure 1C).
Using conventional microbiological ID methods, it was possible to identify at species level only three types of Candida spp. (C. albicans, C. tropicalis and C. krusei), while for the remaining samples the identification was limited to genus Candida. Particularly, 101 out of 156 strains were identified as C. albicans (64.7%), two out of 156 as C. tropicalis (1.2%) and one out of 156 as C. krusei (0.6%) (Figure 1D).
To compare and refine on conventional yeast identification, MALDI-TOF MS biotyper was applied to all strains, allowing the accurate identification at species level (MT score ≥1.7) of 12 different yeast and yeast-like species from 140 samples: C. albicans (63.7%), C. glabrata (13.6%), Saccharomyces cerevisiae (6.5%), C. parapsilosis (5.7%), C. tropicalis (2.1%), C. pararugosa (2.1%), C. guilliermondii (2.1%), C. kefyr (1.4%), C. lusitaniae (0.7%), C. palmioleophila (0.7%), Geotrichum silvicola (0.7%), R. mucilaginosa (0.7%) (MS data are reported in Figure 2).
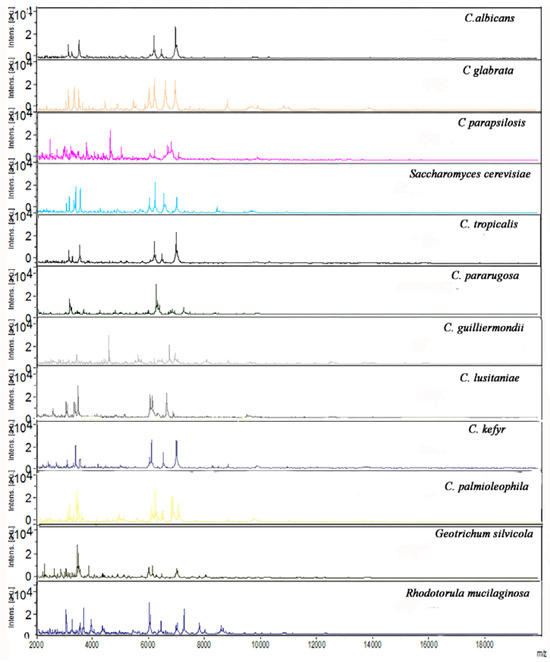
Figure 2.
MALDI-TOF MS spectra of the 12 yeast and yeast-like species identified by MT ID originated by MALDI Biotyper Software analysis. The m/z values are expressed in Da and the amplitudes are reported in a scale of intensity arbitrary units (a.u.). The MT IS score was in all cases >1.7. One-hundred and thirteen strains of the 140 (80.7%) showed a best MT score ≥2.0.
By MALDI-TOF MS we obtained 91 identifications (65.0%) in agreement with the traditional identification method, with a modest degree of concordance (Cohen's kappa coefficient of 0.281). Three strains (2.2%) showed discrepancy between the two methods: a presumptive C. krusei was identified as S. cerevisiae by MALDI TOF MS, while two strains identified as C. albicans by traditional methods were C. kefyr and C. parapsilosis. Moreover, for 46 strains (32.8%), previously identified as Candida spp., the MALDI-TOF MS method overcame the traditional one, since this method was able to give an identification at the species level. The susceptibility distribution toward seven common antifungal agents were assessed for the 140 samples identified by MALDI-TOF. Susceptibility breakpoints and MIC categorization for antifungal susceptibility analysis were performed by applying interpretative criteria as published by EUCAST (Table 1) [18].

Table 1.
Antifungal susceptibility of Candida spp. isolates (N=140).
As showed in Table 1, all C. albicans isolated were sensitive to fluconazole (FLC), voriconazole (VRC), posaconazole (POS) and amphotericin B (AMB), while 20.2% of them showed resistance to micafungin (MCF) and anidulafungin (ANF) with MIC values very close to the clinical breakpoint (CB) (Figure 3). Overall, 5.3% of C. glabrata showed resistance to MCF and 10.5% to ANF, while all of them showed sensitivity against AMB and intermediate sensitivity to FLC; 12.5% and 25% of C. parapsilosis showed resistance to itraconazole (ITC) and POS, respectively. Finally, 66.7% of C. tropicalis were I to FLC and 33.3% were R to ANF. For all the other yeasts and Candida spp. isolated we found that 25% were I and 33% R to FLC. No strain showed resistance against VRC and AMB.
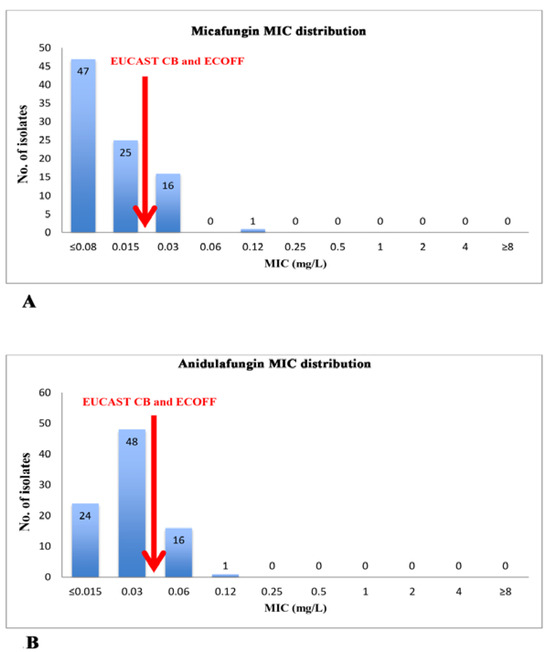
Figure 3.
MIC distribution of echinocandins for Candida albicans isolates (N=89). The arrows indicate EUCAST Clinical Breakpoint (CB) and epidemiological cut-off values (ECOFF) for micafungin (Panel A) and anidulafungin (Panel B).
Moreover, analyzing resistance to antifungals based on the type of yeast and on the type of samples we found a significant association only between resistance to anidulafungin and C. albicans isolates collected from genital samples (vaginal swabs, urethral swabs and vulvar swabs) with a p=0.013, calculated by Pearson's Chi-squared test.
Discussion
In order to correctly and quickly identify the causative agent of yeast infections and to promptly choose the correct therapy it is necessary to set up fast and reliable ID methods that can help to better understand yeast and yeast-like species-specific susceptibility patterns. Chromogenic agar media and biochemical or enzymatic panels may be occasionally disadvantageous due to their limited range of species identification profiles [19]. In fact, using only conventional culture-based ID methods, some less common species (i.e, strains of S. cerevisiae, C. parapsilosis, C. kefyr, C. lusitaniae, and R. mucilaginosa) can be often misidentified or, simply, cannot be identified at all. For this reason, microbiologists are forced to deal with not-identified or misidentified yeast strains with the consequent clinical risk of ineffective treatments for several microorganisms [20].
Since the advent of ionization techniques, MS has become a standard method of protein analysis and TOF instruments have been used for proteomics applications, specifically in combination with MALDI procedures [21,22]. In fact, MALDI-TOF MS appears as an efficient strain ID/epidemiological typing tool, generating data with negligible cost, and allowing the elimination of inappropriate biochemical tests.
Although MALDI TOF MS is actually used predominantly as a rapid identification method and it does not provide information about susceptibility to antimicrobial agents, its contribution to targeted therapy could be essential because a correct identification allows the selection of antifungals based on the knowledge of local epidemiological data regarding antifungal susceptibility of different fungal genera or species and may help in decreasing hospital length of stay and total care costs; for example they could be crucial in clinical management of new emerging multi-drug resistant Candida spp. such as C. auris that is currently causing outbreaks in healthcare settings worldwide [3,23].
In addition to species-specific antifungal resistance patterns (Table 1), in our survey we detected a characteristic MIC distribution of C. albicans isolates for echinocandins: particularly, for both anidulafungin and micafungin, we noticed that 64 isolates out of 89 (71.9%) and 41 strains out of 89 (46.1%), respectively, showed a MIC value close to clinical breakpoint (Figure 3) and in this context it is important to underline that CB is equivalent to epidemiological cut-off (ECOFF) as defined by EUCAST [18,24]. Comparing our study with those of other Italian authors it is interesting to note that we found a higher percentage of C. albicans resistant to echinocandins (according to EUCAST) [25].
Our study has the limitation of being an observational laboratory-based survey, without clinical evidences about type and duration of antifungal therapy, and molecular resistance mechanisms were not actually investigated; for these reasons further studies are needed.
Conclusion
An accurate identification of microorganisms and the study of their antimicrobial susceptibility allow the understanding of the epidemiology of a particular area, permitting the most appropriate choice of early antifungal treatment. Until now, in Italy, studies about in vitro susceptibility of echinocandins against Candida isolates using EUCAST methods are rarely reported [25].
MS-based diagnostic methods, coupled with reasoned antimicrobial strategy, may provide a substantial alternative to conventional laboratory methods allowing advanced clinical management of patients.
Author Contributions
MS designed the study, performed the laboratory experiments, collected the data, interpreted the findings and wrote the paper. AB collected the data, interpreted the findings and wrote the paper. FDC, CA, FDG, AM helped interpret the findings. MM, LP coordinated the study, supervised the laboratory experiments and helped interpret the findings. All authors read and approved the final manuscript.
Funding
None to declare.
Conflicts of interest
All authors – none to disclose.
References
- Pappas, P.G.; Kauffman, C.A.; Andes, D.; et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 2009, 48, 503–535. [Google Scholar] [CrossRef]
- Sullivan, D.J.; Coleman, D.C. Candida dubliniensis: characteristics and identification. J Clin Microbiol 1998, 31, 329–334. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Berman, J.; Novikov, A.; et al. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 2000, 50 Pt 3, 1351–1371. [Google Scholar] [CrossRef]
- Cornely, O.A.; Bassetti, M.; Calandra, T.; et al. ESCMID guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect 2012, 18 (Suppl. 7), 19–37. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Gomez-Lopez, A.; Alastruey-Izquierdo, A.; et al. Comparison of the Vitek 2 Antifungal Susceptibility System with the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution reference methods and with the Sensititre YeastOne and Etest techniques for in vitro detection of antifungal resistance in yeast isolates. J Clin Microbiol 2010, 48, 1782–1786. [Google Scholar] [PubMed]
- Rodriguez-Tudela, J.L.; Alcazar-Fuoli, L.; Cuesta, I.; et al. Clinical relevance of resistance to antifungals. Int J Antimicrob Agents 2008, 32 (Suppl. 2), S111–S113. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Torelli, R.; De Carolis, E.; Posteraro, P.; Sanguinetti, M. Update on the laboratory diagnosis of invasive fungal infections. Mediterr J Hematol Infect Dis 2011, 3, e2011002. [Google Scholar] [CrossRef]
- Risch, M.; Radjenovic, D.; Han, J.N.; Wydler, M.; Nydegger, U.; Risch, L. Comparison of MALDI TOF with conventional identification of clinically relevant bacteria. Swiss Med Wkly 2010, 140, w13095. [Google Scholar] [CrossRef]
- Dworzanski, J.P.; Snyder, A.P. Classification and identification of bacteria using mass spectrometry-based proteomics. Expert Rev Proteomics 2005, 2, 863–878. [Google Scholar] [CrossRef]
- Claydon, M.A.; Davey, S.N.; Edwards-Jones, V.; Gordon, D.B. The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol 1996, 14, 1584–1586. [Google Scholar] [CrossRef] [PubMed]
- Marinach-Patrice, C.; Fekkar, A.; Atanasova, R.; et al. Rapid species diagnosis for invasive candidiasis using mass spectrometry. PLoS One 2010, 5, e8862. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, N.; Hall, L.; Wohlfiel, S.L.; Buckwalter, S.P.; Wengenack, N.L. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 2011, 49, 1614–1616. [Google Scholar] [CrossRef]
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’hom, G. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol 2010, 48, 1549–1554. [Google Scholar] [CrossRef]
- Hsieh, S.Y.; Tseng, C.L.; Lee, Y.S.; et al. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol Cell Proteomics 2008, 7, 448–456. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Huston, A.; Coffman, S. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J Clin Microbiol 1996, 34, 58–61. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Meletiadis, J.; Mouton, K.W.; et al. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts; EUCAST definitive document E.DEF 7.3.1.; 2017. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST): Clinical Breakpoints. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 26 May 2018).
- Murray, C.K.; Beckius, M.L.; Green, J.A.; Hospenthal, D.R. Use of chromogenic medium for the isolation of yeasts from clinical specimens. J Med Microbiol 2005, 54, 981–985. [Google Scholar] [CrossRef]
- Zaragoza, O.; Mesa-Arango, A.C.; Gómez-López, A.; Bernal-Martínez, L.; Rodríguez-Tudela, J.L.; Cuenca-Estrella, M. Process analysis of variables for standardization of antifungal susceptibility testing of nonfermentative yeasts. Antimicrob. Agents Chemother 2011, 55, 1563–1570. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem 1991, 63, 1193A–2203A. [Google Scholar] [CrossRef]
- Amiri-Eliasi, B.; Fenselau, C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Anal Chem 2001, 73, 5228–5231. [Google Scholar] [CrossRef] [PubMed]
- Perez, K.K.; Olsen, R.J.; Musick, W.L.; et al. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med 2013, 137, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST): MIC Distributions and ECOFFs. Available online: http://www.eucast.org/mic_distributions_and_ecoffs/ (accessed on 28 August 2017).
- Montagna, M.T.; Lovero, G.; Coretti, C.; et al. Susceptibility to echinocandins of Candida spp. strains isolated in Italy assessed by European Committee for Antimicrobial Susceptibility Testing and Clinical Laboratory Standards Institute broth microdilution methods. BMC Microbiol 2015, 15, 106. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2018.