Introduction
COVID-19 (coronavirus disease-2019), caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), emerged in Wuhan in December 2019 and was declared a pandemic by the World Health Organization (WHO) in March 2020. Italy, reporting its first cases in January 2020, has seen 25.6 million cases due to different waves and variants. Transmission mainly occurs through saliva droplets, causing symptoms such as fatigue, headache, fever, anosmia, cough and breathing difficulties. Realtime polymerase chain reaction (RT-PCR) on nasopharyngeal swabs, the gold standard for diagnosing patients with COVID-19, faces limitations, straining global healthcare in certain setting.
This study explores saliva as an alternative to RT-PCR, assessing SARS-CoV-2 presence during Italy's pandemic. A comparison with nasopharyngeal swabs aims to validate saliva's reliability, offering similar sensitivity and specificity, simplifying collection and potentially addressing challenges like overcrowding, infectious risks, and patient discomfort.
Methods
The study included 109 non-consecutive adult patients sent by Italian Health Authorities to the Emergency Department and the COVID Test Point of our Center with symptoms compatible with SARS-CoV2 infection, but without a diagnosis. The study protocol was approved by the Institutional Review Board – Monza and Brianza Province (IRB No: 3405). Informed consent was obtained from all subjects involved in the study. The study was conducted in accordance with the Declaration of Helsinki and registered on ClinicalTrials.gov (NCT04953039).
Saliva and nasopharyngeal swabs were collected from symptomatic patients between November 2020 and October 2021. Patients independently collected saliva under supervision, generating sufficient volume (2 mL) by relaxing, massaging cheeks, and expelling it directly into a sterile container.
Nurses collected nasopharyngeal swabs by inserting a swab into both nostrils, reaching the nasopharynx with a rotary motion. The collected sample was placed in a special container with 3 cL of viral transport medium (VTM). Nasopharyngeal swabs were promptly tested for SARS-CoV-2 RNA using RT-PCR after collection. Simultaneously, saliva samples were stored at -80°C and later analyzed.
For both saliva and nasopharyngeal swabs RNA was extracted form 200 µL (saliva or universal transport medium) using the STARMag extraction kit (Seegene, Korea) following the manufacturer instructions. The extraction process was performed automatically on Microlab Nimbus instrument (Seegene). The detection of SARS-CoV-2 RNA was performed with Allplex SARS-CoV-2 kit (Seegene, Korea). In detail, 200 µL of saliva or universal transport medium (UTM) were mixed with proteinase K and lysis buffer; following the lysis step the samples were purified using magnetic beads and ethanol. The PCR setup was also performed automatically by Microlab Nimbus instrument (Seegene).
The amplification step was performed with CFX96 Real-Time System C100 Thermal Cycler (Bio-Rad, California, USA). The combination of targets/fluorescent reporters were E gene-FAM, RdRp/S-Cal Red 610, N gene-Quasar 670 and IC-HEX. Positive samples were detected when cycle threshold was below 40.
Sample description used median, interquartile range, mean, standard deviation (SD) for continuous variables. Diagnostic performance of the salivary test compared to the swab included sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy, and Cohen's Kappa with 95% confidence interval.
Results
Our sample consisted of 56 males (51%) and 53 females (49%) with a mean±SD age of 48.1±17.2 years. In
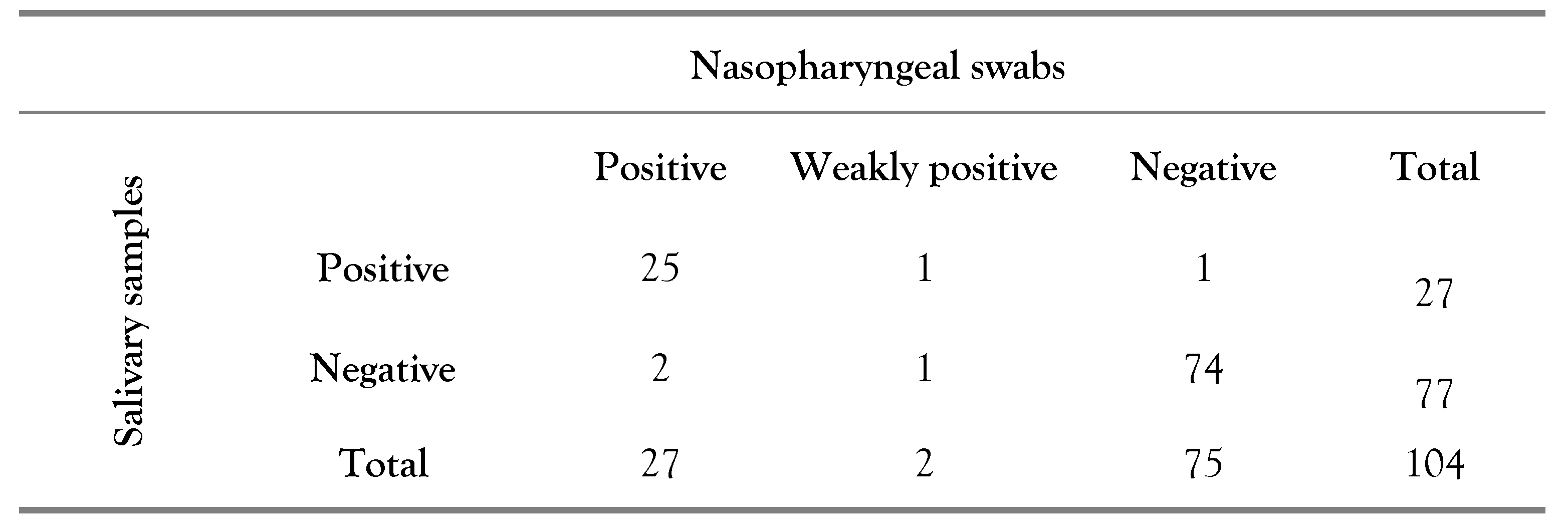
Table 1 the results of the study are reported: nasopharyngeal and salivary samples simultaneously collected from 104 patients (95.4%) were analyzed. Five patients (4.6%) were excluded owing to the inability to proceed with the RT-PCR due to sample collection or storage problems. Consequently, it can be seen that in 4 cases (3.9%) there was no agreement between the results of the analyses performed on nasopharyngeal swabs and salivary samples.
Specifically, in 2 cases (50.0%) the nasopharyngeal swab was positive while the salivary sample negative; in 1 case (25.0%) the nasopharyngeal swab was weakly positive and the salivary sample negative and in 1 case (25.0%) the salivary sample was positive while the nasopharyngeal swab negative. Finally,
Table 2 shows the diagnostic performances, evaluated with a 95% confidence interval, of the RT-PCR on salivary samples.
Discussion
The use of saliva for medical diagnostics is well-established, extending to the detection of various pathologies. In infectious diseases, salivary samples can reveal systemic infections, including those caused by pathogens like EBV, HCV, HIV, HPV, HSV, rabies virus and norovirus. Saliva emerged as a potential alternative for diagnosing SARS-CoV-2, supported by studies during the pandemic: this choice is grounded in the physiological rationale that the target of SARS-CoV-2, the angiotensin II converting enzyme (ACE-2), is expressed in the oral mucosa and salivary glands [
1].
Scientific publications throughout the pandemic have sought to validate saliva as a suitable sample for diagnosing SARS-CoV-2 infections. Early studies focused on clinically evident cases reported high viral presence in salivary samples [
2], while subsequent research explored the feasibility of salivary analysis as a screening tool for pauci- or asymptomatic patients, yielding varying results [
3].
Recent meta-analyses reveal a sensitivity range of 83% to 87% and a specificity of 99% for salivary samples [
4,
5]. Our results align with this data, demonstrating a sensitivity of 89.7% and a specificity of 98.7%. Notably, our study encompasses the collection of salivary samples across Italy's three main pandemic waves, unaffected by viral variants like Delta.
The protocol's advantages include patient comfort, as it eliminates the need to abstain from food, drink, or smoking. It allows for easy, autonomous, and risk-free sample collection directly by the patient in sterile containers without the use of VTM. This simplicity contrasts with nasopharyngeal swabs, burdened by logistical challenges, trained personnel requirements, and higher consumption of personal protective equipment.
Saliva's characteristics make patients more compliant, expedite procedures, and reduce the need for large collection centers, especially in resource-limited settings. The absence of VTM lowers costs, easing emergency container supply. These features contribute to reduced economic impact on healthcare organizations, allowing focused investments in direct patient care.
While our study highlights the potential of saliva-based testing, it acknowledges limitations, including a small sample size, limited tracking of symptom evolution, lack of understanding regarding the effect of sample storage on virus identification, absence of test repetition in case of discordant results and enrollment of only adults.
Conclusions
This approach offers reliability and comparability with the current gold standard for diagnosing SARS-CoV-2 infections, making it a valuable tool in both emergency and endemic conditions.
Author Contributions
Conceptualization: GN and MM. Methodology: DPB and MCM. Validation: DPB. Formal analysis: DPB. Investigation: GN, MM, LAP, SMIM and MR. Resources: GN, CMAC and DS. Data curation: MM and SMIM. Writing—original draft preparation: GN and MM. Writing—review and editing: GN, MCM and MM. Visualization: GN, MM and LAP. Supervision: GN, MCM, DS and MM. Project administration: GN and MM. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the University of Milano Bicocca (Funds 2016-CONT-0449). Financial support was provided only for the purchase of the reagents needed for the described analyses. Apart from this, the funding source had no role in: study design; collection, management, analysis, and interpretation of data; writing of the report; and the decision to submit the report for publication.
Availability of data
The data supporting the findings of this study are available from the corresponding author, upon reasonable request.
Acknowledgments
The authors acknowledge Serena Citriniti and the nursing personnel working at the COVID Test Point of the ASST Monza - San Gerardo Hospital for their precious contribution.
Conflicts of Interest
All authors – none to declare.
References
- Xu, H.; Zhong, L.; Deng, J.; et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Azzi, L.; Carcano, G.; Gianfagna, F.; et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, K.A.F.; Nonaka, C.K.V.; de Ávila Mendonça, R.N.; et al. SARS-CoV-2 detection via RT-PCR in matched saliva and nasopharyngeal samples reveals high concordance in different commercial assays. Diagnostics 2023, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Duncan, D.B.; Mackett, K.; Ali, M.U.; Yamamura, D.; Balion, C. Performance of saliva compared with nasopharyngeal swab for diagnosis of COVID-19 by NAAT in crosssectional studies: systematic review and meta-analysis. Clin Biochem. 2023, 117, 84–93. [Google Scholar] [CrossRef] [PubMed]
Table 1.
Results: concordance between salivary and nasopharyngeal samples.
Table 1.
Results: concordance between salivary and nasopharyngeal samples.
Table 2.
Diagnostic performance of the salivary test.
Table 2.
Diagnostic performance of the salivary test.