Genetic Diversity of SARS-CoV-2 over a One-Year Period of the COVID-19 Pandemic: A Global Perspective
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Multiple Sequence Alignment
2.3. Protein Sequence Diversity
2.4. Selective Pressure Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. Basic Characteristics of SARS-CoV-2 Genome Sequences
3.2. Amino Acid Diversity of SARS-CoV-2 Proteins
3.3. Positive Selection Is Rare in SARS-CoV-2 Protein-Coding Genes
3.4. Circulating Variants of SARS-CoV-2
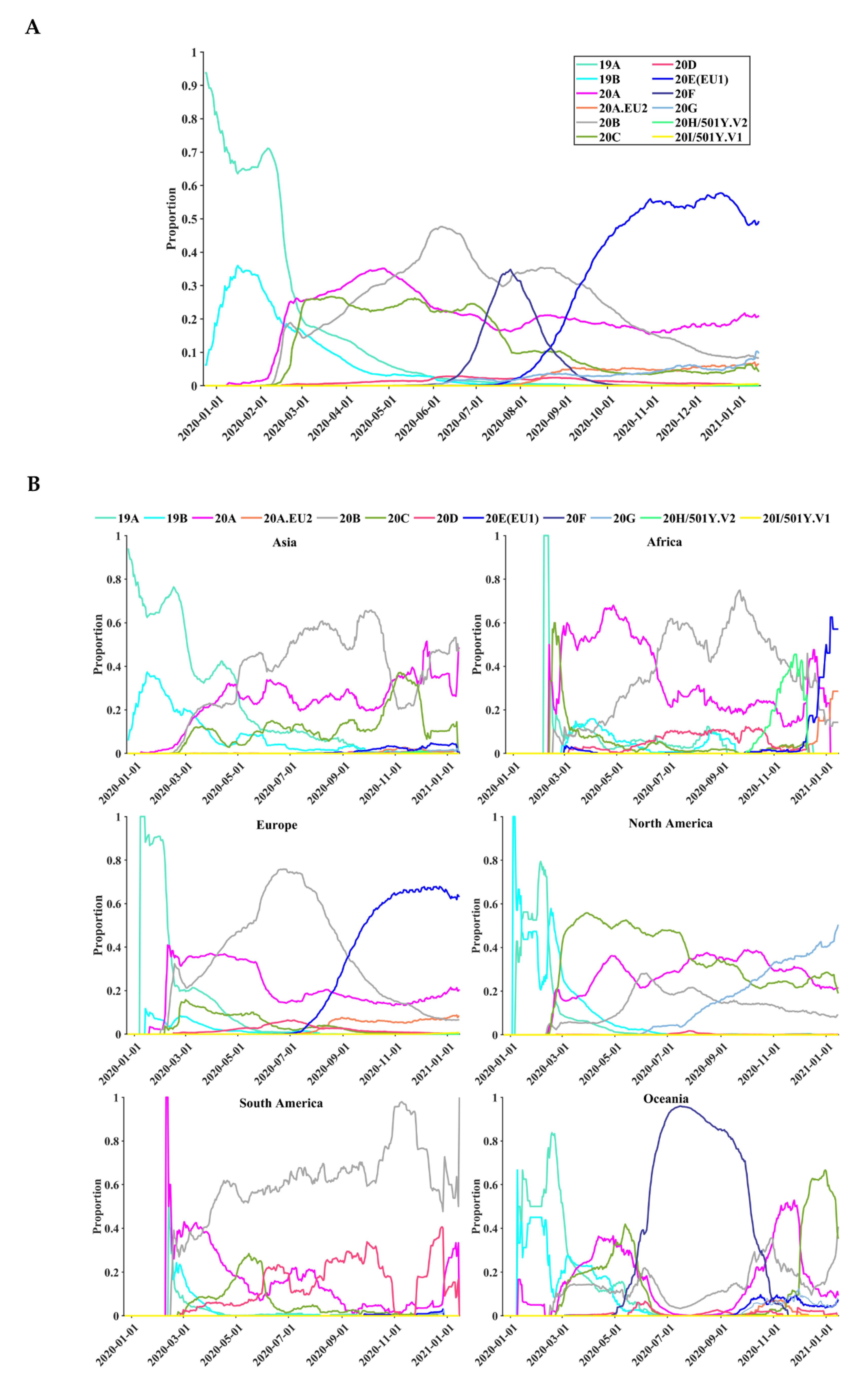
3.5. Geographical and Temporal Trends of the Frequent Variants
3.6. Global Surveillance of SARS-CoV-2 Clades
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shang, J.; Han, N.; Chen, Z.; Peng, Y.; Li, L.; Zhou, H.; Ji, C.; Meng, J.; Jiang, T.; Wu, A. Compositional diversity and evolutionary pattern of coronavirus accessory proteins. Briefings Bioinform. 2021, 22, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Weeraratne, D.; Snowdon, J.L.; Parida, L. Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens 2020, 9, 324. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.; Jia, N.; Zhang, Y.W.; Shum, M.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef]
- Duchene, S.; Featherstone, L.; Haritopoulou-Sinanidou, M.; Rambaut, A.; Lemey, P.; Baele, G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020, 6, veaa061. [Google Scholar] [CrossRef]
- Jia, J.S.; Lu, X.; Yuan, Y.; Xu, G.; Jia, J.; Christakis, N.A. Population flow drives spatio-temporal distribution of COVID-19 in China. Nature 2020, 582, 389–394. [Google Scholar] [CrossRef]
- Popa, A.; Genger, J.-W.; Nicholson, M.D.; Penz, T.; Schmid, D.; Aberle, S.W.; Agerer, B.; Lercher, A.; Endler, L.; Colaço, H.; et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020, 12, eabe2555. [Google Scholar] [CrossRef]
- du Plessis, L.; McCrone, J.T.; Zarebski, A.E.; Hill, V.; Ruis, C.; Gutierrez, B.; Raghwani, J.; Ashworth, J.; Colquhoun, R.; Connor, T.R.; et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021, 371, 708–712. [Google Scholar] [CrossRef]
- Candido, D.S.; Claro, I.M.; De Jesus, J.G.; Souza, W.M.; Moreira, F.R.R.; Dellicour, S.; Mellan, T.A.; Du Plessis, L.; Pereira, R.H.M.; Sales, F.C.S.; et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 2020, 369, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Gu, W.; Federman, S.; Du Plessis, L.; Pybus, O.G.; Faria, N.R.; Wang, C.; Yu, G.; Bushnell, B.; Pan, C.-Y.; et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science 2020, 369, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Ding, N.; Li, J.; Zhang, F.; Wang, Q.; Chen, Z.; Song, C.; Han, K.; Xie, W.; Liu, J.; et al. Genomic surveillance of COVID-19 cases in Beijing. Nat. Commun. 2020, 11, 5503. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Giorgi, F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef] [PubMed]
- Elizondo, V.; Harkins, G.W.; Mabvakure, B.; Smidt, S.; Zappile, P.; Marier, C.; Maurano, M.T.; Perez, V.; Mazza, N.; Beloso, C.; et al. SARS-CoV-2 genomic characterization and clinical manifestation of the COVID-19 outbreak in Uruguay. Emerg. Microbes Infect. 2021, 10, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-C.; Chen, C.-H.; Wang, J.-H.; Liao, H.-C.; Yang, C.-T.; Chen, C.-W.; Lin, Y.-C.; Kao, C.-H.; Lu, M.-Y.J.; Liao, J.C. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. Proc. Natl. Acad. Sci. USA 2020, 117, 30679–30686. [Google Scholar] [CrossRef] [PubMed]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; Du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Gomez-Carballa, A.; Bello, X.; Pardo-Seco, J.; Martinon-Torres, F.; Salas, A. Mapping genome variation of SARS-CoV-2 worldwide highlights the impact of COVID-19 super-spreaders. Genome Res. 2020, 30, 1434–1448. [Google Scholar] [CrossRef]
- Digard, P.; Lee, H.M.; Sharp, C.; Grey, F.; Gaunt, E. Intra-genome variability in the dinucleotide composition of SARS-CoV-2. Virus Evol. 2020, 6, veaa057. [Google Scholar] [CrossRef]
- Zeng, H.-L.; Dichio, V.; Horta, E.R.; Thorell, K.; Aurell, E. Global analysis of more than 50,000 SARS-CoV-2 genomes reveals epistasis between eight viral genes. Proc. Natl. Acad. Sci. USA 2020, 117, 31519–31526. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.-W.; Chan, Y.-H.; Mak, T.-M.; Ang, L.W.; E Anderson, D.; Lee, C.Y.-P.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Crooke, S.N.; Poland, G.A.; Kennedy, R.B. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol. Rev. 2020, 296, 205–219. [Google Scholar] [CrossRef]
- Zhang, F.; Gan, R.; Zhen, Z.; Hu, X.; Li, X.; Zhou, F.; Liu, Y.; Chen, C.; Xie, S.; Zhang, B.; et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020, 5, 156. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Zeng, L.; Li, D.; Tong, W.; Shi, T.; Ning, B. Biochemical features and mutations of key proteins in SARS-CoV-2 and their impacts on RNA therapeutics. Biochem. Pharmacol. 2021, 114424. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef]
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 80, 1136–1138. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2020, 592, 116–121. [Google Scholar] [CrossRef]
- Moore, J.P.; Offit, P.A. SARS-CoV-2 Vaccines and the Growing Threat of Viral Variants. JAMA 2021, 325, 821–822. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, G. Vaccine makers in Asia rush to test jabs against fast-spreading COVID variant. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Peng, P.; Wang, K.; Fang, L.; Luo, F.-Y.; Jin, A.-S.; Liu, B.-Z.; Tang, N.; Huang, A.-L. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021, 18, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Drosten, C. Authors’ response: SARS-CoV-2 detection by real-time RT-PCR. Eurosurveillance 2020, 25, 2001035. [Google Scholar] [CrossRef] [PubMed]
- Artesi, M.; Bontems, S.; Göbbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.-P.; Bours, V.; et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, 58, e01598-20. [Google Scholar] [CrossRef] [PubMed]
- Issa, E.; Merhi, G.; Panossian, B.; Salloum, T.; Tokajian, S. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. mSystems 2020, 5, e00266-20. [Google Scholar] [CrossRef] [PubMed]
- Pancer, K.; Milewska, A.; Owczarek, K.; Dabrowska, A.; Kowalski, M.; Łabaj, P.P.; Branicki, W.; Sanak, M.; Pyrc, K. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog. 2020, 16, e1008959. [Google Scholar] [CrossRef]
- Motayo, B.O.; Oluwasemowo, O.O.; Olusola, B.A.; Akinduti, P.A.; Arege, O.T.; Obafemi, Y.D.; Faneye, A.O.; Isibor, P.O.; Aworunse, O.S.; Oranusi, S.U. Evolution and genetic diversity of SARS-CoV-2 in Africa using whole genome sequences. Int. J. Infect. Dis. 2021, 103, 282–287. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Lessells, R.J.; Giandhari, J.; Pillay, S.; Msomi, N.; Mlisana, K.; Bhiman, J.N.; von Gottberg, A.; Walaza, S.; et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021, 27, 440–446. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Baric, R.S. Emergence of a Highly Fit SARS-CoV-2 Variant. N. Engl. J. Med. 2020, 383, 2684–2686. [Google Scholar] [CrossRef]
- Zhang, M.; Li, G.; Shang, J.; Pan, C.; Zhang, M.; Yin, Z.; Xie, Q.; Peng, Y.; Mao, Q.; Xiao, X.; et al. Rapidly decreased HBV RNA predicts responses of pegylated interferons in HBeAg-positive patients: A longitudinal cohort study. Hepatol. Int. 2020, 14, 212–224. [Google Scholar] [CrossRef]
- Li, G.; Liu, Y.; Jing, X.; Wang, Y.; Miao, M.; Tao, L.; Zhou, Z.; Xie, Y.; Huang, Y.; Lei, J.; et al. Mortality risk of COVID-19 in elderly males with comorbidities: A multi-country study. Aging 2020, 13, 27–60. [Google Scholar] [CrossRef]
- Miao, M.; Jing, X.; De Clercq, E.; Li, G. Danoprevir for the Treatment of Hepatitis C Virus Infection: Design, Development, and Place in Therapy. Drug Des. Dev. Ther. 2020, 14, 2759–2774. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, Y.; Hu, M.; Wen, L.; Wen, C.; Wang, Y.; Zhu, W.; Tai, S.; Jiang, Z.; Xiao, K.; et al. Antibody seroconversion in asymptomatic and symptomatic patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Transl. Immunology 2020, 9, e1182. [Google Scholar] [CrossRef]
- Miao, M.; De Clercq, E.; Li, G. Clinical significance of chemokine receptor antagonists. Expert Opin. Drug Metab. Toxicol. 2020, 16, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, M.; Wang, Y.; Zheng, F.; Huang, Y.; Huang, K.; Yu, Q.; Cai, C.; Chen, D.; Tian, Y.; et al. Clinical characteristics of older and younger patients infected with SARS-CoV-2. Aging 2020, 12, 11296–11305. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.O.; Wiehe, K.; Tian, M.; Acharya, P.; Bradley, T.; Alam, S.M.; Go, E.P.; Scearce, R.; Sutherland, L.; Henderson, R.; et al. Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 2019, 366, eaay7199. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wa Wang, Y.; De Clercq, E.; Li, G. Current and emerging non-nucleoside reverse transcriptase inhibitors (NNRTIs) for HIV-1 treatment. Expert Opin. Drug Metab. Toxicol. 2019, 15, 813–829. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Z.; Wang, C.; Ren, H.; Gao, L.; Peng, H.; Niu, Z.; Ren, H.; Huang, H.; Sun, Q. Bimodular effects of D614G mutation on the spike glycoprotein of SARS-CoV-2 enhance protein processing, membrane fusion, and viral infectivity. Signal Transduct. Target. Ther. 2020, 5, 268. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Quinn, T.P.; Heng, X.; Lorson, C.L.; Sönnerborg, A.; Byrareddy, S.N.; Singh, K. Infectivity of SARS-CoV-2: There Is Something More than D614G? J. Neuroimmune Pharmacol. 2020, 15, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Garvin, M.R.; Prates, E.T.; Pavicic, M.; Jones, P.; Amos, B.K.; Geiger, A.; Shah, M.B.; Streich, J.; Gazolla, J.G.F.M.; Kainer, D.; et al. Potentially adaptive SARS-CoV-2 mutations discovered with novel spatiotemporal and explainable AI models. Genome Biol. 2020, 21, 304. [Google Scholar] [CrossRef] [PubMed]
- Chand, G.B.; Banerjee, A.; Azad, G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ 2020, 8, e9492. [Google Scholar] [CrossRef]
- Xu, Y.; Kang, L.; Shen, Z.; Li, X.; Wu, W.; Ma, W.; Fang, C.; Yang, F.; Jiang, X.; Gong, S.; et al. Dynamics of severe acute respiratory syndrome coronavirus 2 genome variants in the feces during convalescence. J. Genet. Genom. 2020, 47, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Jarvis, C.I.; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021. [Google Scholar] [CrossRef]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; Filipe, A.D.S.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e20. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Xu, M.; Yue, T.; Gu, W.; Tan, L. Life-long passion for antiviral research and drug development: 80th birthday of Prof. Dr. Erik De Clercq. Biochem. Pharmacol. 2021, 185, 114485. [Google Scholar] [CrossRef]
- Li, G.; Yue, T.; Zhang, P.; Gu, W.; Gao, L.J.; Tan, L. Drug Discovery of Nucleos(t)ide Antiviral Agents: Dedicated to Prof. Dr. Erik De Clercq on Occasion of His 80th Birthday. Molecules 2021, 26, 923. [Google Scholar] [CrossRef] [PubMed]
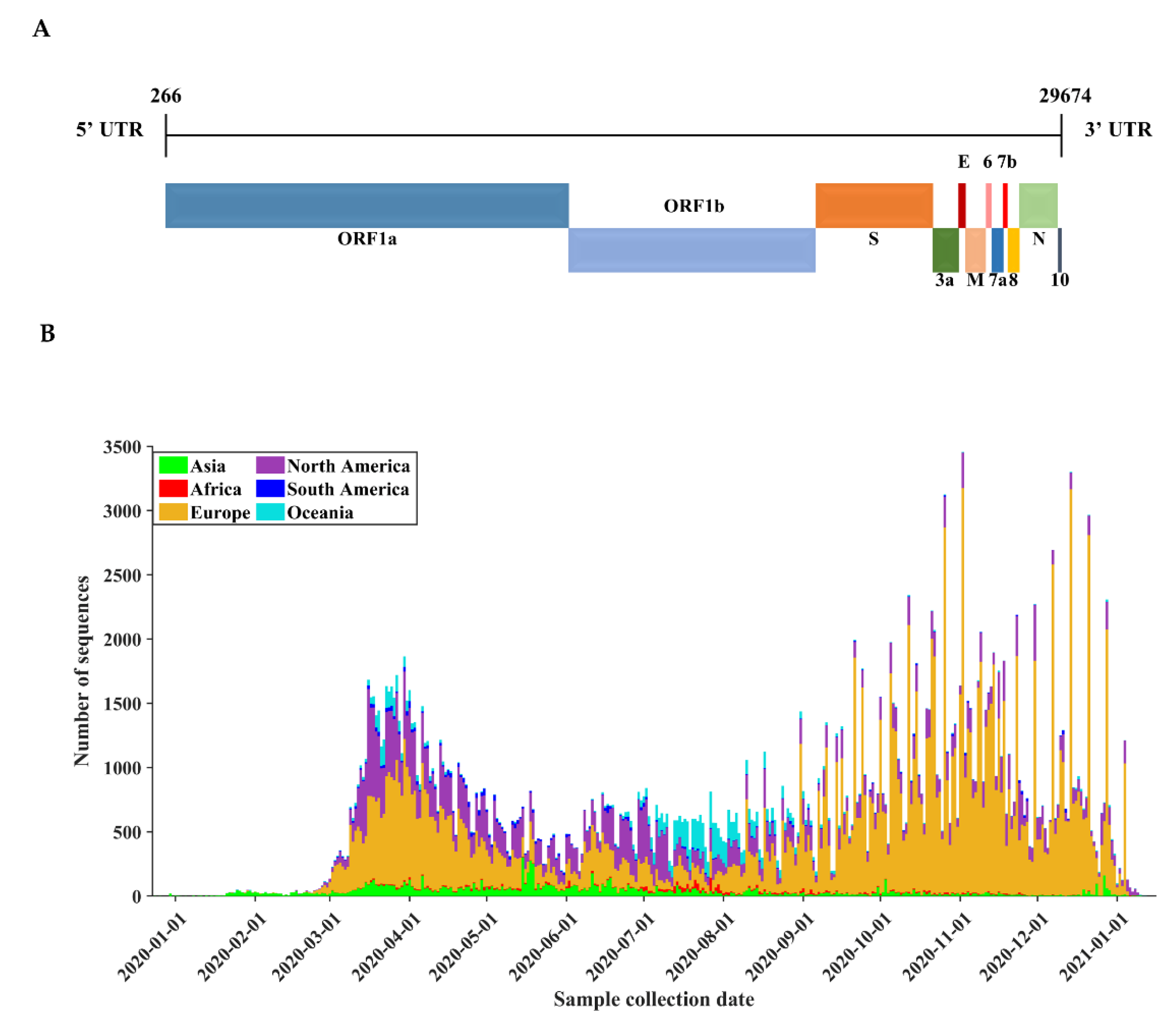

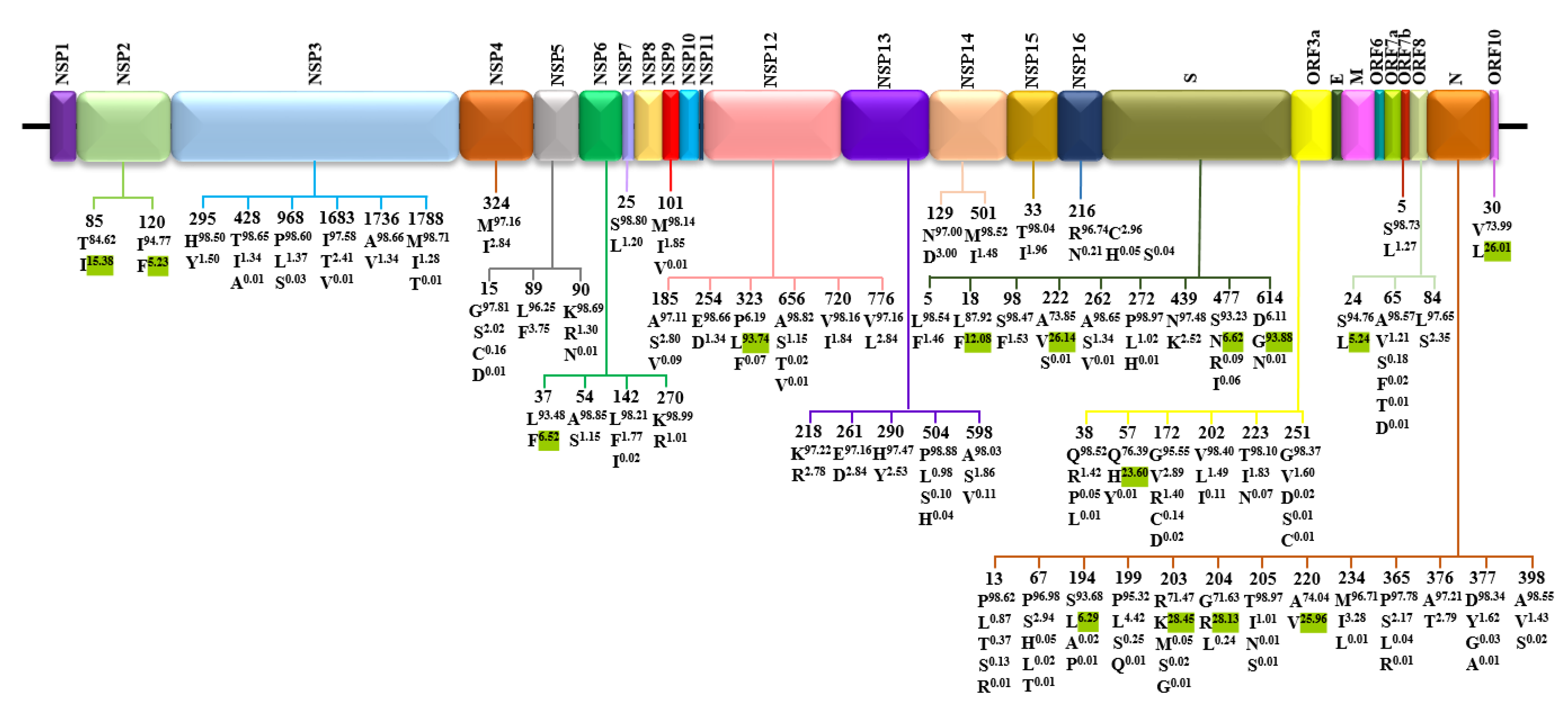
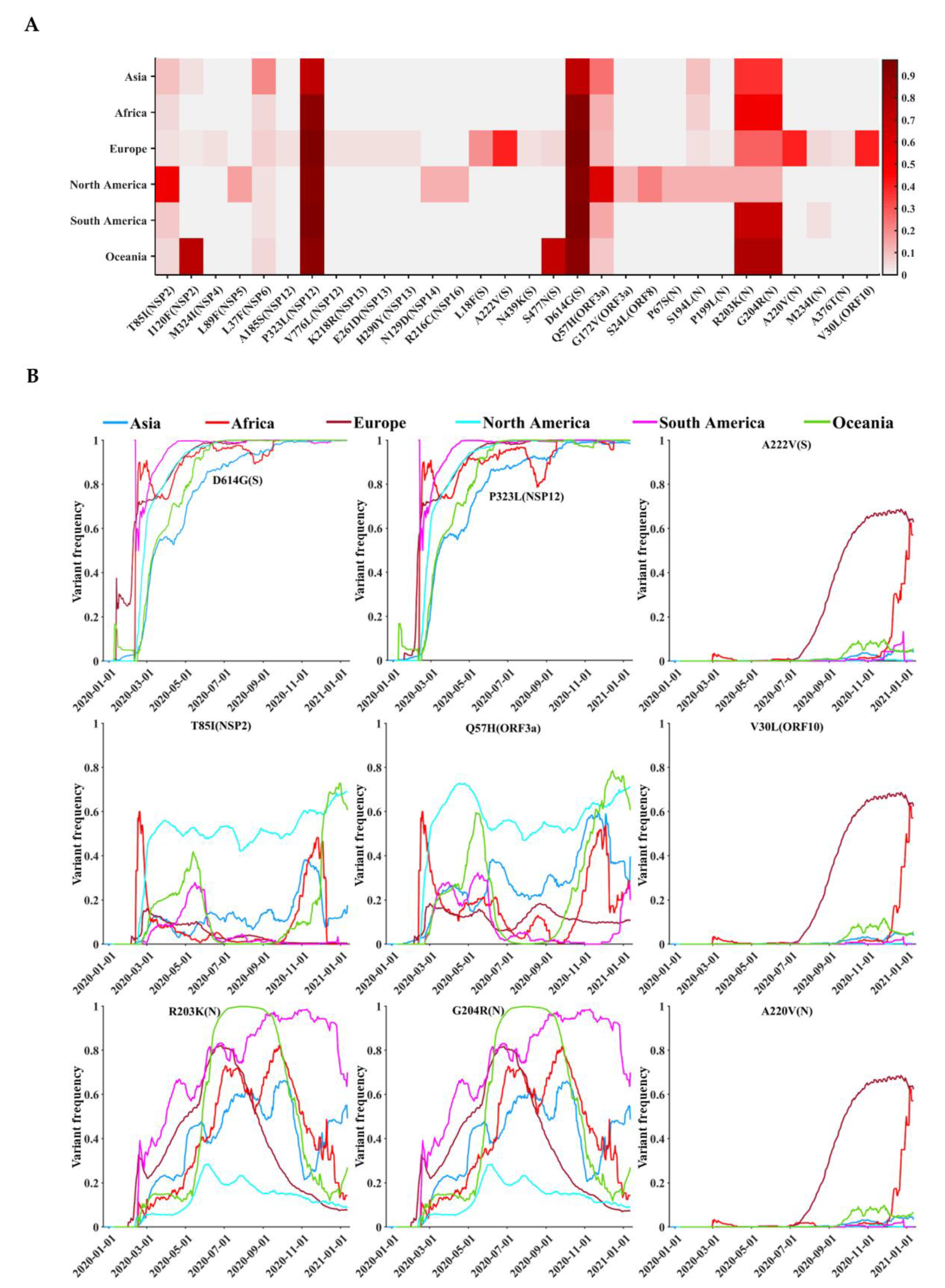
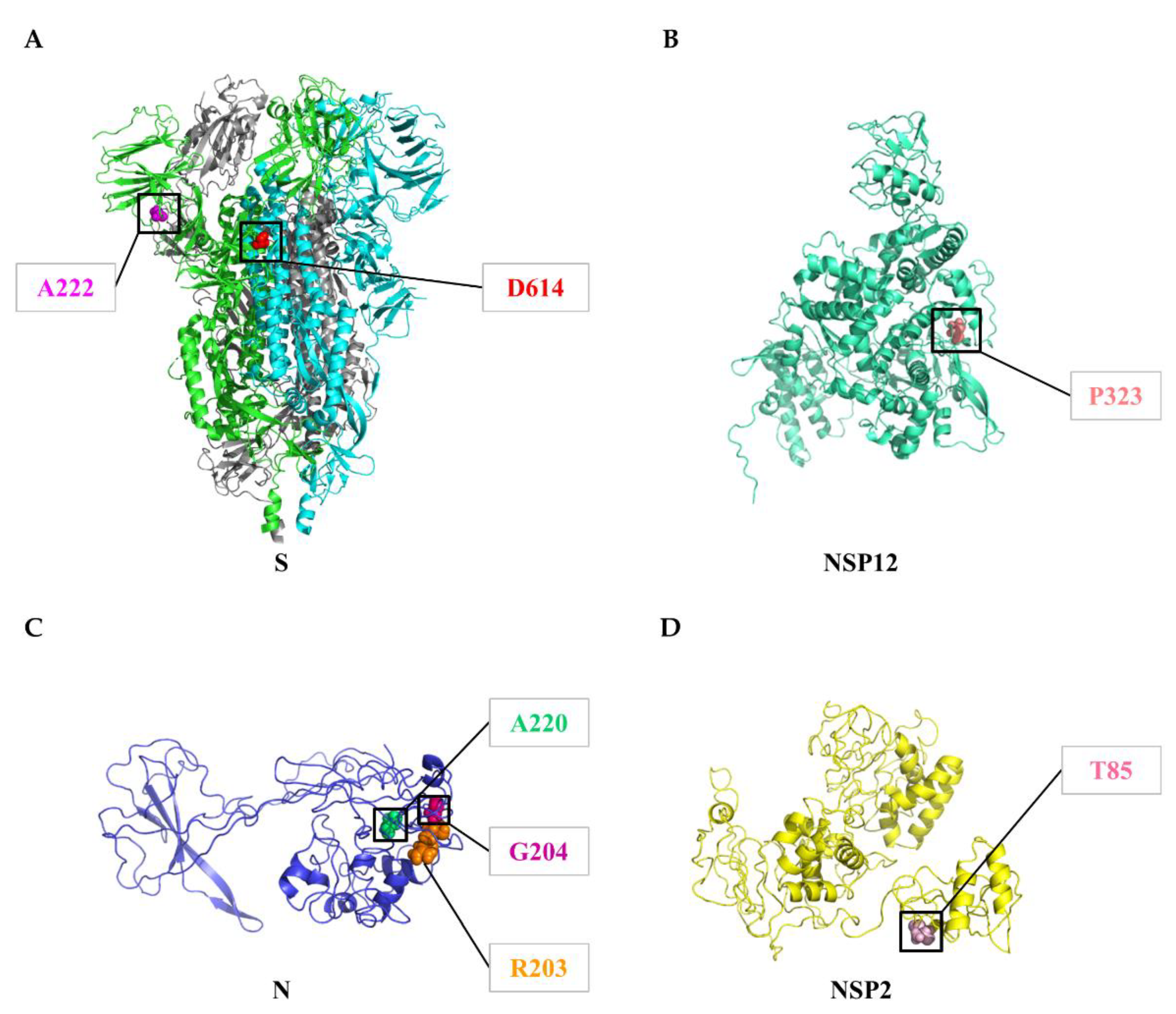
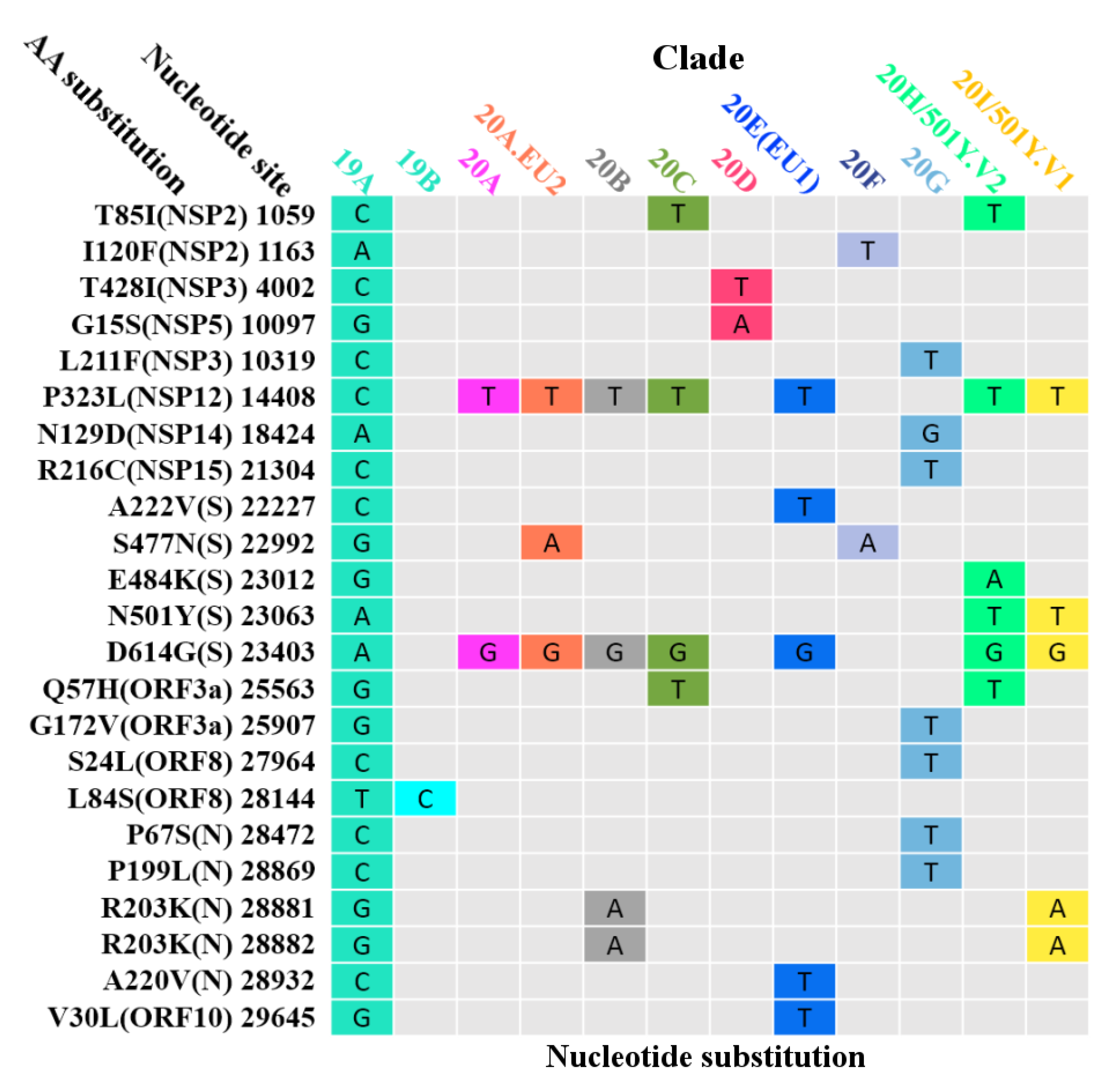
| Protein | Gene | Protein Location | Protein Length | Polymorphic Sites | Substitution Rate (%) |
|---|---|---|---|---|---|
| NSP1 | ORF1a | 266–805 | 180 | 177 | 0.03 |
| NSP2 | ORF1a | 806–2719 | 638 | 608 | 0.07 |
| NSP3 | ORF1a | 2720–8554 | 1945 | 1752 | 0.03 |
| NSP4 | ORF1a | 8555–10,054 | 500 | 419 | 0.02 |
| 3C-like protease (NSP5) | ORF1a | 10,055–10,972 | 306 | 251 | 0.04 |
| NSP6 | ORF1a | 10,973–11,842 | 290 | 256 | 0.06 |
| NSP7 | ORF1a | 11,843–12,091 | 83 | 72 | 0.04 |
| NSP8 | ORF1a | 12,092–12,685 | 198 | 176 | 0.03 |
| NSP9 | ORF1a | 12,686–13,024 | 113 | 93 | 0.03 |
| NSP10 | ORF1a | 13,025–13,441 | 139 | 105 | 0.01 |
| NSP11 | ORF1a | 13,442–13,480 | 13 | 11 | 0.01 |
| RNA-dependent RNA polymerase (NSP12) | ORF1b | 13,442–13,468 13,468–16,236 | 932 | 715 | 0.13 |
| Helicase (NSP13) | ORF1b | 16,237–18,039 | 601 | 478 | 0.04 |
| 3′-to-5′ exonuclease (NSP14) | ORF1b | 18,040–19,620 | 527 | 444 | 0.03 |
| endoRNAse (NSP15) | ORF1b | 19,621–20,658 | 346 | 313 | 0.04 |
| 2′-O-ribose methyltransferase (NSP16) | ORF1b | 20,659–21,552 | 298 | 245 | 0.03 |
| Spike glycoprotein (S) | ORF2 | 21,563–25,384 | 1273 | 1096 | 0.14 |
| ORF3a | ORF3a | 25,393–26,220 | 275 | 273 | 0.20 |
| Envelope protein (E) | ORF4 | 26,245–26,472 | 75 | 72 | 0.02 |
| Membrane protein (M) | ORF5 | 26,523–27,191 | 222 | 168 | 0.02 |
| ORF6 | ORF6 | 27,202–27,387 | 61 | 60 | 0.03 |
| ORF7a | ORF7a | 27,394–27,759 | 121 | 121 | 0.04 |
| ORF7b | ORF7b | 27,756–27,887 | 43 | 43 | 0.07 |
| ORF8 | ORF8 | 27,894–28,259 | 121 | 121 | 0.12 |
| Nucleocapsid protein (N) | ORF9 | 28,274–29,533 | 419 | 378 | 0.31 |
| ORF10 | ORF10 | 29,558–29,674 | 38 | 37 | 0.72 |
| Genetic Variant | Variant Frequency (%) | ||||||
|---|---|---|---|---|---|---|---|
| Asia | Africa | Europe | North America | South America | Oceania | Total | |
| T85I(NSP2) | 10.23 | 5.72 | 3.26 | 52.63 | 8.84 | 5.96 | 15.38 |
| I120F(NSP2) | 3.43 | 0.00 | 2.12 | 0.02 | 0.00 | 72.98 | 5.23 |
| M324I(NSP4) | 0.25 | 0.41 | 4.38 | 0.08 | 0.09 | 0.11 | 2.84 |
| L89F(NSP5) | 0.23 | 0.03 | 0.08 | 15.73 | 0.06 | 0.33 | 3.75 |
| L37F(NSP6) | 19.93 | 4.93 | 6.84 | 3.28 | 3.09 | 4.78 | 6.52 |
| A185S(NSP12) | 0.15 | 0.34 | 4.35 | 0.04 | 0.00 | 0.12 | 2.81 |
| P323L(NSP12) | 71.54 | 91.83 | 96.04 | 92.87 | 97.29 | 91.22 | 93.74 |
| V776L(NSP12) | 0.18 | 0.34 | 4.33 | 0.24 | 0.00 | 0.12 | 2.84 |
| K218R(NSP13) | 0.15 | 0.34 | 4.32 | 0.02 | 0.00 | 0.11 | 2.78 |
| E261D(NSP13) | 0.20 | 0.58 | 4.39 | 0.08 | 0.03 | 0.11 | 2.84 |
| H290Y(NSP13) | 0.55 | 0.17 | 3.80 | 0.25 | 0.03 | 0.22 | 2.53 |
| N129D(NSP14) | 0.17 | 0.00 | 0.05 | 12.61 | 0.03 | 0.32 | 3.00 |
| R216C(NSP16) | 0.20 | 0.00 | 0.07 | 12.40 | 0.03 | 0.33 | 2.96 |
| L18F(S) | 0.32 | 0.28 | 18.78 | 0.30 | 0.34 | 0.12 | 12.08 |
| A222V(S) | 0.50 | 0.79 | 40.78 | 0.17 | 0.34 | 0.53 | 26.14 |
| N439K(S) | 0.45 | 0.03 | 3.90 | 0.01 | 0.06 | 0.06 | 2.52 |
| S477N(S) | 0.19 | 0.66 | 4.82 | 0.14 | 0.12 | 71.22 | 6.62 |
| D614G(S) | 71.91 | 93.91 | 96.11 | 93.10 | 97.26 | 91.26 | 93.88 |
| Q57H(ORF3a) | 24.97 | 12.34 | 11.86 | 59.86 | 14.94 | 8.08 | 23.59 |
| G172V(ORF3a) | 0.20 | 0.07 | 0.07 | 12.07 | 0.06 | 0.31 | 2.89 |
| S24L(ORF8) | 0.23 | 0.17 | 0.15 | 21.78 | 0.03 | 0.90 | 5.24 |
| P67S(N) | 0.17 | 0.03 | 0.11 | 12.21 | 0.06 | 0.33 | 2.94 |
| S194L(N) | 9.64 | 6.54 | 4.27 | 12.50 | 0.19 | 1.19 | 6.29 |
| P199L(N) | 0.29 | 0.21 | 2.29 | 12.49 | 0.06 | 0.40 | 4.42 |
| R203K(N) | 36.96 | 52.15 | 28.16 | 13.35 | 69.12 | 78.12 | 28.45 |
| G204R(N) | 36.73 | 51.62 | 27.71 | 13.31 | 69.07 | 78.08 | 28.13 |
| A220V(N) | 0.45 | 1.10 | 40.53 | 0.13 | 0.09 | 0.35 | 25.96 |
| M234I(N) | 0.50 | 0.55 | 4.77 | 0.61 | 3.64 | 0.30 | 3.28 |
| A376T(N) | 0.15 | 0.35 | 4.33 | 0.02 | 0.00 | 0.11 | 2.78 |
| V30L(ORF10) | 0.58 | 0.69 | 40.64 | 0.08 | 0.13 | 0.35 | 26.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, M.; Clercq, E.D.; Li, G. Genetic Diversity of SARS-CoV-2 over a One-Year Period of the COVID-19 Pandemic: A Global Perspective. Biomedicines 2021, 9, 412. https://doi.org/10.3390/biomedicines9040412
Miao M, Clercq ED, Li G. Genetic Diversity of SARS-CoV-2 over a One-Year Period of the COVID-19 Pandemic: A Global Perspective. Biomedicines. 2021; 9(4):412. https://doi.org/10.3390/biomedicines9040412
Chicago/Turabian StyleMiao, Miao, Erik De Clercq, and Guangdi Li. 2021. "Genetic Diversity of SARS-CoV-2 over a One-Year Period of the COVID-19 Pandemic: A Global Perspective" Biomedicines 9, no. 4: 412. https://doi.org/10.3390/biomedicines9040412
APA StyleMiao, M., Clercq, E. D., & Li, G. (2021). Genetic Diversity of SARS-CoV-2 over a One-Year Period of the COVID-19 Pandemic: A Global Perspective. Biomedicines, 9(4), 412. https://doi.org/10.3390/biomedicines9040412







