Abstract
Introduction: Listeria monocytogenes is one of the most pathogenic bacteria related to the consumption of contaminated food. This study aims to determine the prevalence of L. monocytogenes in raw beef meat in Meknes city of Morocco, to evaluate its pathogenicity and resistance to antimicrobials. Methods: During four seasons, a total of 140 samples were collected from supermarkets, butcheries and Souk (weekly traditional market). The PCR method was used to examine the presence of specific and virulence genes in the isolated strains, and also to identify their serotypes. The antimicrobial resistance was determined. Results: The results show a prevalence of 7.14% which depends on retail sites and also on the season’s variation. The majority of the strains were detected in butcheries (6 strains), and supermarkets (4 strains). Moreover, the majority of strains were detected during summer (50%). Concerning virulence genes, the seven researched genes were detected in 100% of isolated strains. The majority of strains were of the (1/2a, 1/2c, 3a and 3c) serogroup (70%), while two of them were of the (1/2b, 3b, 4b and 4d) serogroup (20%). All isolates were resistant to at least one antimicrobial, while three strains were resistant to nine tested antimicrobials. However, they were highly susceptible to amikacin, imipenem, gentamicin, sulfamethoxazole and chloramphenicol. Conclusions: According to results, isolated L. monocytogenes from analyzed beef meat shows a high level of pathogenicity and resistance to the most used antimicrobials in listeriosis therapy, which calls for the severe application of quality systems at the slaughterhouses and retail sites level.
Introduction
Listeriosis is a serious infection around the world. It results from consumption of food contaminated with Listeria monocytogenes and is considered among the major causes of death related to foodborne illnesses worldwide [1]. According to records, the infection risk is higher for the immunocompromised, the pregnant women, the elderly and the newborns [2]. In Europe, the frequency of reported cases has increased from 1581 to 2480 cases during 2007-2017 [3]. In Morocco, despite the low frequency of detection and the lack of data and studies about this illness, pregnant women represent 1/3 of affected patients with listeriosis [4].
L. monocytogenes is a Gram positive, facultative anaerobic, non-spore-forming, motile at 20-25°C, coccobacillus, and catalase-positive pathogenic bacterium. The optimal temperature for growth of this bacterium is between 30-37°C. However, its danger increases with its ability to survive and grow at low temperatures (refrigeration temperatures) that can reach -2°C [5]. Three phylogenetic divisions of L. monocytogenes have been determined including 14 serotypes of which three (1/2a, 1/2b, 4b) are considered the most pathogenic on the human health and account globally for more than 90% of human infections related to germs worldwide [6].
Beef meat occupies a large part of meat consumption around the world and the first position as the most consumed red meat in Morocco [7]. Because of its richness in water and proteins that can enrich approximately 65% of the fresh meat’s weight, and its high rate of water activity (aw=0.99), fresh beef carcass can be contaminated by several types of pathogenic germs including L. monocytogenes during slaughter operations because of its natural presence in soil and water [8]. Deficiencies in respecting hygienic standards during the slaughter process increase the risk of contamination. In Morocco, the national office of food safety has declared the acute insufficiency of certified red meat’s slaughterhouses in the country [9]. Furthermore, an epidemiological study shows that the majority of listeriosis cases detected in Morocco were related to meat products consumption outside home [4]. In order to determine the risk of infection with L. monocytogenes related to beef consumption, this study aims to determine its contamination level with this germ, virulence genes, serogroups and also its multiresistance to antibiotics.
Methods
Sampling
A total of 140 samples of raw beef meat were collected during four seasons: from 20 December 2016 to 28 February 2017 (winter), from 1 June to 31 August 2017 (summer), from 1 October to 30 November (autumn) and from 1 March to 30 April 2018 (spring). The retail sites were distributed on three types: 40 from supermarkets, 60 from butcheries and 40 from Souk (weekly market). These retail sites were distributed geographically over twelve different regions on the city of Meknes, Morocco, which have a direct relation with the compliance level with hygienic conditions. The samples were kept at 4°C and analyzed after a maximum of 24 hours.
Isolation and detection of Listeria monocytogenes
The isolation of L. monocytogenes was carried out following the routine method for the Research of L. monocytogenes NF V08-055, 1997 (French standard, 1997) [10]. This method consists of a pre-enrichment of 25 g of raw beef meat into 225 mL of half Fraser (BIOKAR, Beauvais, France) with an incubation at 30°C during 24 h. Enrichment of 0.1 mL of pre-enrichment into 10 mL of Fraser (BIOKAR) with an incubation at 37°C during 48h. The enrichment cultures were streaked on PALCAM Agar (BIOKAR) and incubated at 37°C during 24 h to 48 h. After incubation, suspect colonies of L. monocytogenes (olive green with black center hollow and surrounded by black areas) were cultured on TSYEA agar (BIOKAR). Presumptive strains of L. monocytogenes were subjected to the biochemical identification tests: catalase, Gram stain, hemolysis, CAMP test, carbohydrate and API tests.
Detection of virulence genes and serogroups of Listeria monocytogenes
Following the conventional polymerase chain reaction (PCR) described by Bouymajane et al. [11]. The DNA extraction was performed using thermal lysis for a bacterial suspension of isolated strain in 50 μL of H2O that was subjected three times to 15 min of freezing at -20°C followed by thawing for 3 min at 94°C. The quantification of the extracted DNA was carried out using the Nanodrop 8000 (Thermo Fisher Scientific, Waltham, MA, USA), while the 1% agarose gel was used for its control. The isolates which presented a phenotypic character of L. monocytogenes were analyzed for the hly specific genes for this bacterium. After that, confirmed strains by PCR were tested for the virulence genes act A, hly A, inl J, inl A, inl C, plc B and prf A. The different serovars of L. monocytogenes strains were determined also by the PCR method using the Imo 0737 and ORF 2819 genes, corresponding to eight serotypes: 1/2a, 1/2c, 3a, 3c, 1/2b, 3b, 4b and 4d. In a PCR reaction mixture of 25 μL, the PCR mix of specific genes was composed of 13.9 μL of H2O, 2.5 μL of buffer (10×), 2 μL of 25 mM MgCl2, 2.5 μL of 1 μM dNTP mix (KAPA Biosystems, Wilmington, USA), 0.1 μL of Taq DNA polymerase (1 U/μL, KAPA Biosystems), 2 μL of template DNA and 1 μL of each 10 μM primer. Concerning the act A, hly A and inl J virulence genes, the PCR mix was composed of 8.71 μL of H2O, 2.5 μL of buffer (10×), 3 μL of 25 mM MgCl2, 2.5 μL of 1 μM dNTP mix (KAPA Biosystems), 0.3 μL of Taq DNA polymerase (1 U/μL, KAPA Biosystems), 2 μL of template DNA and 1 μL of each 10 μM primer. While for the Inl d, Inl C, plc B and prf A virulence genes and serotyping genes Imo 0737 and ORF 2819, the PCR mix was composed of 11.8 μL of H2O, 2.5 μL of buffer (10×), 2 μL of 25 mM MgCl2, 2.5 μL of 1 μM dNTP mix (KAPA Biosystems), 0.2 μL of Taq DNA polymerase (1 U/μL, KAPA Biosystems), 2 μL of template DNA and 1 μL of each 10 μM primer. Temperature profiles differed according to the researched gene (Table 1).

Table 1.
Specific, virulence and serovars genes of Listeria monocytogenes.
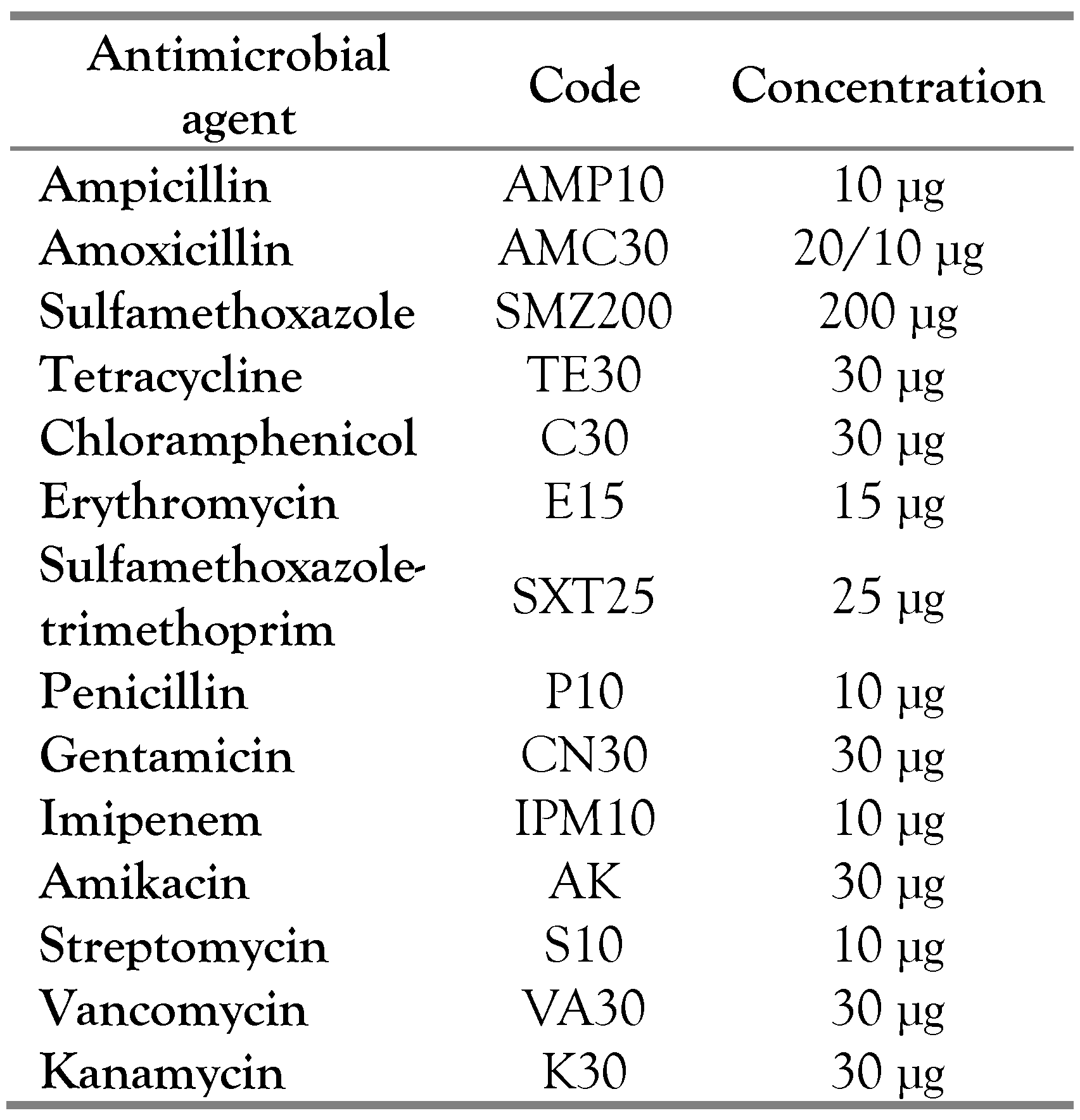
Antimicrobial susceptibility testing
In order to evaluate the level of sensitivity of L. monocytogenes isolates to antibiotics, the isolated strains were cultured on TSYE agar medium and tested for 14 different antimicrobials commonly used for human listeriosis therapy (Table 2). After incubation at 37°C during 48 h, the measurement of the inhibition zone allows to conclude the level of the bacterium’s resistance to the antimicrobial according to the CLSI guidelines [12].

Table 2.
Antimicrobial agents used for susceptibility testing of Listeria monocytogenes.
Results
Isolation and detection of L. monocytogenes
From 140 analyzed beef meat samples, 48 strains had the morphological aspect of L. monocytogenes, in which ten have been confirmed by PCR (Figure 1). L. monocytogenes was detected in 7.14% of analyzed samples (10/140). The L. monocytogenes strain ATCC 19112 was used as a reference.
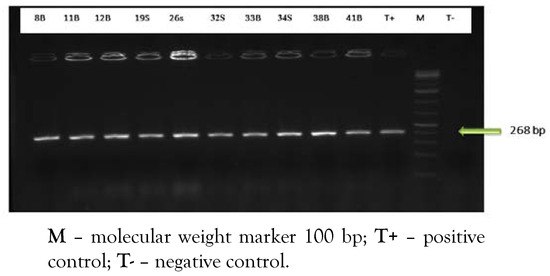
Figure 1.
PCR amplification of specific hly gene.
Beef meat retailed in butcheries was the more contaminated with this germ with 6/10 strains, followed by retail meat in supermarkets with 4/10 strains, while no strain of L. monocytogenes was isolated from Souk’s beef meat. The majority of isolated strains were detected during summer (5 strains) and winter (3 strains), while 2 strains were detected in the autumn season.
Detection of virulence genes and serogroups of L. monocytogenes
The results show that the seven researched virulence genes were detected for all isolated strains of L. monocytogenes (Figure 2).
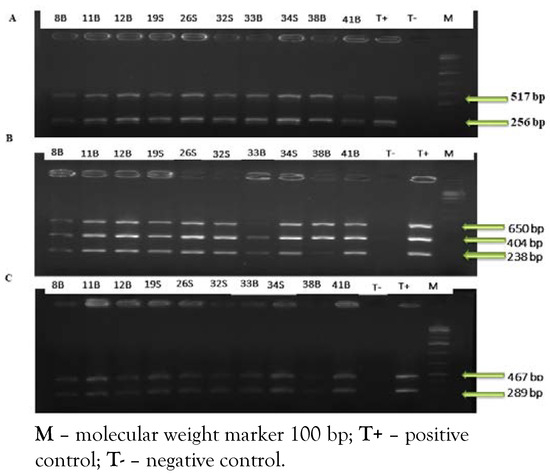
Figure 2.
A) PCR amplification of virulence inl A and inl C genes; B) PCR amplification of virulence act A, hly A and inl J genes. C) PCR amplification of virulence plc B, prf A genes.
Concerning serotyping, eight serotypes were detected in this study (Figure 3). According to results, seven strains were from the Imo 0737 serogroup (3 from supermarkets and 4 from butcheries) while two strains were from the ORF 2819 serogroup (1 from supermarket and 1 from butchery). The variation of retail sites has an effect on the distribution of serotypes of the studied strains.
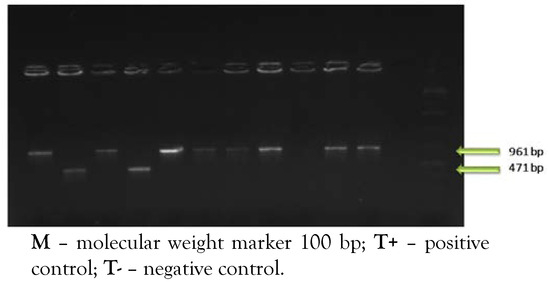
Figure 3.
PCR amplification of serovars genes of Listeria monocytogenes.
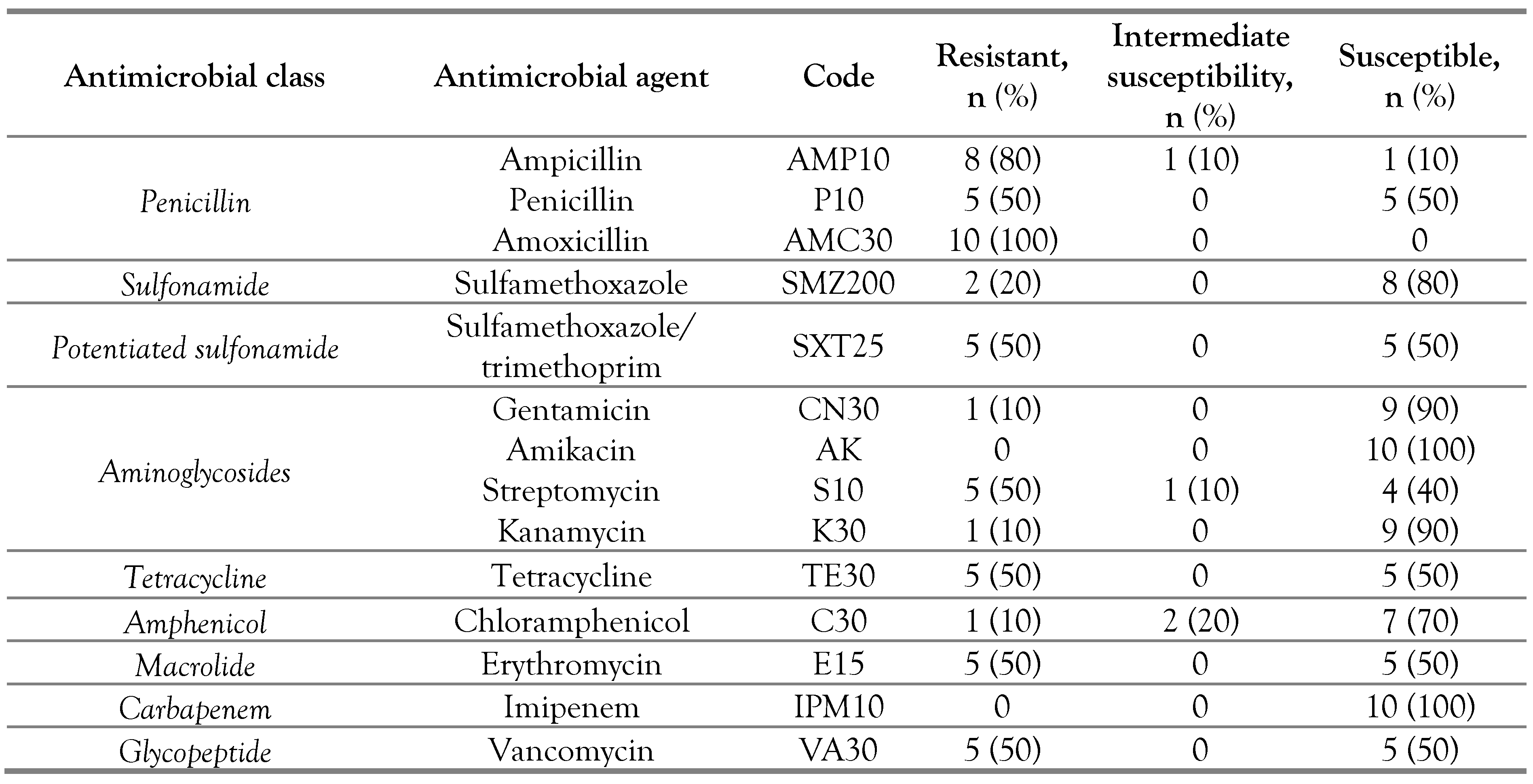
Antimicrobial susceptibility of isolated strains
The ten isolated strains of L. monocytogenes showed a resistance to at least one antimicrobial agent. Four strains (40%) were resistant to at least 2 antimicrobials while two strains (20%) were resistant to 7 antimicrobials; also 30% of L. monocytogenes showed a resistance to three or more antimicrobials (Table 3).

Table 3.
Antimicrobial susceptibility of Listeria monocytogenes isolated from beef meat.
Discussion
L. monocytogenes was detected in 7.14% of analyzed beef meat samples (10/140). This prevalence is higher than that found in Egypt (6.67%) [13], and lower than that found in Poland (19.4%) [14]. Our previous study showed that the prevalence of these bacteria was high during summer and winter seasons (8.8%) with an increase in summer (5 strains), which can be explained by the availability of the optimum temperature and humidity conditions for the growth of L. monocytogenes in the slaughterhouses and retail sites during this season [15].
Collected beef meat from butcheries and supermarkets were the most contaminated with L. monocytogenes (with 10% for each site), however any strain was detected from Souk’s beef meat. The high hygienic quality of Souk’s beef meat compared to that of the other retail sites was related with the storage duration of meat in butcheries and supermarkets in comparison with that of Souk which is selling during the same slaughter day [15]. Regarding virulence genes, the seven researched genes were detected in 100% of L. monocytogenes strains. Act A, prf A, hly A, plc B genes of the prfA virulence genes group (pVGC) that represents the most pathogenic genes group of L. monocytogenes were detected in this study, as well as the inl A, inl C and inl J virulence genes, which are considered among the most essential tools in the adhesion and invasion of L. monocytogenes [16]. In this study, the isolates predominantly belonged to the serogroup (1/2a, 1/2c, 3a and 3c) (70%), which includes one of the most pathogenic serotypes (1/2a), in addition to three others (1/2c, 3a, 3c), while two strains (20%) belonged to the serogroup (1/2b, 3b, 4b and 4d) including two of the most pathogenic serotypes (1/2b and 4b) with two others (3b and 4d). In order to determine the prevalence and different serotypes of L. monocytogenes in bovine hides and carcasses, a study performed in Poland indicates that 87% of isolated strains were of the 1/2a serotype [14]. Regarding the resistance of L. monocytogenes, all isolated strains were resistant to at least one tested antimicrobial (100%). The isolates showed a very high resistance to amoxicillin (100%) and ampicillin (80%). Moreover, they were moderately resistant to streptomycin (50%), penicillin (50%), erythromycin (50%), vancomycin (50%), tetracycline (50%) and sulfamethoxazole/trimethoprim (50%). Amoxicillin as an amino-penicillin, ampicillin and penicillin are among the beta-lactam antibiotics which have a high inhibitory effect on the Gram-positive bacterial cells in general including L. monocytogenes [17]. For this reason, they are frequently used in therapy for the infection with L. monocytogenes whereas the standard therapy is formed from amoxicillin or ampicillin in combination with an aminoglycoside like streptomycin [18].
However, isolated strains were very highly susceptible to imipenem (100%), amikacin (100%), gentamicin (90%), kanamycin (90%), sulfamethoxazole (80%) and chloramphenicol (70%). This result is compatible with a study that was conducted in Romania in which all isolates of L. monocytogenes from meat, dairy products and clinical samples were very susceptible for imipenem, amikacin, gentamicin and chloramphenicol [18]. In the present study as in many other recent studies, Listeria monocytogenes is becoming increasingly resistant to a wide number of antimicrobials including the beta-lactam antibiotics [18], which indicates that its danger on the human health is also becoming more and more preoccupying.
Conclusions
In this study, the prevalence of L. monocytogenes was 7.14%. This prevalence was moderately high compared to that found in many countries. From forty-eight suspect strains, just ten were confirmed by PCR technique, which indicates the importance of genetic identification tests to identify isolated strains. On the other hand, researched virulence genes in this study play a key role in the multiplication, propagation and pathogenicity of L. monocytogenes, while they were totally detected in all isolated strains. Furthermore, the isolates showed a resistance against the most used antimicrobials for human listeriosis therapy (amoxicillin, ampicillin, penicillin, streptomycin, erythromycin, tetracycline and vancomycin) hence the need to develop more effective antibiotics against this bacterium. These results call for the great necessity to the severe application of the quality systems at the slaughterhouses and retail sites level in Morocco, in order to fight against listeriosis.
Author Contributions
MB prepared the manuscript and conducted the analysis. FRF ensured the necessary equipments and edited the article. NL conducted the PCR amplification. AB prepared the PCR protocol. MS and MM revised and approved the article. All authors read and approved the final version of the manuscript.
Funding
None to declare.
Acknowledgments
All authors are acknowledged for their efforts and contributions to this work.
Conflicts of Interest
All authors – none to declare.
References
- Barbau-Piednoir, E.; Botteldoorn, N.; Yde, M.; Mahillon, J.; Roosens, N.H. Development and validation of qualitative SYBR®Green real-time PCR for detection and discrimination of Listeria spp. and Listeria monocytogenes. Appl Microbiol Biotechnol 2013, 97, 4021–4037. [Google Scholar] [CrossRef] [PubMed]
- Hof, H. History and epidemiology of listeriosis. FEMS Immunol Med Microbiol 2003, 35, 199–202. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC). Disease Data from ECDC Surveillance Atlas—Listeriosis. August 2018. Available online: https://ecdc.europa.eu/en/listeriosis/surveillance-and- disease-data/atlas (accessed on 1 September 2019).
- Benabdejlil, Y.; Chergi, M.; Kouach, J.; Massaoui, D.; Dehayni, M. Listeriosis in pregnant women in Morocco : A case report. J Gynecol Obstet 2015, 3, 18–20. [Google Scholar] [CrossRef]
- National Agency for Food Safety, Food, Environment, Work (anses). Listeria monocytogenes. 2011. Available online: https://www.pasteur.fr/fr/file/3252/download?token=nuSFUB9S (accessed on 1 September 2019).
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect 2007, 9, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Breeding Institute (idele) & National Interprofessional Association for Livestock and Meat (Interbev). The Beef Sector in Morocco, How to Reconcile Growth and Self-Sufficiency? Ministry of Agriculture, Agri-Food and Forestry: France, 2014. [Google Scholar]
- Fox, E.; Hunt, K.; O'Brien, M.; Jordan, K. Listeria monocytogenes in Irish farmhouse cheese processing environments. Int J Food Microbiol 2011, 145 (Suppl 1), S39–S45. [Google Scholar] [CrossRef] [PubMed]
- National Office of Food Safety (ONSSA). List of Establishments for the Preparation of Approved Meat and Meat Products; ONSSA: Morocco, 2018. [Google Scholar]
- French Standard (NF V08-055); Microbiology of Food—Research of Listeria monocytogenes—Routine Method. AFNOR publication: France, 1997.
- Bouymajane, A.; Rhazi Filali, F.; Oulghazi, S.; et al. Occurrence, molecular and antimicrobial resistance of Enterococcus spp. isolated from raw cow’s milk trade by street trading in Meknes city, Morocco. Germs. 2018, 8, 77–84. [Google Scholar] [CrossRef] [PubMed]
- M100-S25; Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement. Clinical and Laboratory Standards Institute (CLSI): USA, 2015.
- Ismaiel, A.A.R.; Ali, A.E.S.; Enan, G. Incidence of Listeria in Egyptian meat and dairy samples. Food Sci Biotechnol 2014, 23, 179–185. [Google Scholar] [CrossRef]
- Wieczorek, K.; Dmowska, K.; Osek, J. Characterization and antimicrobial resistance of Listeria monocytogenes isolated from retail beef meat in Poland. Foodborne Pathog Dis 2012, 9, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Boukili, M.; Filali, F.R.; Aboulkacem, A.; Sefiani, M. Assessment of factors influencing the hygienic quality of retail beef meat in Meknes City, Morocco. Int J Vet Sci 2019, 8, 43–48. [Google Scholar]
- Poimenidou, S.V.; Dalmasso, M.; Papadimitriou, K.; Fox, E.M.; Skandamis, P.N.; Jordan, K. Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front Microbiol 2018, 9, 1103. [Google Scholar] [CrossRef] [PubMed]
- Hof, H. Chemotherapy of Listeria infections. GMS Infect Dis 2013, 1, Doc06. [Google Scholar] [CrossRef]
- Caplan, M.E.; Mateescu, L.A.; Dimov, T.V.; Rafila, A.; Borcan, A.M. Antibiotic susceptibility profiles of Listeria monocytogenes strains isolated from food products and clinical samples. Rom Rev Lab Med 2014, 22, 255–261. [Google Scholar] [CrossRef][Green Version]
- Meng, X.; Li, F.; Li, F.; Xiong, Y.; Xu, H. Vancomycin modified PEGylated-magnetic nanoparticles combined with PCR for efficient enrichment and detection of Listeria monocytogenes. Sens Actuators B Chem 2017, 247, 546–555. [Google Scholar] [CrossRef]
- Coroneo, V.; Carraro, V.; Aissani, N.; Sanna, A.; Ruggeri, A.; Succa, S.; et al. Detection of virulence genes and growth potential in Listeria monocytogenes strains isolated from ricotta salata cheese. J Food Sci 2016, 81, M114–M120. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Austin, F.W.; Ainsworth, A.J. A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J Microbiol Methods 2007, 71, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Lomonaco, S.; Patti, R.; Knabel, S.J.; Civera, T. Detection of virulence associated genes and epidemic clone markers in Listeria monocytogenes isolates from PDO Gorgonzola cheese. Int J Food Microbiol 2012, 160, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Bubert, A.; Sokolovic, Z.; Chun, S.K.; Papatheodorou, L.; Simm, A.; Goebel, W. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol Gen Genet 1999, 261, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Du, X.J.; Zhang, X.; Wang, X.Y.; Su, Y.L.; Li, P.; Wang, S. Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Control 2017, 74, 9–16. [Google Scholar] [CrossRef]
- Lotfollahi, L.; Chaharbalesh, A.; Ahangarzadeh Rezaee, M.; Hasani, A. Prevalence, antimicrobial susceptibility and multiplex PCR-serotyping of Listeria monocytogenes isolated from humans, foods and livestock in Iran. Microb Pathog 2017, 107, 425–429. [Google Scholar] [CrossRef]
© GERMS 2020.