Abstract
Introduction: This study aimed to examine the effect of commonly used non-antibiotic drugs (dexamethasone and tenoxicam), on treatment of Pseudomonas aeruginosa infections, antibiotic resistance and virulence in this pathogen. Methods: Four antibiotics (gentamicin, cefepime, ciprofloxacin and meropenem) were investigated. The proteolysis and hemolysis were selected as virulence factors for investigation. In this work, we selected the following final concentrations: dexamethasone (0.0052 µg/mL) and tenoxicam (2.7 µg/mL) to be used in combination with antibiotics or alone for investigation of their effects on antibiotic resistance and virulence in P. aeruginosa isolates. Results: The drugs either increased or decreased antibiotic resistance in only 0-3 isolates, which indicates that the investigated drugs did not significantly affect the antibiotic resistance. Interestingly, our study demonstrated that both dexamethasone and tenoxicam increased the hemolytic activity of the investigated isolates. On the other hand, our results indicated that no overall final increasing or decreasing effect could be observed for dexamethasone on the proteolytic activity, while tenoxicam increased the proteolytic activity of the investigated isolates. Interestingly, by real-time PCR dexamethasone has shown significant down-regulation of virulence genes namely algD, plcH and toxA, apparently, in case of combination with ciprofloxacin and with gentamicin in one isolate. However, a negative influence was observed in another isolate. Unfortunately, in the case of tenoxicam the only positive effect was observed in the combination with gentamicin in one isolate. Conclusions: Resistance of P. aeruginosa against gentamicin and ciprofloxacin may be affected by combining these antibiotics with dexamethasone or tenoxicam.
Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic bacterium, particularly affecting immunocompromised patients and capable of causing many life-threatening infections.[1] This infection is usually associated with high mortality and morbidity owing to increasingly frequent infections caused by multidrug resistant (MDR) strains with limited therapeutic options.[2] Intrinsic resistance is conferred by production of antibiotic inactivating enzymes, efflux pumps expression and low outer membrane permeability. Acquired resistance may be related to mutational changes or resistance genes acquisition through horizontal transfer by mobile genetic elements, such as plasmids, transposons or integrons.[3] For these reasons, since 2017, the World Health Organization has listed P. aeruginosa as one of the critical priority pathogens to encourage development and research of new anti-bacterials.[4]
For the treatment of infectious diseases which associate an extensive and strong inflammatory process, the combined use of corticosteroids and antimicrobials is commonly adopted by prescribers.[5] Dexamethasone is a synthetic glucocorticoid widely used in infectious diseases treatment, in order to modulate immune responses triggered by microbial varied toxins, lipopolysaccharide and extracellular DNA.[6] Beyond this, dexamethasone also has the ability to penetrate the central nervous system, being used in the treatment of encephalitis and bacterial meningitis.[6]
Similarly, tenoxicam is part of the class of nonsteroidal anti-inflammatory drugs (NSAIDs), which have been reported to exhibit a certain degree of antimicrobial activity with growth inhibition of S. aureus and P. aeruginosa.[7,8] Recently, synergistic drug screening and drug repurposing have become promising ways to combat infections caused by MDR pathogens.[9] For the above observations, this work aimed to evaluate the impact of combination of dexamethasone and tenoxicam with different antimicrobial drugs (gentamicin, ciprofloxacin, cefepime and meropenem) and whether this combination in vitro improves or abrogates the activity of these antimicrobial drugs against P. aeruginosa strains.
Methods
Collection, isolation, and identification of P. aeruginosa clinical isolates
Sixty-five clinical isolates of P. aeruginosa (P165) were obtained, during 2017-2018, from Mansoura International Hospital, University Hospital, Burns and Cosmetics Centre, Chest Hospital, Urology and Nephrology Centre and Paediatric University Hospital, Mansoura, Egypt. All isolates were identified as P. aeruginosa, based mainly on their growth on cetrimide agar plate (0.03% cetrimide), oxidase test, motility test, pyocyanin pigment production, in addition to the ability of the isolates to grow at 42°C. A reference strain PAO1, obtained from the MIRCENS center (Faculty of Agriculture, Ain Shams University, Egypt), was included as a positive control in all biochemical tests. After the identification and purification of isolates, they were cultured on nutrient broth and stored in 20% v/v glycerol stock at -80°C until further use. The experimental protocol designed in this study complies with the ethical guidelines adopted by the Ethics Committee in the Faculty of Pharmacy, Mansoura University, which approved this study.
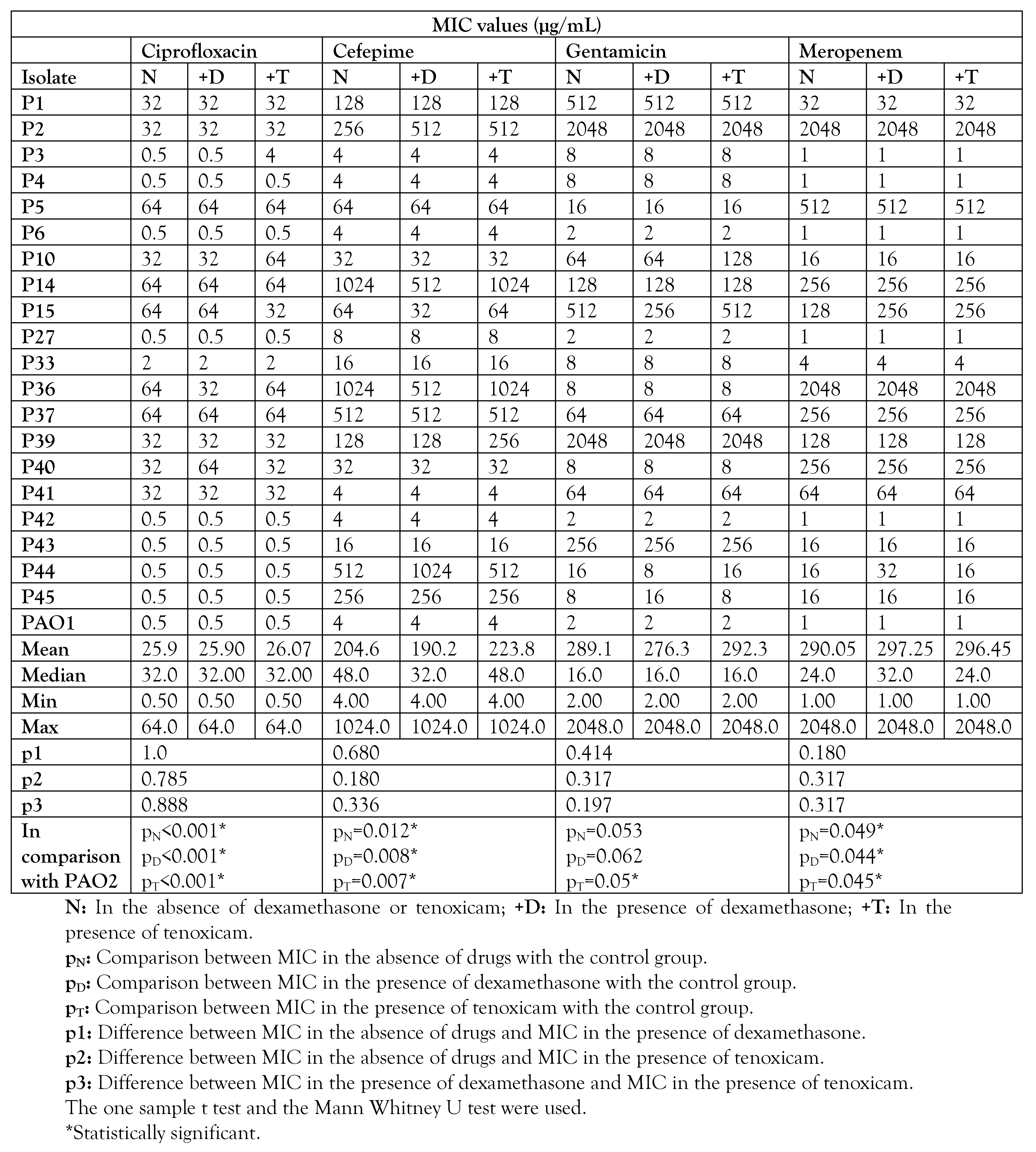
MIC for different antibiotics in the presence or absence of dexamethasone and tenoxicam
MICs of the antibiotics against all P. aeruginosa isolates and the standard PAO1 strain were determined by the broth microdilution method (CLSI, 2016). Overnight culture of isolated colonies in Muller-Hinton broth (MHB, Oxoid, UK) was prepared and the turbidity was adjusted to 0.5 McFarland. The prepared suspension was diluted with MHB to have an approximate cell density of 5×106 CFU/mL then 50 µL of the prepared suspensions were transferred to each well of a microtiter plate containing 50 µL of two-fold serial dilutions of each antibiotic in MHB. The final starting concentration of each antibiotic was 2048, 4096, 512, and 1024 µg/mL for gentamicin, cefepime, ciprofloxacin and meropenem respectively, while the final concentration of each antibiotic in the last dilution for each antibiotic was 2, 4, 0.5, and 1 µg/mL for gentamicin, cefepime, ciprofloxacin and meropenem respectively. The plates were incubated at 37°C for 24 hours and the MIC was determined as the lowest concentration of antibiotic that inhibited any visible growth. Each experiment was performed as triplicate wells in each plate. Positive control wells comprised the culture of each isolate in MHB while negative control wells harbored 100 µL MHB.
In this study, only 20 P. aeruginosa isolates were selected for evaluating the effect of dexamethasone and tenoxicam on antibiotic sensitivity and bacterial virulence factors. The selection criterion was that the isolates showed high resistance rates (resistant to 3/4 of the antibiotics). Either dexamethasone (0.0052 µg/mL) or tenoxicam (2.7 µg/mL) were included in the previous experiment for broth microdilution together with each antibiotic. Each experiment was performed as triplicate wells in each plate. In addition to positive control wells (culture of each isolate in MHB) and negative control wells (MHB medium only), another control of either dexamethasone or tenoxicam together with each bacterial isolate was also included to make sure that these drugs did not affect bacterial growth. The plates were incubated at 37°C for 24 hours, then the MIC was determined as the lowest concentration of antibiotic that inhibited any visible growth in presence or absence of these drugs.
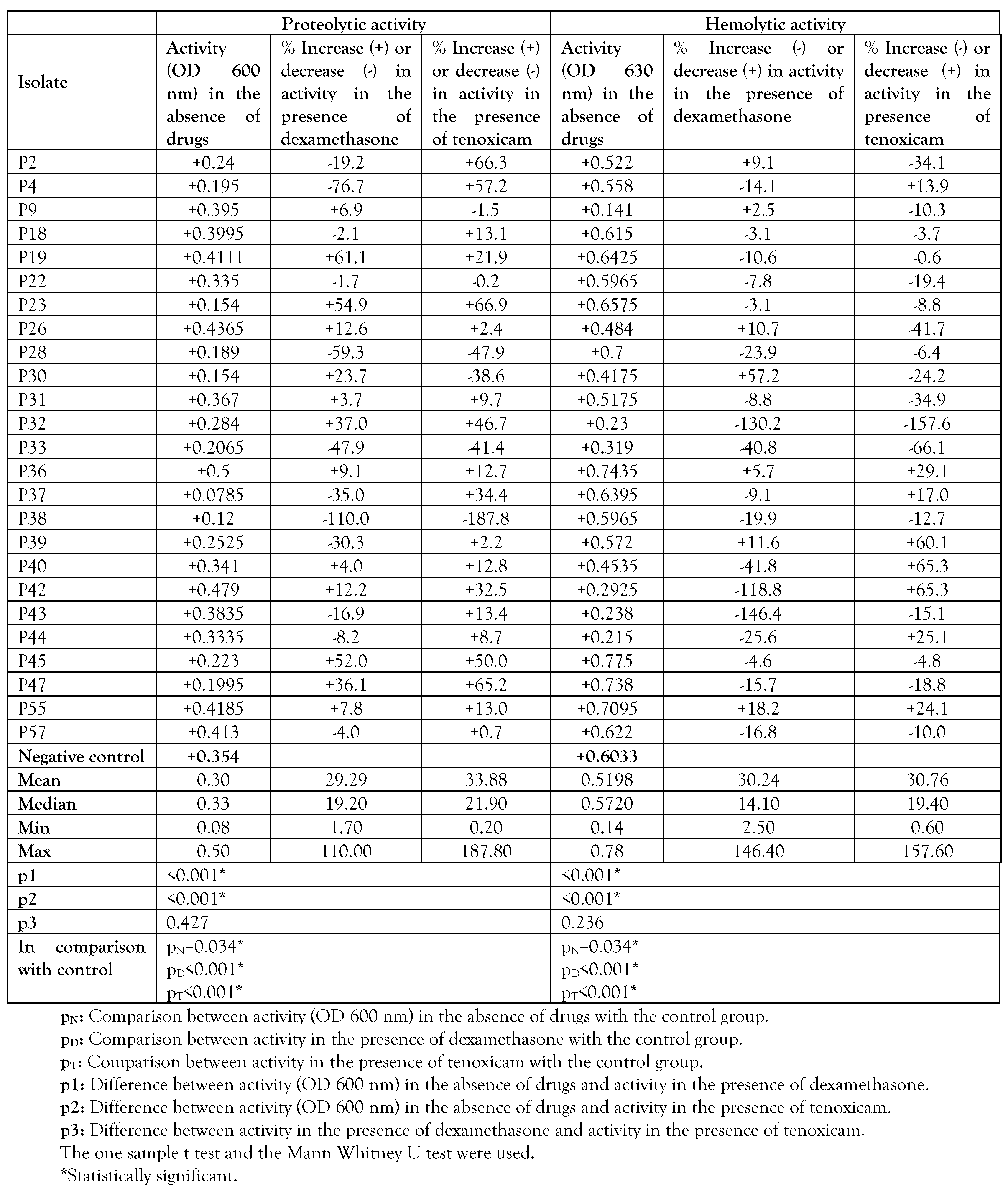
Phenotypic impact of dexamethasone and tenoxicam on certain P. aeruginosa virulence factors
To evaluate the effect of dexamethasone and tenoxicam on the protease activity of P. aeruginosa isolates, the skim milk agar method was prepared as described previously.[10] The turbidity of overnight cultures for each isolate in MHB was adjusted to match that of 0.5 McFarland. The turbidity-adjusted cultures were centrifuged and the supernatant of each bacterial isolate (20 µL) was incubated with skim milk (1.25%, 100 µL) and 1.5 µL of either dexamethasone (final concentration 0.0052 µg) or tenoxicam (final concentration 2.7µg/mL) in Eppendorf tube at 37°C for 15 min. The degree of clearance of skim milk indicated the protease activity and it was calculated by measuring the turbidity at OD 600 nm. The value was taken by the average of two readings for each isolate and compared to the negative control tube without drug.
In the hemolysin test, the turbidity of overnight cultures for each isolate in MHB was adjusted to match that of 0.5 McFarland. The turbidity-adjusted cultures were centrifuged and the supernatant of each bacterial isolate (75 µL) was incubated with 75 µL fresh sheep red blood cells (RBCs) in the wells of microtiter plate for 2 hours at 37°C. Before incubation of the plate, 1.5 µL of either dexamethasone (0.0052 µg/mL) or tenoxicam (2.7 µg/mL) were added to the reaction mixture for each isolate. Positive control wells were composed of 75 µL SDS (0.2% in MHB) + 75 µL fresh sheep RBCs, while negative control ones comprised 75 µL MHB +75 µL fresh sheep RBCs. Each experiment was performed as three readings in each plate. To ensure that the drugs did not exhibit any hemolytic activity, 75 µL of MHB containing each drug (at the same final concentrations) were added to 75 µL fresh sheep RBCs then inoculated at 37°C for 2 hours. In all experiments, hemolytic activity was determined by measuring hemoglobin release at OD 630 nm after incubation of the plates.[11]
Detection of virulence genes using polymerase chain reaction
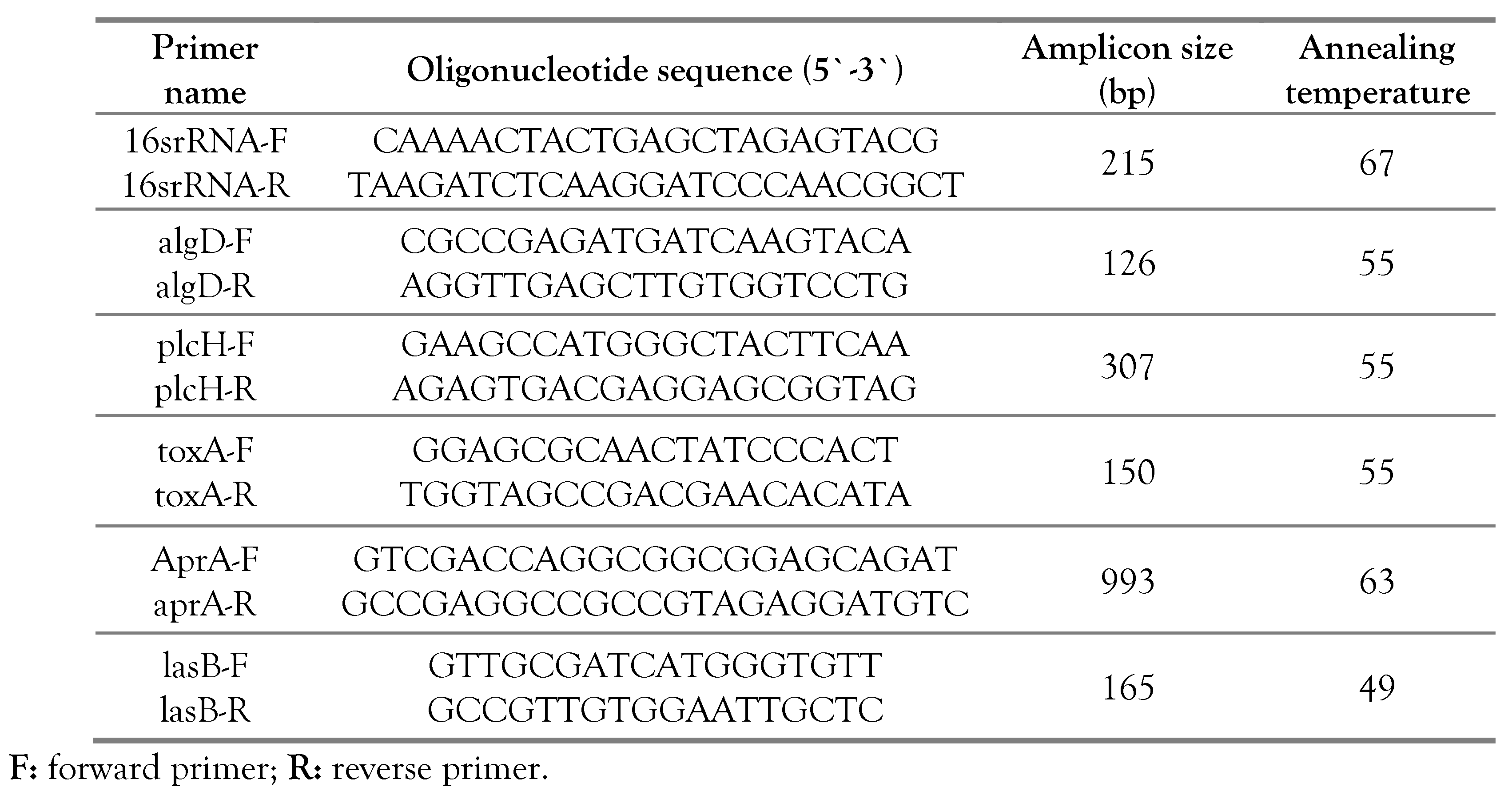
PCR amplification of P. aeruginosa genes, namely algD, plcH, toxA, aprA and lasB was performed using the following reaction mixture: 12.5 µL Dream Taq Green PCR Master Mix (2X), 1 µL of forward primer (10 µM), 1 µL of reverse primer (10 µM), 1 µL of bacterial DNA template and 9.5 µL of nuclease free water were added for a total of 25 µL per reaction. Negative control reaction (without bacterial DNA templates) was also performed. The cycling conditions included: initial denaturing at 95°C for 5 min, followed by denaturation at 95°C for 30 sec, annealing for 30 sec (Table 1) and extension at 72°C for 1 min) for 40 cycles, and a final extension cycle at 72°C for 5 min.

Table 1.
Amplification primers used in this study for amplifying different genes among Pseudomonas aeruginosa isolates.
Determination of bacterial growth curve:
In order to determine the most suitable conditions for RNA isolation, a bacterial growth curve was first established using three different Pseudomonas species (stain 39, stain 41, strain 42), in addition to the standard PAO. The maximum growth rate was fitted with the exponential stage (4-6 hours) after incubation at 37°C.
Preparation of inocula:
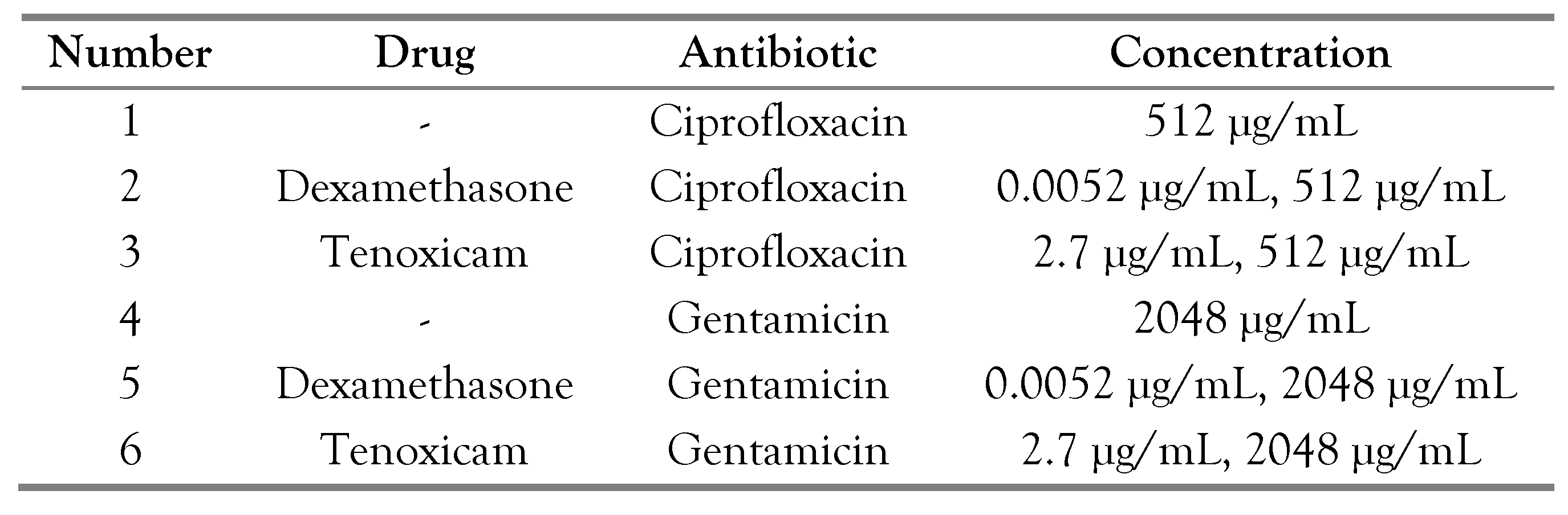
Pure colonies of three bacterial isolates were inoculated into 3 mL broth and left for growth overnight at 37°C with shaking. About 100 µL of each overnight broth culture were sub-cultured into 5 mL of broth containing the calculated amount of antibiotic (gentamicin and ciprofloxacin), in addition to the calculated amount of dexamethasone and tenoxicam, as presented in Table 2, followed by incubation at 37°C for 4-5 hours with shaking at 110-115 rpm). Bacterial cells were harvested in the middle of exponential phase OD 630 nm (0.1-0.2).

Table 2.
Different drugs and antimicrobial combinations tested in this study.
Similarly, P. aeruginosa PAO was grown under the same conditions then cell pellets were collected by cooling centrifugation at 5000 rpm and the supernatant was decanted before storing at -80°C until use for RNA extraction.
Bacteria total RNA purification and cDNA synthesis:
RNA extraction was performed by using a GeneJET RNA Purification Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. Purified RNAs were frozen at −80°C. For further analysis, the concentration of purified RNA was determined using a NanoDrop instrument (OPTIZEN NanoQ, Mecasys,
Republic of Korea). First-strand cDNA synthesis was performed using a Revert Aid H Minus FirstStrand cDNA Synthesis Kit (Thermo Scientific), according to the manufacturer’s instructions. The synthesized first-strand cDNAs were stored at −80°C until use.
Quantitative polymerase chain reaction
The extracted cDNA was subjected to real time PCR reaction in a reaction mix containing 1.5 μL of each forward and reverse primer (10 μm each), 12.5 μL (2×) SYBR Green PCR master mix (Willowfort Co., UK) and nuclease-free H2O were added to complete the final volume of 25 μL. All real-time reactions were carried out using
MyGo real-time PCR machine under the following conditions: 95°C for 5 min, followed by 45 cycles of 95°C for 20 sec, annealing temperature for 20 sec, and finally 72°C for 40 sec. Ct values and melting curve detection were obtained using the MyGo real-time PCR machine software. The relative abundance of each microbial species was calculated as a relative unit normalized to the total bacteria of the corresponding sample, using the 2-ΔCt method (where ΔCt = the average Ct value of each target - the average Ct value of total bacteria).[12] Different strains in addition to the PAO1 standard strain were selected in this study. The strains were tested under 6 different conditions as presented in Table 2.
Statistical analysis and data interpretation:
Data analysis was performed with the SPSS software, version 25 (SPSS Inc., USA). Qualitative data are presented using numbers and percentages. Quantitative data are presented using the median (minimum and maximum) for non-normally distributed data and the mean ± standard deviation (SD) for normally distributed data after testing for normality using the Shapiro Wilk test. Significance of the obtained results was judged at the (≤0.05) level. The Mann Whitney U test was used to compare non-normally distributed data between 2 studied groups. The student t test was used to compare normally distributed data between 2 independent groups. The one sample t test was used to compare values of each reading with the standard value.
Results
Effect on antibiotic susceptibility of P. aeruginosa clinical isolates
Using different antibiotic-drug combinations against resistant P. aeruginosa isolates, dexamethasone and tenoxicam showed different changes in antimicrobial activity. As shown in Table 3, neither dexamethasone nor tenoxicam significantly increased the sensitivity to antibiotics. In terms of MIC, the significant effect was observed in only 0 to 3 isolates of the 20 isolates, which indicates that the investigated drugs did not significantly affect the MICs of the antibiotics.

Table 3.
MIC values of different antibiotics in the presence or absence of dexamethasone or tenoxicam.
Phenotypic effect of dexamethasone and tenoxicam on certain virulence factors of P. aeruginosa
Although it did not significantly affect the MICs of antibiotics, dexamethasone was found to decrease the proteolytic activity in 12 isolates of the investigated strains, and to increase it in 13 isolates. Also, dexamethasone was found to decrease the hemolytic activity in 7 isolates, and to increase it in 18 isolates. Similarly, tenoxicam was found to decrease the proteolytic activity in 6 of the investigated isolates, and to increase it in 19 isolates. Also, tenoxicam was found to decrease the hemolytic activity in 8 isolates and to increase it in 17 isolates (Table 4).

Table 4.
Impact of dexamethasone or tenoxicam on proteolytic and hemolytic activity.
Molecular identification of certain virulence factors
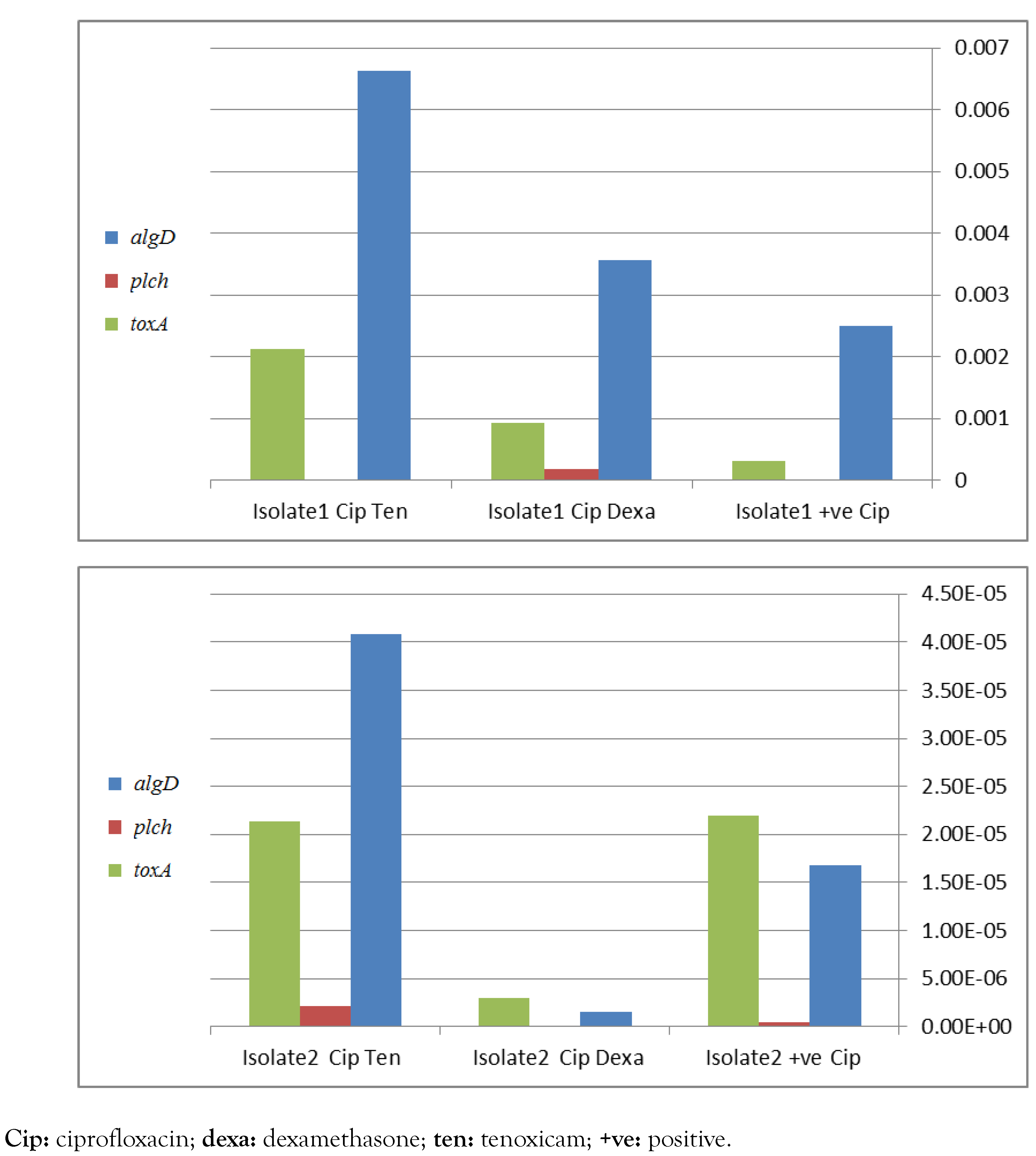
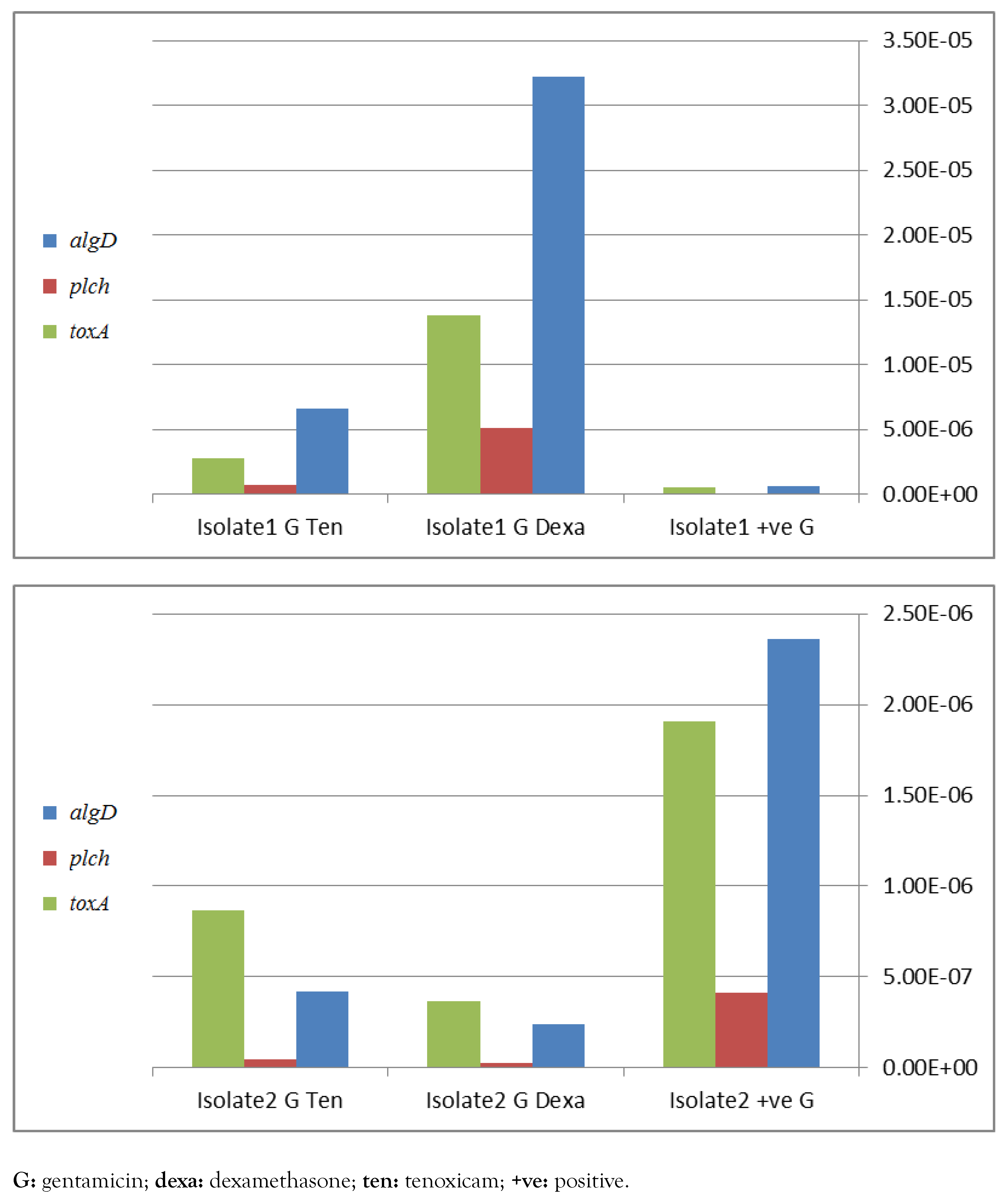
Detection of algD, plcH, toxA and lasB genes by PCR is recommended for molecular identification of P. aeruginosa. These genes were detected at different percentages in the selected 20 isolates in 88%, 94%, 93.7%, 82% and 93.7% in case of algD, plcH, toxA, aprA and lasB, respectively. Real time PCR has shown a significant down-regulation of virulence genes, namely algD, plcH and toxA, apparently in case of combination of dexamethasone with ciprofloxacin (Figure 1) and with gentamicin (Figure 2) in isolate 2. In contrast, in isolate 1, a negative influence was detected. However, the only positive effect in tenoxicam was observed in combination with gentamicin for isolate 2.
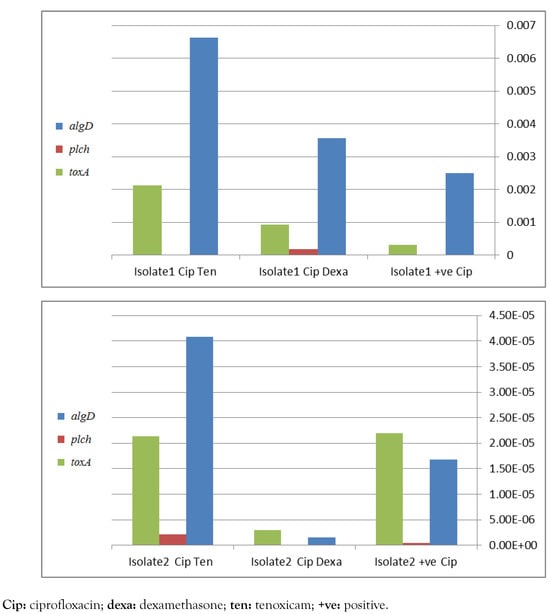
Figure 1.
Effect of different drugs in addition to ciprofloxacin on P. aeruginosa virulence factors genes toxA, plcH and algD, assessed by real-time PCR for 2 different isolates 1 and 2, figures a and b respectively.
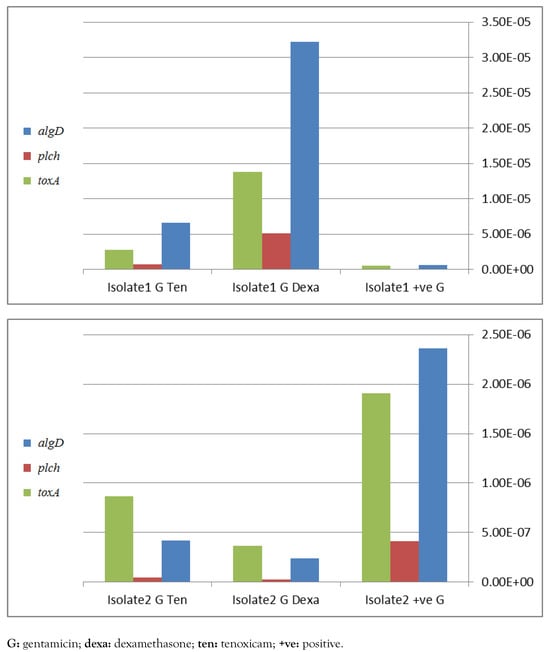
Figure 2.
Effect of different drugs in addition to gentamicin on P. aeruginosa virulence factors genes toxA, plcH and algD, assessed by real-time PCR for 2 different strains 1 and 2, figures a and b respectively.
Discussion
By combining with non-antibiotics, antibiotics can re-become active against MDR bacteria.[13] In this study, we were interested to examine the effect of commonly used nonantibiotic drugs, dexamethasone and tenoxicam, on infections treatment, antibiotic resistance and virulence factors of P. aeruginosa. The investigated antibiotics were gentamicin, cefepime, ciprofloxacin and meropenem, while proteolysis and hemolysis were selected as virulence factors for investigation.
In this work we selected the following final concentrations 0.0052 µg/mL for dexamethasone and 2.7 µg/mL for tenoxicam. These concentrations were peaks of the plasma active metabolite hydroxyl.[14,15]
Although dexamethasone has not been frequently recommended for clinical antiinfection therapy, previous studies have shown that this glucocorticoid can enhance the activity of antimicrobial drugs against different microbes including P. aeruginosa.[6] Results of these studies showed that dexamethasone potentiates drug antimicrobial effect against resistant bacteria, which is related to inhibiting the virulence of the bacteria.[6]
However, the exact mechanism is uncertain, as several studies have suggested that NSAIDs including tenoxicam exhibit some antibacterial activity.[8] Some synergistic effects were reported when resistant antibiotics were combined with the NSAID tenoxicam.[8]
Unexpectedly, the impact of drugs on antibiotic resistance did not show sound results as only few isolates (0-3 isolates) were affected in each combination, which indicates that the investigated drugs did not significantly affect the antibiotic resistance when used in combinations. Previous reports demonstrated the effect of drugs on antibiotic resistance of P. aeruginosa.[6,8]
Interestingly, dexamethasone decreased the proteolytic activity in 12 isolates of the investigated strains by 5-100%. On the other hand, it increased the proteolytic activity in 13 isolates of the investigated strains by 5-100%. Regarding tenoxicam, it was found to decrease the proteolytic activity in 6 isolates of the investigated strains by 5-100%. On the other hand, tenoxicam increased the proteolytic activity in 19 isolates of the investigated strains by 5100%. These results indicated that no overall final increasing or decreasing effect could be observed regarding the effect of these drugs on the proteolytic activity of the investigated isolates.
Concerning the effect of drugs on hemolytic activity in the investigated strains, dexamethasone decreased the hemolytic activity in 7 isolates of the investigated strains by 5-100%. On the other hand, dexamethasone increased the hemolytic activity in 18 isolates of the investigated strains by 5-100%. Regarding tenoxicam, it was found to decrease the hemolytic activity in 8 isolates of the investigated strains by 10-100%. On the other hand, tenoxicam increased the hemolytic activity in 17 isolates of the investigated strains by 5-00%.
These results demonstrate that either dexamethasone or tenoxicam increased the hemolytic activity of the investigated isolates. Similarly, RNA based studies have shown apparent reduced expression of virulence genes namely algD, plcH and toxA and lasB, in case of combination of dexamethasone with ciprofloxacin or with gentamicin in isolate 2, compared to isolate 1. Other studies demonstrated the effect of drugs on virulence factors of P. aeruginosa. In a previous study,[6] impairment of antimicrobial activity was detected when combined with dexamethasone. However a significant role of tenoxicam was observed with reduced quorum sensing virulence activity.[8] This study was limited by its preliminary nature, by the fact that only in vitro studies were performed, and by limited resources (no funding), but these findings pave the way for further in vivo pharmacological funded studies, which can lead to clinical trials of the use of glucocorticoids and
NSAIDs in combination with traditional antibiotics for treatment of P. aeruginosa infection.
Conclusions
The impact of anti-inflammatory drugs on antibiotic resistance did not show consistent results. Variable results were obtained when dexamethasone and tenoxicam when combined with antibiotics. We recommend future studies to be done to study the effect of other nonantibiotic drugs on antibiotic resistance and virulence of P. aeruginosa.
Author Contributions
Supervision: EEH and MMAE. Study planning: MMAE. Methodology: MMAE and HEA. Formal analysis: MMAE. Resources: MMAE. Software: MMAE. Data analysis: EEH, MMAE and HEA. Writing of manuscript: HEA, EEH and MMAE. Validation: EEH and MMAE. Reviewing and editing of manuscript: EEH and MMAE. All authors read and approved the final version of the manuscript.
Funding
None to declare.
Data Availability Statement
Data supporting the results are available from the corresponding author upon reasonable request.
Conflicts of Interest
All authors – none to declare.
Ethics Approval and Consent to Participate
The study has been approved by the Research Ethics Committee, Faculty of Pharmacy, Mansoura University. The experimental protocol conducted in this study complies with the ethical guidelines adopted by the Code of Ethics of the World Medical Association (the 1975 Declaration of Helsinki with subsequent revisions).
References
- Esposito, F.; Cardoso, B.; Fontana, H.; et al. Genomic analysis of carbapenem-resistant Pseudomonas aeruginosa isolated from urban rivers confirms spread of clone sequence type 277 carrying broad resistome and virulome beyond the hospital. Front. Microbiol. 2021, 12, 701921. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.A.; et al. Genetic diversity of multidrug-resistant Pseudomonas aeruginosa isolates carrying blaVIM-2 and blaKPC-2 genes that spread on different genetic environment in Colombia. Front. Microbiol. 2021, 12, 663020. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.M.; Faustino, A.; Pastrana, L.M.; Bañobre-López, M.; Sillankorva, S. Pseudomonas aeruginosa PAO 1 in vitro time-kill kinetics using single phages and phage formulations-modulating death, adaptation, and resistance. Antibiotics 2021, 10, 877. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Semino, M.; Picciolli, I.; Principi, N. Should corticosteroids be used in bacterial meningitis in children? Eur. J. Paediatr. Neurol. 2013, 17, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.; Gomes, A.; Marçal, P.H.F.; Dias-Souza, M.V. Dexamethasone abrogates the antimicrobial and antibiofilm activities of different drugs against clinical isolates of Staphylococcus aureus and Pseudomonas aeruginosa. J. Adv. Res. 2017, 8, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.R.; Pruthvishree, B.S.; Yadav, A.; Ravichandran, K.; Vinodhkumar, O.R.; Sinha, D.K. Comparative antimicrobial activity of aspirin, paracetamol, flunixin meglumine, tolfenamic acid, diclofenac sodium and pheniramine maleate. Acta Sci. Vet. Sci. 2021, 3, 30–42. [Google Scholar]
- Askoura, M.; Saleh, M.; Abbas, H. An innovative role for tenoxicam as a quorum sensing inhibitor in Pseudomonas aeruginosa. Arch. Microbiol. 2020, 202, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tong, Z.; Shi, J.; Li, R.; Upton, M.; Wang, Z. Drug repurposing for next-generation combination therapies against multidrug-resistant bacteria. Theranostics. 2021, 11, 4910–4928. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; et al. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, G.; Merieau, A.; Guerillon, J.; et al. Involvement of a phospholipase C in the hemolytic activity of a clinical strain of Pseudomonas fluorescens. BMC Microbiol. 2008, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Kwong, T.N.Y.; Chow, T.C.; et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017, 66, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.; Ijaz, M.; Ghaffar, A.; Rehman, A.; Avais, M. Antibiotic susceptibility profile and synergistic effect of non-steroidal anti-inflammatory drugs on antibacterial activity of resistant antibiotics (oxytetracycline and gentamicin) against methicillin resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2019, 137, 103755. [Google Scholar] [CrossRef] [PubMed]
- Weijtens, O.; Schoemaker, R.C.; Cohen, A.F.; et al. Dexamethasone concentration in vitreous and serum after oral administration. Am. J. Ophthalmol. 1998, 125, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, O.G. Clinical pharmacokinetics of tenoxicam. Clin. Pharmacokinet. 1998, 26, 16–43. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2023.