Unraveling Molecular Pathways Altered in MeCP2-Related Syndromes, in the Search for New Potential Avenues for Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. mRNA Expression Assays in Whole Blood
2.3. miRNA Expression Assays in Plasma
2.4. Methodologic Considerations
2.5. Brain-Derived Neurotrophic Factor (BDNF) Protein Detection
2.6. Fibroblasts Primary Cultures
2.7. Statistical Analysis
3. Results
3.1. Patients
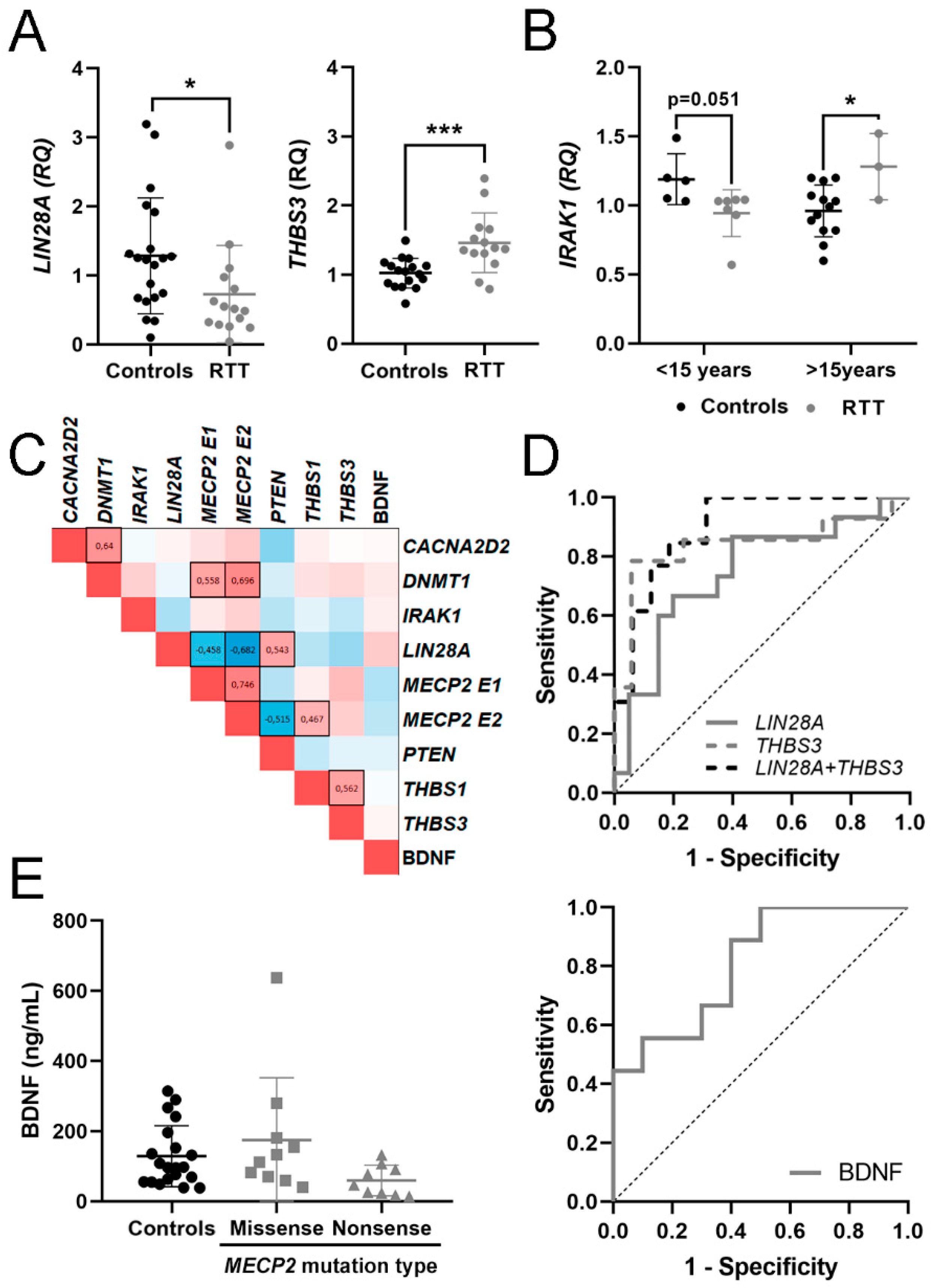
3.2. Expression of mRNA in Blood and BDNF Protein in Plasma
3.3. Evaluation of mRNA in Blood and BDNF Protein in Plasma as Potential Molecular Biomarkers for RTT
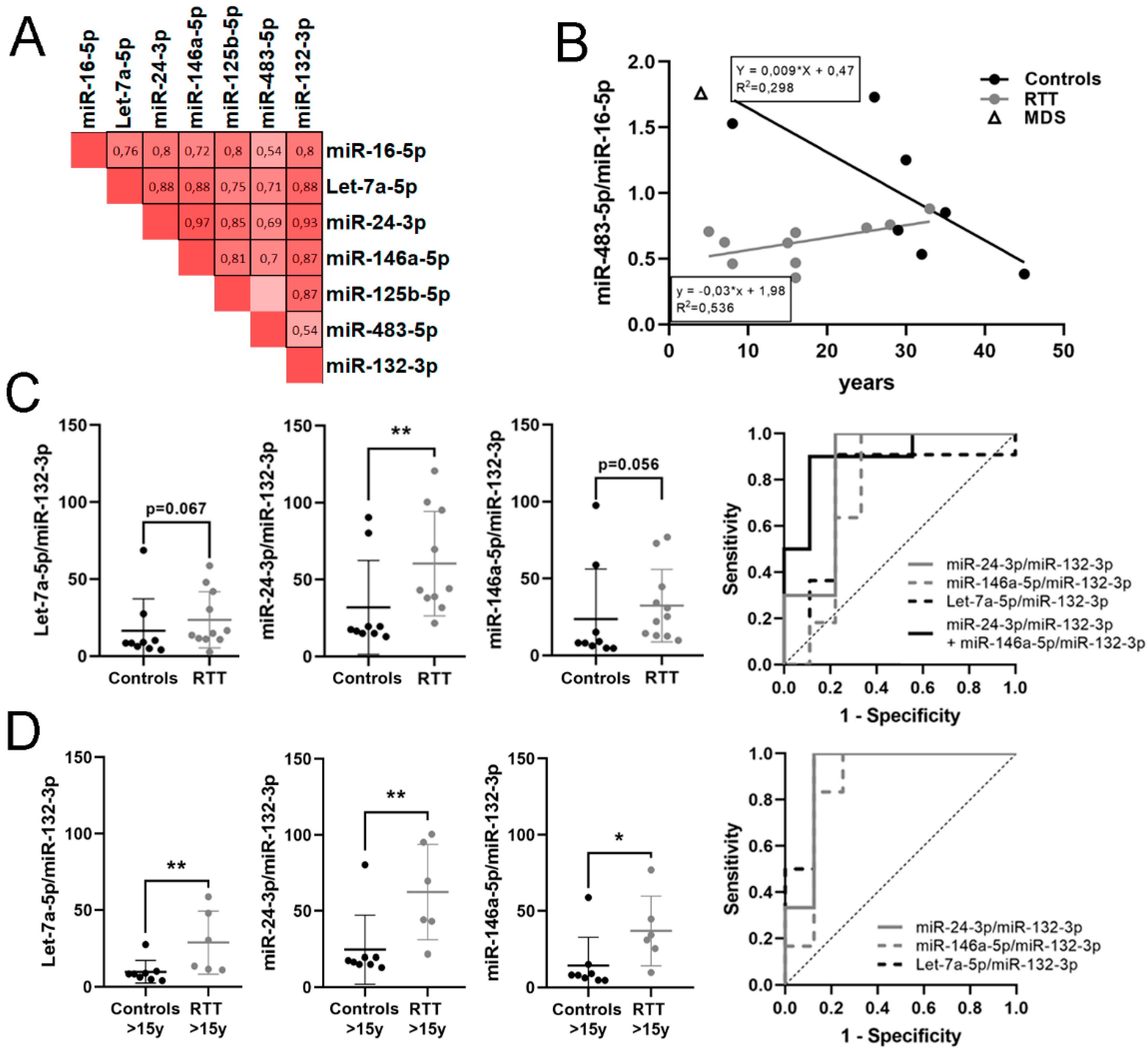
3.4. Identification of a Potential miRNA Biomarker Signature for RTT in Plasma
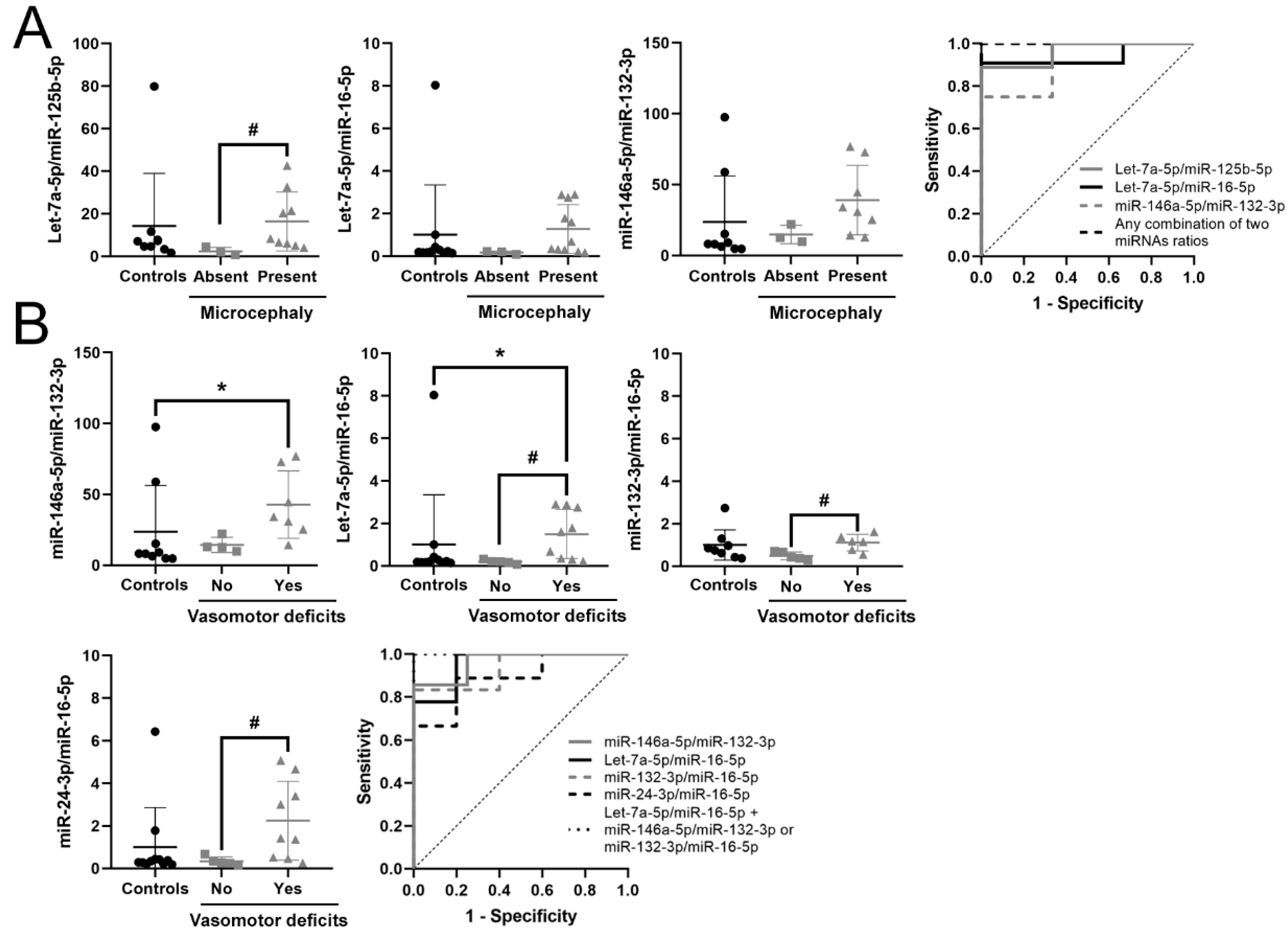
3.5. Identification of a Potential Biomarker Signature for Clinical Parameters in RTT
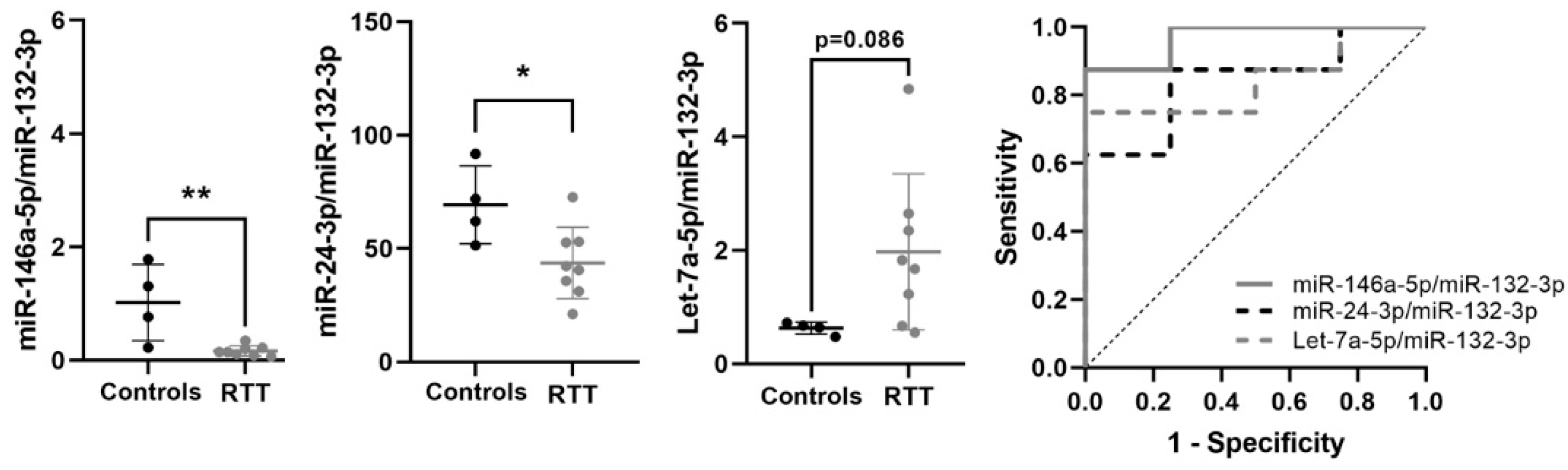
3.6. Expression of Potential Biomarkers in RTT Fibroblasts Primary Cultures
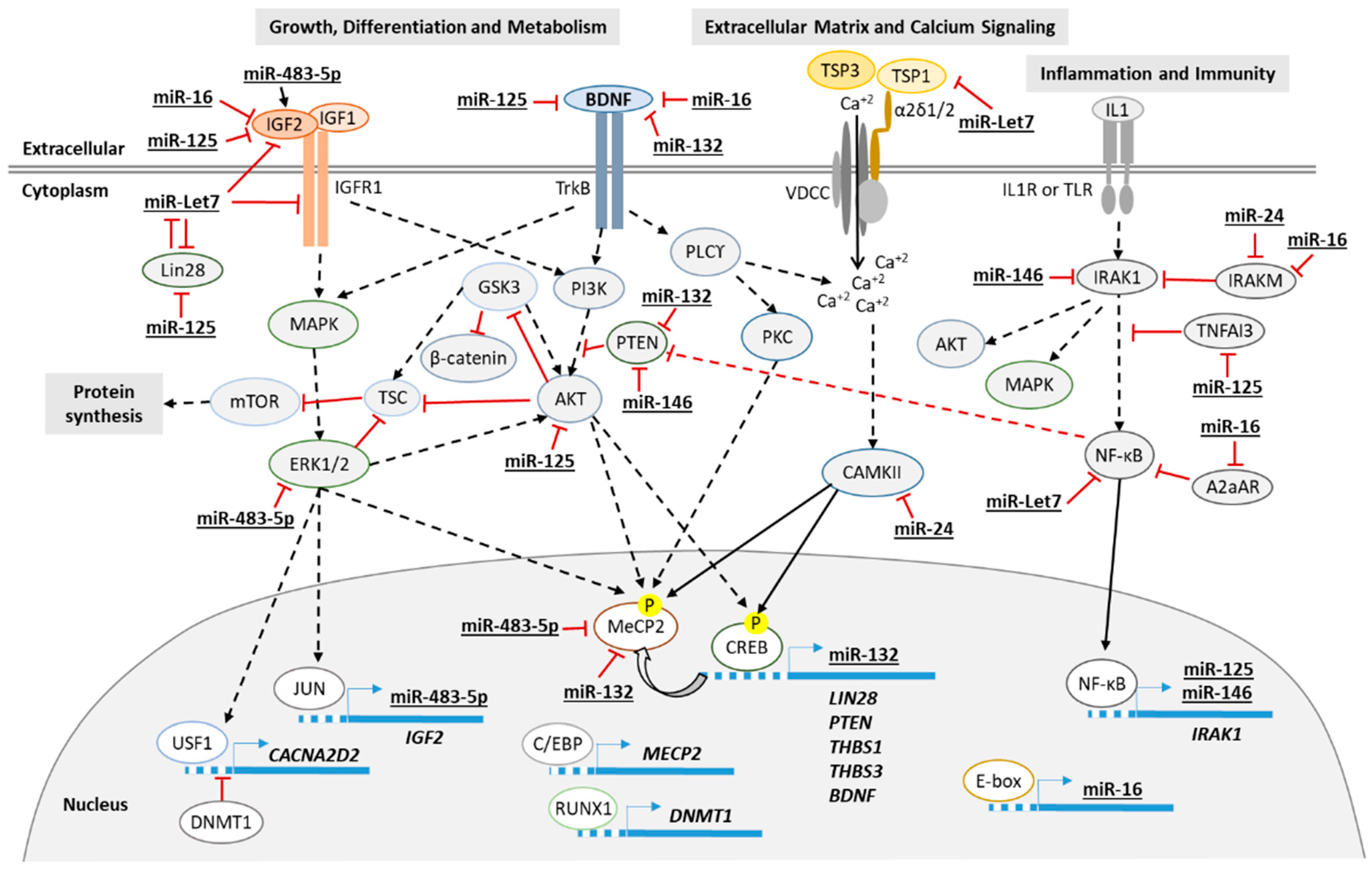
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Georgel, P.T.; Horowitz-Scherer, R.A.; Adkins, N.; Woodcock, C.L.; Wade, P.A.; Hansen, J.C. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003, 278, 32181–32188. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Paz, A.; Ausio, J. MeCP2, A Modulator of Neuronal Chromatin Organization Involved in Rett Syndrome. Adv. Exp. Med. Biol. 2017, 978, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Campoy, F.J.; Bird, A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997, 88, 471–481. [Google Scholar] [CrossRef]
- Yasui, D.H.; Peddada, S.; Bieda, M.C.; Vallero, R.O.; Hogart, A.; Nagarajan, R.P.; Thatcher, K.N.; Farnham, P.J.; Lasalle, J.M. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl. Acad. Sci. USA 2007, 104, 19416–19421. [Google Scholar] [CrossRef] [PubMed]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010, 189, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, D.; Kleefstra, T.; Oudakker, A.R.; Nillesen, W.M.; Yntema, H.G.; Tzschach, A.; Raynaud, M.; Rating, D.; Journel, H.; Chelly, J.; et al. Structural variation in Xq28: MECP2 duplications in 1% of patients with unexplained XLMR and in 2% of male patients with severe encephalopathy. Eur. J. Hum. Genet. EJHG 2009, 17, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Ehrhart, F.; Sangani, N.B.; Curfs, L.M.G. Current developments in the genetics of Rett and Rett-like syndrome. Curr. Opin. Psychiatry 2018, 31, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Trappe, R.; Laccone, F.; Cobilanschi, J.; Meins, M.; Huppke, P.; Hanefeld, F.; Engel, W. MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. Am. J. Hum. Genet. 2001, 68, 1093–1101. [Google Scholar] [CrossRef]
- Amir, R.E.; Van den Veyver, I.B.; Schultz, R.; Malicki, D.M.; Tran, C.Q.; Dahle, E.J.; Philippi, A.; Timar, L.; Percy, A.K.; Motil, K.J.; et al. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann. Neurol. 2000, 47, 670–679. [Google Scholar] [CrossRef]
- Van Esch, H.; Bauters, M.; Ignatius, J.; Jansen, M.; Raynaud, M.; Hollanders, K.; Lugtenberg, D.; Bienvenu, T.; Jensen, L.R.; Gecz, J.; et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 2005, 77, 442–453. [Google Scholar] [CrossRef]
- Stallworth, J.L.; Dy, M.E.; Buchanan, C.B.; Chen, C.F.; Scott, A.E.; Glaze, D.G.; Lane, J.B.; Lieberman, D.N.; Oberman, L.M.; Skinner, S.A.; et al. Hand stereotypies: Lessons from the Rett Syndrome Natural History Study. Neurology 2019, 92, e2594–e2603. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Banerjee, A.; Sur, M. Developmental Dynamics of Rett Syndrome. Neural Plast. 2016, 2016, 6154080. [Google Scholar] [CrossRef] [PubMed]
- Jian, L.; Nagarajan, L.; de Klerk, N.; Ravine, D.; Bower, C.; Anderson, A.; Williamson, S.; Christodoulou, J.; Leonard, H. Predictors of seizure onset in Rett syndrome. J. Pediatrics 2006, 149, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Mount, R.H.; Hastings, R.P.; Reilly, S.; Cass, H.; Charman, T. Behavioural and emotional features in Rett syndrome. Disabil. Rehabil. 2001, 23, 129–138. [Google Scholar] [PubMed]
- Nomura, Y. Early behavior characteristics and sleep disturbance in Rett syndrome. Brain Dev. 2005, 27 (Suppl. 1), S35–S42. [Google Scholar] [CrossRef]
- Kyle, S.M.; Vashi, N.; Justice, M.J. Rett syndrome: A neurological disorder with metabolic components. Open Biol. 2018, 8. [Google Scholar] [CrossRef]
- Buchanan, C.B.; Stallworth, J.L.; Scott, A.E.; Glaze, D.G.; Lane, J.B.; Skinner, S.A.; Tierney, A.E.; Percy, A.K.; Neul, J.L.; Kaufmann, W.E. Behavioral profiles in Rett syndrome: Data from the natural history study. Brain Dev. 2019, 41, 123–134. [Google Scholar] [CrossRef]
- Tarquinio, D.C.; Hou, W.; Berg, A.; Kaufmann, W.E.; Lane, J.B.; Skinner, S.A.; Motil, K.J.; Neul, J.L.; Percy, A.K.; Glaze, D.G. Longitudinal course of epilepsy in Rett syndrome and related disorders. Brain 2017, 140, 306–318. [Google Scholar] [CrossRef]
- Mackay, J.; Downs, J.; Wong, K.; Heyworth, J.; Epstein, A.; Leonard, H. Autonomic breathing abnormalities in Rett syndrome: Caregiver perspectives in an international database study. J. Neurodev. Disord. 2017, 9, 15. [Google Scholar] [CrossRef]
- Witt Engerstrom, I. Age-related occurrence of signs and symptoms in the Rett syndrome. Brain Dev. 1992, 14, S11–S20. [Google Scholar]
- Ramocki, M.B.; Tavyev, Y.J.; Peters, S.U. The MECP2 duplication syndrome. Am. J. Med. Genet. Part A 2010, 152A, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Miguet, M.; Faivre, L.; Amiel, J.; Nizon, M.; Touraine, R.; Prieur, F.; Pasquier, L.; Lefebvre, M.; Thevenon, J.; Dubourg, C.; et al. Further delineation of the MECP2 duplication syndrome phenotype in 59 French male patients, with a particular focus on morphological and neurological features. J. Med. Genet. 2018, 55, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Monros, E.; Armstrong, J.; Aibar, E.; Poo, P.; Canos, I.; Pineda, M. Rett syndrome in Spain: Mutation analysis and clinical correlations. Brain Dev. 2001, 23 (Suppl 1), S251–S253. [Google Scholar] [CrossRef]
- Neul, J.L.; Kaufmann, W.E.; Glaze, D.G.; Christodoulou, J.; Clarke, A.J.; Bahi-Buisson, N.; Leonard, H.; Bailey, M.E.; Schanen, N.C.; Zappella, M.; et al. Rett syndrome: Revised diagnostic criteria and nomenclature. Ann. Neurol. 2010, 68, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Colak, D.; Al-Dhalaan, H.; Nester, M.; Albakheet, A.; Al-Younes, B.; Al-Hassnan, Z.; Al-Dosari, M.; Chedrawi, A.; Al-Owain, M.; Abudheim, N.; et al. Genomic and transcriptomic analyses distinguish classic Rett and Rett-like syndrome and reveals shared altered pathways. Genomics 2011, 97, 19–28. [Google Scholar] [CrossRef] [PubMed]
- De Felice, C.; Leoncini, S.; Signorini, C.; Cortelazzo, A.; Rovero, P.; Durand, T.; Ciccoli, L.; Papini, A.M.; Hayek, J. Rett syndrome: An autoimmune disease? Autoimmun. Rev. 2016, 15, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Shulyakova, N.; Andreazza, A.C.; Mills, L.R.; Eubanks, J.H. Mitochondrial Dysfunction in the Pathogenesis of Rett Syndrome: Implications for Mitochondria-Targeted Therapies. Front. Cell Neurosci. 2017, 11, 58. [Google Scholar] [CrossRef]
- Ip, J.P.K.; Mellios, N.; Sur, M. Rett syndrome: Insights into genetic, molecular and circuit mechanisms. Nat. Rev. Neurosci. 2018, 19, 368–382. [Google Scholar] [CrossRef]
- Jorge-Torres, O.C.; Szczesna, K.; Roa, L.; Casal, C.; Gonzalez-Somermeyer, L.; Soler, M.; Velasco, C.D.; Martinez-San Segundo, P.; Petazzi, P.; Saez, M.A.; et al. Inhibition of Gsk3b Reduces Nfkb1 Signaling and Rescues Synaptic Activity to Improve the Rett Syndrome Phenotype in Mecp2-Knockout Mice. Cell Rep. 2018, 23, 1665–1677. [Google Scholar] [CrossRef]
- Kishi, N.; MacDonald, J.L.; Ye, J.; Molyneaux, B.J.; Azim, E.; Macklis, J.D. Reduction of aberrant NF-kappaB signalling ameliorates Rett syndrome phenotypes in Mecp2-null mice. Nat. Commun. 2016, 7, 10520. [Google Scholar] [CrossRef]
- Leonard, H.; Cobb, S.; Downs, J. Clinical and biological progress over 50 years in Rett syndrome. Nat. Rev. Neurol. 2017, 13, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Bhattacharya, U.; Pradana, W.A.; Tarquinio, D.C. Assessment of a Clinical Trial Metric for Rett Syndrome: Critical Analysis of the Rett Syndrome Behavioural Questionnaire. Pediatric Neurol. 2020, 107, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Calin, G.A. MicroRNA identification in plasma and serum: A new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 2009, 9, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Terrinoni, A.; Calabrese, C.; Basso, D.; Aita, A.; Caporali, S.; Plebani, M.; Bernardini, S. The circulating miRNAs as diagnostic and prognostic markers. Clin. Chem. Lab. Med. 2019, 57, 932–953. [Google Scholar] [CrossRef]
- Beveridge, N.J.; Cairns, M.J. MicroRNA dysregulation in schizophrenia. Neurobiol. Dis. 2012, 46, 263–271. [Google Scholar] [CrossRef]
- Wu, H.; Tao, J.; Chen, P.J.; Shahab, A.; Ge, W.; Hart, R.P.; Ruan, X.; Ruan, Y.; Sun, Y.E. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA 2010, 107, 18161–18166. [Google Scholar] [CrossRef]
- Bai, M.; Zhu, X.; Zhang, Y.; Zhang, S.; Zhang, L.; Xue, L.; Yi, J.; Yao, S.; Zhang, X. Abnormal hippocampal BDNF and miR-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS ONE 2012, 7, e46921. [Google Scholar] [CrossRef]
- Han, K.; Gennarino, V.A.; Lee, Y.; Pang, K.; Hashimoto-Torii, K.; Choufani, S.; Raju, C.S.; Oldham, M.C.; Weksberg, R.; Rakic, P.; et al. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes Dev. 2013, 27, 485–490. [Google Scholar] [CrossRef]
- Vo, N.; Klein, M.E.; Varlamova, O.; Keller, D.M.; Yamamoto, T.; Goodman, R.H.; Impey, S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 16426–16431. [Google Scholar] [CrossRef]
- Huang, Y.W.; Ruiz, C.R.; Eyler, E.C.; Lin, K.; Meffert, M.K. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell 2012, 148, 933–946. [Google Scholar] [CrossRef]
- Lyu, J.W.; Yuan, B.; Cheng, T.L.; Qiu, Z.L.; Zhou, W.H. Reciprocal regulation of autism-related genes MeCP2 and PTEN via microRNAs. Sci. Rep. 2016, 6, 20392. [Google Scholar] [CrossRef] [PubMed]
- GEO—Gene expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6955 (accessed on 17 December 2020).
- Zanutto, S.; Pizzamiglio, S.; Ghilotti, M.; Bertan, C.; Ravagnani, F.; Perrone, F.; Leo, E.; Pilotti, S.; Verderio, P.; Gariboldi, M.; et al. Circulating miR-378 in plasma: A reliable, haemolysis-independent biomarker for colorectal cancer. Br. J. Cancer 2014, 110, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhu, Y.; Wang, H.; Khadka, V.S.; Hu, L.; Ai, J.; Dou, Y.; Li, Y.; Dai, S.; Mason, C.E.; et al. Ratio-Based Method To Identify True Biomarkers by Normalizing Circulating ncRNA Sequencing and Quantitative PCR Data. Anal. Chem. 2019, 91, 6746–6753. [Google Scholar] [CrossRef] [PubMed]
- Xiol, C.; Vidal, S.; Pascual-Alonso, A.; Blasco, L.; Brandi, N.; Pacheco, P.; Gerotina, E.; O’Callaghan, M.; Pineda, M.; Armstrong, J.; et al. X chromosome inactivation does not necessarily determine the severity of the phenotype in Rett syndrome patients. Sci. Rep. 2019, 9, 11983. [Google Scholar] [CrossRef]
- Orlic-Milacic, M.; Kaufman, L.; Mikhailov, A.; Cheung, A.Y.; Mahmood, H.; Ellis, J.; Gianakopoulos, P.J.; Minassian, B.A.; Vincent, J.B. Over-expression of either MECP2_e1 or MECP2_e2 in neuronally differentiated cells results in different patterns of gene expression. PLoS ONE 2014, 9, e91742. [Google Scholar] [CrossRef]
- Sheinerman, K.; Djukic, A.; Tsivinsky, V.G.; Umansky, S.R. Brain-enriched microRNAs circulating in plasma as novel biomarkers for Rett syndrome. PLoS ONE 2019, 14, e0218623. [Google Scholar] [CrossRef]
- Kaufmann, W.E.; Jarrar, M.H.; Wang, J.S.; Lee, Y.J.; Reddy, S.; Bibat, G.; Naidu, S. Histone modifications in Rett syndrome lymphocytes: A preliminary evaluation. Brain Dev. 2005, 27, 331–339. [Google Scholar] [CrossRef]
- Cosentino, L.; Vigli, D.; Franchi, F.; Laviola, G.; De Filippis, B. Rett syndrome before regression: A time window of overlooked opportunities for diagnosis and intervention. Neurosci. Biobehav. Rev. 2019, 107, 115–135. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kaab, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef]
- He, K.; Guo, C.; He, L.; Shi, Y. MiRNAs of peripheral blood as the biomarker of schizophrenia. Hereditas 2018, 155, 9. [Google Scholar] [CrossRef]
- Chahine, L.M.; Stern, M.B.; Chen-Plotkin, A. Blood-based biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 2014, 20 (Suppl. 1), S99–S103. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Pisco, A.; Papalopulu, N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 2011, 20, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Wu, Y.T.; Yu, S.L.; Yu, Y.H.; Lee, S.Y.; Liu, C.M.; Hsieh, W.S.; Hwu, H.G.; Chen, P.C.; Jeng, S.F.; et al. Modulated expression of human peripheral blood microRNAs from infancy to adulthood and its role in aging. Aging Cell 2014, 13, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Mellios, N.; Woodson, J.; Garcia, R.I.; Crawford, B.; Sharma, J.; Sheridan, S.D.; Haggarty, S.J.; Sur, M. beta2-Adrenergic receptor agonist ameliorates phenotypes and corrects microRNA-mediated IGF1 deficits in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA 2014, 111, 9947–9952. [Google Scholar] [CrossRef] [PubMed]
- Paco, S.; Casserras, T.; Rodriguez, M.A.; Jou, C.; Puigdelloses, M.; Ortez, C.I.; Diaz-Manera, J.; Gallardo, E.; Colomer, J.; Nascimento, A.; et al. Transcriptome Analysis of Ullrich Congenital Muscular Dystrophy Fibroblasts Reveals a Disease Extracellular Matrix Signature and Key Molecular Regulators. PLoS ONE 2015, 10, e0145107. [Google Scholar] [CrossRef]
- An, T.; Fan, T.; Zhang, X.Q.; Liu, Y.F.; Huang, J.; Liang, C.; Lv, B.H.; Wang, Y.Q.; Zhao, X.G.; Liu, J.X.; et al. Comparison of Alterations in miRNA Expression in Matched Tissue and Blood Samples during Spinal Cord Glioma Progression. Sci. Rep. 2019, 9, 9169. [Google Scholar] [CrossRef]
- Shah, R.; Tanriverdi, K.; Levy, D.; Larson, M.; Gerstein, M.; Mick, E.; Rozowsky, J.; Kitchen, R.; Murthy, V.; Mikalev, E.; et al. Discordant Expression of Circulating microRNA from Cellular and Extracellular Sources. PLoS ONE 2016, 11, e0153691. [Google Scholar] [CrossRef]
- Lewin, G.R.; Barde, Y.A. Physiology of the neurotrophins. Annu. Rev. Neurosci. 1996, 19, 289–317. [Google Scholar] [CrossRef]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in neuronal development and function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar] [CrossRef] [PubMed]
- Bibel, M.; Barde, Y.A. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000, 14, 2919–2937. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.R.; Miller, F.D. Neurotrophin signal transduction in the nervous system. Curr. Opin. Neurobiol. 2000, 10, 381–391. [Google Scholar] [CrossRef]
- Li, W.; Pozzo-Miller, L. BDNF deregulation in Rett syndrome. Neuropharmacology 2014, 76 Pt C, 737–746. [Google Scholar] [CrossRef]
- Fu, Y.; Hou, B.; Weng, C.; Liu, W.; Dai, J.; Zhao, C.; Yin, Z.Q. Functional ectopic neuritogenesis by retinal rod bipolar cells is regulated by miR-125b-5p during retinal remodeling in RCS rats. Sci. Rep. 2017, 7, 1011. [Google Scholar] [CrossRef] [PubMed]
- Burak, K.; Lamoureux, L.; Boese, A.; Majer, A.; Saba, R.; Niu, Y.; Frost, K.; Booth, S.A. MicroRNA-16 targets mRNA involved in neurite extension and branching in hippocampal neurons during presymptomatic prion disease. Neurobiol. Dis. 2018, 112, 1–13. [Google Scholar] [CrossRef]
- Zhu, H.; Shyh-Chang, N.; Segre, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Daley, G.Q. Lin28: Primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013, 12, 395–406. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Juhila, J.; Sipila, T.; Icay, K.; Nicorici, D.; Ellonen, P.; Kallio, A.; Korpelainen, E.; Greco, D.; Hovatta, I. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS ONE 2011, 6, e21495. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, Y.; Yahagi, K.; Kawano, M.; Nishiyori, H.; Kawazu, C.; Suzuki, N.; Manabe, R.; Hirase, H. miRNA profiling of bilateral rat hippocampal CA3 by deep sequencing. Biochem. Biophys. Res. Commun. 2011, 409, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Moss, E.G.; Lee, R.C.; Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997, 88, 637–646. [Google Scholar] [CrossRef]
- Olde Loohuis, N.F.; Kos, A.; Martens, G.J.; Van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. CMLS 2012, 69, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.A.; Thomson, J.M.; Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 2008, 14, 1539–1549. [Google Scholar] [CrossRef]
- Yang, M.; Yang, S.L.; Herrlinger, S.; Liang, C.; Dzieciatkowska, M.; Hansen, K.C.; Desai, R.; Nagy, A.; Niswander, L.; Moss, E.G.; et al. Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development 2015, 142, 1616–1627. [Google Scholar] [CrossRef]
- Polesskaya, A.; Cuvellier, S.; Naguibneva, I.; Duquet, A.; Moss, E.G.; Harel-Bellan, A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007, 21, 1125–1138. [Google Scholar] [CrossRef]
- Shenoy, A.; Danial, M.; Blelloch, R.H. Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J. 2015, 34, 1180–1194. [Google Scholar] [CrossRef]
- miRTarBase. Available online: http://miRTarBAse.cuhk.edu.cn (accessed on 17 December 2020).
- Liu, M.; Roth, A.; Yu, M.; Morris, R.; Bersani, F.; Rivera, M.N.; Lu, J.; Shioda, T.; Vasudevan, S.; Ramaswamy, S.; et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013, 27, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Vahdatpour, C.; Dyer, A.H.; Tropea, D. Insulin-Like Growth Factor 1 and Related Compounds in the Treatment of Childhood-Onset Neurodevelopmental Disorders. Front. Neurosci. 2016, 10, 450. [Google Scholar] [CrossRef] [PubMed]
- Khwaja, O.S.; Ho, E.; Barnes, K.V.; O’Leary, H.M.; Pereira, L.M.; Finkelstein, Y.; Nelson, C.A., 3rd; Vogel-Farley, V.; DeGregorio, G.; Holm, I.A.; et al. Safety, pharmacokinetics, and preliminary assessment of efficacy of mecasermin (recombinant human IGF-1) for the treatment of Rett syndrome. Proc. Natl. Acad. Sci. USA 2014, 111, 4596–4601. [Google Scholar] [CrossRef] [PubMed]
- Riikonen, R. Treatment of autistic spectrum disorder with insulin-like growth factors. Eur. J. Paediatr. Neurol. EJPN Off. J. Eur. Paediatr. Neurol. Soc. 2016, 20, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, M.R.; Ward, C.S.; Arvide, E.M.; Chapleau, C.A.; Pozzo-Miller, L.; Hoeflich, A.; Sivaramakrishnan, M.; Saenger, S.; Metzger, F.; Neul, J.L. Insulinotropic treatments exacerbate metabolic syndrome in mice lacking MeCP2 function. Hum. Mol. Genet. 2013, 22, 2626–2633. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, B. Clinical manifestations and stages of Rett syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2002, 8, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Tarquinio, D.C.; Motil, K.J.; Hou, W.; Lee, H.S.; Glaze, D.G.; Skinner, S.A.; Neul, J.L.; Annese, F.; McNair, L.; Barrish, J.O.; et al. Growth failure and outcome in Rett syndrome: Specific growth references. Neurology. 2012, 79, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Long, Z.; Chen, Z.; Li, J.; Hu, Z.; Qiu, R.; Zhuang, W.; Tang, B.; Xia, K.; Jiang, H. Investigation of Gene Regulatory Networks Associated with Autism Spectrum Disorder Based on MiRNA Expression in China. PLoS ONE 2015, 10, e0129052. [Google Scholar] [CrossRef] [PubMed]
- Cortelazzo, A.; De Felice, C.; De Filippis, B.; Ricceri, L.; Laviola, G.; Leoncini, S.; Signorini, C.; Pescaglini, M.; Guerranti, R.; Timperio, A.M.; et al. Persistent Unresolved Inflammation in the Mecp2-308 Female Mutated Mouse Model of Rett Syndrome. Mediat. Inflamm. 2017, 2017, 9467819. [Google Scholar] [CrossRef]
- Cortelazzo, A.; De Felice, C.; Guerranti, R.; Signorini, C.; Leoncini, S.; Pecorelli, A.; Zollo, G.; Landi, C.; Valacchi, G.; Ciccoli, L.; et al. Subclinical inflammatory status in Rett syndrome. Mediat. Inflamm. 2014, 2014, 480980. [Google Scholar] [CrossRef]
- O’Driscoll, C.M.; Lima, M.P.; Kaufmann, W.E.; Bressler, J.P. Methyl CpG binding protein 2 deficiency enhances expression of inflammatory cytokines by sustaining NF-kappaB signaling in myeloid derived cells. J. Neuroimmunol. 2015, 283, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Alonso, A.; Blasco, L.; Vidal, S.; Gean, E.; Rubio, P.; O’Callaghan, M.; Martinez-Monseny, A.F.; Castells, A.A.; Xiol, C.; Catala, V.; et al. Molecular characterization of Spanish patients with MECP2 duplication syndrome. Clin. Genet. 2020, 97, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-kappaB signaling. J. Mol. Cell Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Chen, K.; Yuan, R.; Peng, L.; Maitra, U.; Diao, N.; Chen, C.; Zhang, Y.; Hu, Y.; Qi, C.F.; et al. The persistence of low-grade inflammatory monocytes contributes to aggravated atherosclerosis. Nat. Commun. 2016, 7, 13436. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hernandez, L.D.; Galan, J.E.; Janeway, C.A., Jr.; Medzhitov, R.; Flavell, R.A. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef]
- Kim, S.W.; Ramasamy, K.; Bouamar, H.; Lin, A.P.; Jiang, D.; Aguiar, R.C. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc. Natl. Acad. Sci. USA 2012, 109, 7865–7870. [Google Scholar] [CrossRef]
- Tian, T.; Zhou, Y.; Feng, X.; Ye, S.; Wang, H.; Wu, W.; Tan, W.; Yu, C.; Hu, J.; Zheng, R.; et al. MicroRNA-16 is putatively involved in the NF-kappaB pathway regulation in ulcerative colitis through adenosine A2a receptor (A2aAR) mRNA targeting. Sci. Rep. 2016, 6, 30824. [Google Scholar] [CrossRef]
- Lopez, J.P.; Fiori, L.M.; Cruceanu, C.; Lin, R.; Labonte, B.; Cates, H.M.; Heller, E.A.; Vialou, V.; Ku, S.M.; Gerald, C.; et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat. Commun. 2017, 8, 15497. [Google Scholar] [CrossRef]
- Adams, J.C.; Lawler, J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef]
- Garcia, O.; Torres, M.; Helguera, P.; Coskun, P.; Busciglio, J. A role for thrombospondin-1 deficits in astrocyte-mediated spine and synaptic pathology in Down’s syndrome. PLoS ONE 2010, 5, e14200. [Google Scholar] [CrossRef]
- Cheng, C.; Lau, S.K.; Doering, L.C. Astrocyte-secreted thrombospondin-1 modulates synapse and spine defects in the fragile X mouse model. Mol. Brain 2016, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Bauman, M.L.; Kemper, T.L.; Arin, D.M. Microscopic observations of the brain in Rett syndrome. Neuropediatrics 1995, 26, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Belichenko, N.P.; Belichenko, P.V.; Mobley, W.C. Evidence for both neuronal cell autonomous and nonautonomous effects of methyl-CpG-binding protein 2 in the cerebral cortex of female mice with Mecp2 mutation. Neurobiol. Dis. 2009, 34, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Belichenko, P.V.; Wright, E.E.; Belichenko, N.P.; Masliah, E.; Li, H.H.; Mobley, W.C.; Francke, U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: Evidence for disruption of neuronal networks. J. Comp. Neurol. 2009, 514, 240–258. [Google Scholar] [CrossRef]
- Dogar, A.M.; Semplicio, G.; Guennewig, B.; Hall, J. Multiple microRNAs derived from chemically synthesized precursors regulate thrombospondin 1 expression. Nucleic Acid. Ther. 2014, 24, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Castells, A.A.; Gueraldi, D.; Balada, R.; Tristan-Noguero, A.; Cortes-Saladelafont, E.; Ramos, F.; Meavilla, S.; De Los Santos, M.; Garcia-Volpe, C.; Colome, R.; et al. Discovery of Biomarker Panels for Neural Dysfunction in Inborn Errors of Amino Acid Metabolism. Sci. Rep. 2019, 9, 9128. [Google Scholar] [CrossRef]
- Lioy, D.T.; Garg, S.K.; Monaghan, C.E.; Raber, J.; Foust, K.D.; Kaspar, B.K.; Hirrlinger, P.G.; Kirchhoff, F.; Bissonnette, J.M.; Ballas, N.; et al. A role for glia in the progression of Rett’s syndrome. Nature 2011, 475, 497–500. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, H.; Wu, D.; Ni, H.; Chen, Z.; Chen, C.; Xiang, Y.; Dai, K.; Chen, X.; Li, X. MicroRNA let-7a regulates angiogenesis by targeting TGFBR3 mRNA. J. Cell Mol. Med. 2019, 23, 556–567. [Google Scholar] [CrossRef]
- Sun, L.L.; Li, W.D.; Lei, F.R.; Li, X.Q. The regulatory role of microRNAs in angiogenesis-related diseases. J. Cell Mol. Med. 2018, 22, 4568–4587. [Google Scholar] [CrossRef]
- Che, F.; Du, H.; Zhang, W.; Cheng, Z.; Tong, Y. MicroRNA-132 modifies angiogenesis in patients with ischemic cerebrovascular disease by suppressing the NFkappaB and VEGF pathway. Mol. Med. Rep. 2018, 17, 2724–2730. [Google Scholar] [CrossRef]
| Patient | Age (y) | Sex | MeCP2 Mutation/Duplication | ICX | Age First Sign in Months | Microcephaly | Sitting Alone | Ambulation | Language | Epilepsy (y) | Respiratory Function | Hands Use | Onset of Stereotypies (m) | Score (y) | Kyphoscoliosis (y) | Growth Failure | Peripheral Vasomotor Deficits (y) | Bowel Control (y) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 19 | F | c.916C>T (p.R306C) (MS) | 63:37 | 12–24 | Yes | Yes | Acquired < 18 months | Lost | Controlled (8) | Hyperventilation | Partially conserved | 18–36 | 8 (18.5) | No | Yes | Yes (6) | Yes (9,33) |
| 2 | 32 | F | deletion exon 1&2 (NS) | 73:27 | 0–12 | Yes | Seat & lost | Never | Never | Controlled (0.42) | Apnea | Lost | >36 | 13 (23.9) | Yes (10) | No | Yes (6) | No |
| 3 | 7 | F | c.473C>T (p.T158M) (MS) | 64:36 | 12–24 | Yes | Yes | Acquired < 30 months | Lost | Controlled (7) | Hyperventilation | Lost | >36 | 8 (13.7) | Yes (5,75) | Yes | Yes (7) | No |
| 4 | 16 | F | c.473C>T (p.T158M) (MS) | 67:33 | 0–12 | Yes | Yes | Acquired < 18 months | Lost | Controlled | Hyperventilation | Lost | 18–36 | 8 (2.2) | No | Yes | Yes (6) | No |
| 5 | 23 | F | c.473C>T (p.T158M) (MS) | 77:23 | 0–12 | Yes | Yes | Lost | Never | Refractory (2) | Hyperventilation | Lost | <18 | 14 (6.3) | Yes (6) | - | Yes (3,5) | No |
| 6 | 33 | F | c.916C>T (p.R306C) (MS) | 79:21 | 0–12 | Yes | Yes | Acquired > 30 months | Never | Controlled (8) | Hyperventilation + apnea | Lost | >36 | 10 (23.7) | Yes (9) | Yes | Yes (9) | No |
| 7 | 28 | F | c.763C>T (p.R255X) (NS) | 73:27 | 0–12 | Yes | Yes | Lost | Lost | Refractory (7) | Hyperventilation | Lost | 18–36 | 14 (19.5) | Yes (12) | Yes | Yes (10) | No |
| 8 | 25 | F | c.880C>T (p.R294X) (NS) | 59:41 | 12–24 | Yes | Yes | Acquired < 30 months | Lost | Controlled (6) | Hyperventilation | Lost | 18–36 | 11 (22.1) | Yes (9) | Yes | No | No |
| 9 | 10 | F | c.473C>T (p.T158M) (MS) | 65:35 | 0–12 | Yes | Seat & lost | Lost | Never | Controlled (5) | Hyperventilation | Never | <18 | 11(9.1) | No | No | Yes (5) | No |
| 10 | 14 | F | deletion exon 3&4 (NS) | 62:38 | 0–12 | No | Yes | Acquired < 18 months | Lost | Controlled (4.5) | Hyperventilation | Lost | 18–36 | 7 (13) | No | No | No | Yes (5) |
| 11 | 11 | F | c.808C>T (p.R270X) (NS) | 88:12 | 0–12 | Yes | Yes | Acquired < 18 months | Conserved | Controlled (3,25) | Hyperventilation | Partially conserved | >36 | 7(3.5) | No | - | No | No |
| 12 | 15 | F | c.961_1188delins220 (p.K321G fs X6) (NS) | 68:32 | >24 | No | Yes | Acquired <18 months | Lost | No | No dysfunction | Partially conserved | >36 | 5 (14.7) | Yes (14) | No | No | Yes (5,75) |
| 13 | 16 | F | c.916C>T (p.R306C) (MS) | 78:22 | >24 | No | Yes | Acquired < 18 months | Conserved | No | No dysfunction | Lost | >36 | 3 (4.7) | No | - | No | No |
| 14 | 13 | F | c.1189G>T (p.Q397X) (NS) | 68:32 | 12–24 | No | Yes | Acquired < 18 months | Lost | No | No dysfunction | Lost | 18–36 | 5 (5.2) | No | No | No | No |
| 15 | 16 | F | c.276_281del4 (p.P93fs) (N) | 81:19 | 12–24 | Yes | Yes | Lost | Lost | Controlled (3) | Hyperventilation | Lost | 18–36 | 8 (15.1) | No | No | Yes (6) | No |
| 16 | 5 | F | c.473C>T (p.T158M) (MS) | 70:30 | 0–12 | Yes | Yes | Lost | Lost | Refractory (3) | Hyperventilation | Lost | <18 | 12 (4.6) | Yes (2) | No | Yes (4) | Yes (4) |
| 17 | 5 | F | c.808C>T (p.R270X) (NS) | 52:48 | 0–12 | Yes | Yes | Never | Lost | Controlled (2) | Hyperventilation | Lost | 18–36 | 15 (4.5) | No | - | No | No |
| 18 | 8 | F | c.916C>T (p.R306C) (MS) | 66:34 | 0–12 | No | Yes | Acquired < 30 months | Conserved | Controlled (6) | Hyperventilation | Lost | 18–36 | 10 (6.8) | No | No | No | No |
| 19 | 8 | F | c.502C>T (p.R168X) (NS) | 54:46 | 0–12 | Yes | Yes | Acquired <18 months | Conserved | Controlled (5) | No dysfunction | Lost | 18–36 | 8 (7.2) | No | No | Yes (5) | Yes (7) |
| 20 | 8 | F | c.316C>T (p.R106W) (MS) | 59:41 | 0–12 | Yes | Yes | Never | Never | Refractory (3) | Hyperventilation | Lost | <18 | 15 (6.8) | Yes (5) | No | Yes (4,5) | No |
| 21 | 1 | M | Duplication chrX: 149116213–154929279 | - | 0–12 | No | No | - | Never | Yes (1) | - | - | <18 | - | No | - | - | No |
| 22 | 4 | M | Duplication chrX: 153101077–153565901 | - | 0–12 | No | Yes | Acquired <18 months | Never | Yes (6) | - | Partially conserved | <18 | - | No | - | - | - |
| 23 | >30 | F | Duplication chrX: 153101077–153565901 | - | Asymptomatic | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Controls (n = 21) | RTT (n= 16) | MDS (n = 2) | MDC (n = 1) | |
|---|---|---|---|---|
| mRNA (RQ) | ||||
| CACNA2D2 | 1.07 ± 0.36 [0.88–1.25] | 1.20 ± 0.62 [0.84–1.56] | 0.85 ± 0.55 | 1.37 |
| DNMT1 | 1.03 ± 0.24 [0.91–1.15] | 1.11 ± 0.32 [0.94–1.28] | 1.17 ± 0.20 | 1.12 |
| IRAK1 | 1.02 ± 0.21 [0.92–1.13] | 1.04 ± 0.24 [0.87–1.22] | 2.52 ± 0.25 * | 0.97 |
| LIN28A | 1.28 ± 0.84 [0.89–1.68] | 0.73 ± 0.70 [0.34–1.12] * | 0.75 ± 0.02 | 1.33 |
| MECP2 E1 | 1.04 ± 0.29 [0.89–1.20] | 1.78 ± 0.91 [1.02–2.54] * | 1.85 ± 0.38 * | 0.94 |
| MECP2 E2 | 1.03 ± 0.26 [0.90–1.17] | 1.58 ± 0.87 [0.96–2.21] ** | 1.71 ± 0.51 * | 1.01 |
| PTEN | 1.04 ± 0.31 [0.87–1.22] | 0.91 ± 0.50 [0.55–1.27] | 0.84 ± 0.14 | 0.92 |
| THBS1 | 1.22 ± 0.88 [0.76–1.67] | 1.67 ± 1.54 [0.81–2.52] | 1.22 ± 0.81 | 1.39 |
| THBS3 | 1.02 ± 0.21 [0.91–1.13] | 1.46 ± 0.43 [1.21–1.71] *** | 0.87 ± 0.08 | - |
| Protein (ng/mL) | ||||
| BDNF | 128.68 ± 87.25 [87.85–169-51] | 80.55 ± 50.55 [52.11–188.55] | - | - |
| AUC | DE | p Value | 95% CI | J | Trend | n | |
|---|---|---|---|---|---|---|---|
| RTT vs. Controls | |||||||
| THBS3 | 0.840 | 0.083 | 0.001 | 0.678–1 | 0.727 | ↑ | 14 vs. 17 |
| LIN28A | 0.747 | 0.087 | 0.014 | 0.576–0.917 | 0.467 | ↓ | 15 vs. 20 |
| LIN28A + THBS3 | 0.899 | 0.057 | 0.0003 | 0.787–1 | 0.687 | 13 vs. 16 | |
| MECP2 mutation type (MS vs. NS) | |||||||
| BDNF | 0.811 | 0.099 | 0.022 | 0.617–1 | 0.500 | ↓ | 10 vs. 9 |
| Ratio | FC | p-Value | AUC | p-Value | 95% CI | J | Trend | n | |
|---|---|---|---|---|---|---|---|---|---|
| RTT vs. Controls | |||||||||
| miR-24-3p/miR-132-3p | 1.143 | 2.209 | 0.01 | 0.844 | 0.011 | 0.64–1 | 0.778 | ↑ | 10 vs. 9 |
| miR-146a-5p/miR-132-3p | 1.046 | 2.065 | 0.056 | 0.758 | 0.053 | 0.505–1 | 0.667 | ↑ | 11 vs. 9 |
| Let-7a-5p/miR-132-3p | 0.74 | 1.670 | 0.067 | 0.747 | 0.063 | 0.495–1 | 0.687 | ↑ | 11 vs. 9 |
| miR-24-3p/miR-132-3p + miR-146a-5p/miR-132-3p | 0.9 | 0.003 | 0.954–1 | 1 | 10 vs. 9 | ||||
| RTT vs. Controls >15 years | |||||||||
| Let-7a-5p/miR-132-3p | 1.471 | 2.772 | 0.02 | 0.938 | 0.007 | 0.804–1 | 0.875 | ↑ | 6 vs. 8 |
| miR-24-3p/miR-132-3p | 1.453 | 2.737 | 0.005 | 0.917 | 0.01 | 0.751–1 | 0.875 | ↑ | 6 vs. 8 |
| miR-146a-5p/miR-132-3p | 1.685 | 3.214 | 0.008 | 0.875 | 0.02 | 0.671–1 | 0.75 | ↑ | 6 vs. 8 |
| Ratio | FC | p-Value | AUC | p-Value | 95% CI | J | Trend | n | |
|---|---|---|---|---|---|---|---|---|---|
| Microcephaly (Yes vs. No) | |||||||||
| Let-7a-5p/miR-125b-5p | 2.693 | 6.466 | 0.025 | 0.963 | 0.021 | 0.857–1 | 0.889 | ↑ | 9 vs. 3 |
| Let-7a-5p/miR-16-5p | 2.397 | 5.266 | 0.022 | 0.924 | 0.029 | 0.779–1 | 0.818 | ↑ | 11 vs. 3 |
| miR-146a-5p/miR-132-3p | 1.22 | 2.33 | 0.003 | 0.917 | 0.041 | 0.731–1 | 0.750 | ↑ | 8 vs. 3 |
| Any combination of two miRNA ratios | 1 | 0.013–0.02 | 1–1 | 1 | |||||
| Vasomotor deficits (Yes vs. No) | |||||||||
| miR-146a-5p/miR-132-3p | 1.438 | 2.709 | 0.002 | 0.964 | 0.014 | 0.859–1 | 0.857 | ↑ | 7 vs. 4 |
| Let-7a-5p/ miR-16-5p | 2.501 | 5.659 | 0.004 | 0.944 | 0.008 | 0.827–1 | 0.778 | ↑ | 9 vs. 5 |
| miR-132-3p /miR-16-5p | 1.187 | 2.278 | 0.017 | 0.933 | 0.018 | 0.78–1 | 0.833 | ↑ | 6 vs. 5 |
| miR-24-3p /miR-16-5p | 2.244 | 4.736 | 0.019 | 0.889 | 0.020 | 0.713–1 | 0.689 | ↑ | 9 vs. 5 |
| Let-7a-5p/miR-125b-5p | 2.232 | 4.696 | 0.04 | 0.857 | 0.042 | 0.638–1 | 0.714 | ↑ | 7 vs. 5 |
| miR-16-5p/ miR-146a-5p | -2.266 | 0.208 | 0.042 | 0.844 | 0.039 | 0.623–1 | 0.778 | ↓ | 9 vs. 5 |
| Let-7a-5p/ miR-16-5p + miR-146a-5p/miR-132-3p or miR-132-3p /miR-16-5p or Let-7a-5p/miR-125b-5p | 1 | 0.004–0.008 | 1–1 | 1 |
| Ratio | FC | p-Value | AUC | p-Value | 95% CI | J | Trend | n | |
|---|---|---|---|---|---|---|---|---|---|
| RTT vs. Controls | |||||||||
| miR-146a-5p/miR-132-3p | −2.461 | 0.182 | 0.004 | 0.969 | 0.011 | 0.876–1 | 0.875 | ↓ | 8 vs. 4 |
| miR-24-3p/miR-132-3p | −0.719 | 0.608 | 0.027 | 0.844 | 0.0617 | 0.612–1 | 0.625 | ↓ | 8 vs. 4 |
| Let-7a-5p/miR-132-3p | 1.388 | 2.636 | 0.086 | 0.844 | 0.062 | 0.617–1 | 0.75 | ↑ | 8 vs. 4 |
| Trend | |
|---|---|
| RTT | |
| miR-24-3p/miR-132-3p + miR-146a-5p/miR-132-3p | ↑ |
| Microcephaly | |
| Let-7a-5p/miR-125b-5p + Let-7a-5p/miR-16-5p | ↑ |
| Let-7a-5p/ miR-16-5p + miR-146a-5p/miR-132-3p | ↑ |
| Let-7a-5p/miR-125b-5p + miR-146a-5p/miR-132-3p | ↑ |
| Vasomotor deficits | |
| Let-7a-5p/miR-16-5p + miR-132-3p/miR-16-5p | ↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castells, A.-A.; Balada, R.; Tristán-Noguero, A.; O’Callaghan, M.; Cortès-Saladelafont, E.; Pascual-Alonso, A.; Garcia-Cazorla, À.; Armstrong, J.; Alcántara, S. Unraveling Molecular Pathways Altered in MeCP2-Related Syndromes, in the Search for New Potential Avenues for Therapy. Biomedicines 2021, 9, 148. https://doi.org/10.3390/biomedicines9020148
Castells A-A, Balada R, Tristán-Noguero A, O’Callaghan M, Cortès-Saladelafont E, Pascual-Alonso A, Garcia-Cazorla À, Armstrong J, Alcántara S. Unraveling Molecular Pathways Altered in MeCP2-Related Syndromes, in the Search for New Potential Avenues for Therapy. Biomedicines. 2021; 9(2):148. https://doi.org/10.3390/biomedicines9020148
Chicago/Turabian StyleCastells, Alba-Aina, Rafel Balada, Alba Tristán-Noguero, Mar O’Callaghan, Elisenda Cortès-Saladelafont, Ainhoa Pascual-Alonso, Àngels Garcia-Cazorla, Judith Armstrong, and Soledad Alcántara. 2021. "Unraveling Molecular Pathways Altered in MeCP2-Related Syndromes, in the Search for New Potential Avenues for Therapy" Biomedicines 9, no. 2: 148. https://doi.org/10.3390/biomedicines9020148
APA StyleCastells, A.-A., Balada, R., Tristán-Noguero, A., O’Callaghan, M., Cortès-Saladelafont, E., Pascual-Alonso, A., Garcia-Cazorla, À., Armstrong, J., & Alcántara, S. (2021). Unraveling Molecular Pathways Altered in MeCP2-Related Syndromes, in the Search for New Potential Avenues for Therapy. Biomedicines, 9(2), 148. https://doi.org/10.3390/biomedicines9020148






