Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy
Abstract
1. Introduction
2. Experimental Section
2.1. Cell Culturing
2.2. Vesicle Collection
2.3. Differential Ultracentrifugation (UC)
2.4. Quantification of Microvesicles by Nanoparticle Tracking Analysis (NTA)
2.5. Sample Preparation & Nano-LC-MS/MS
2.6. MS Data Processing
2.7. Bioinformatics Analysis
2.8. Data Availability
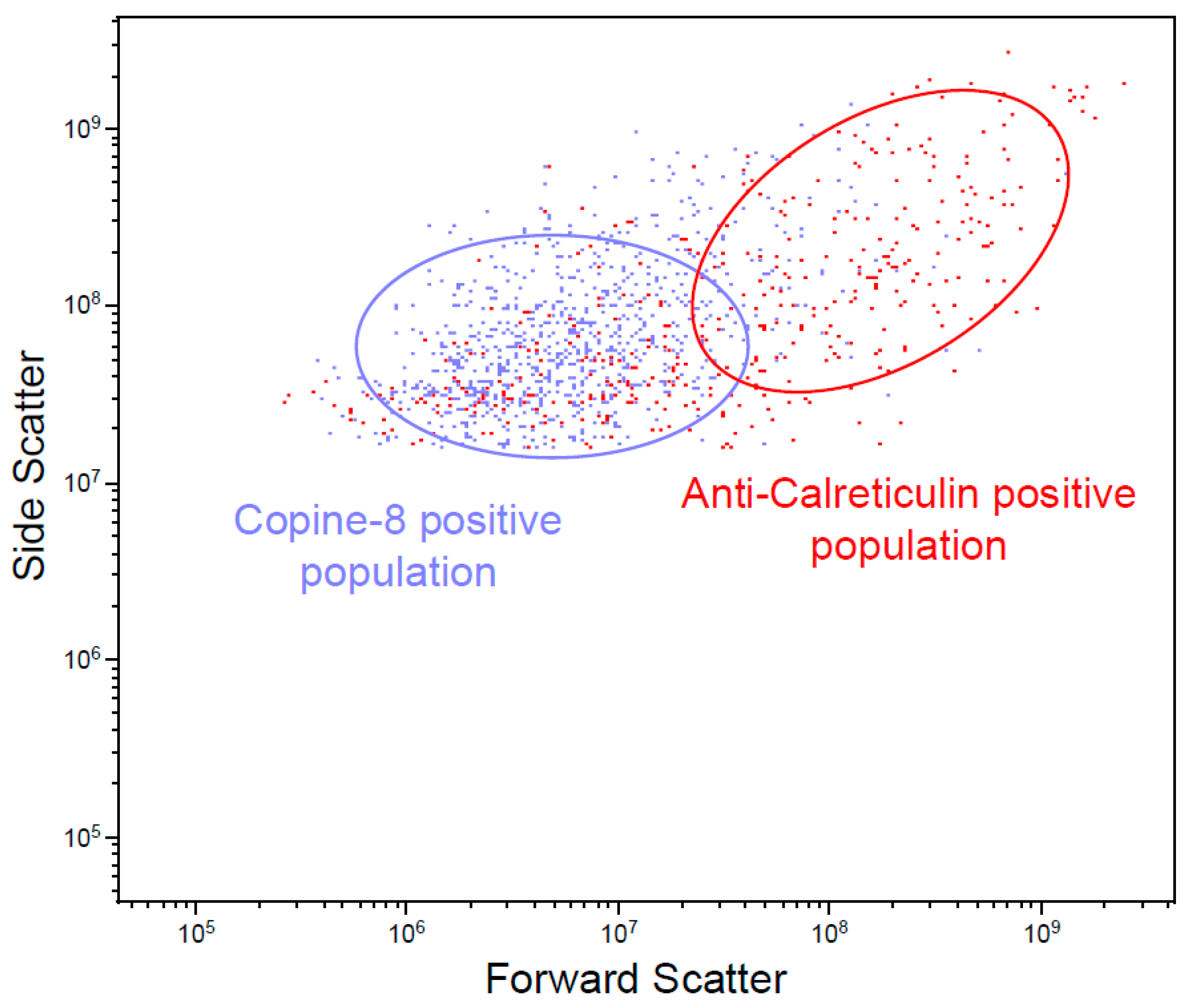
2.9. Flow Cytometry
2.10. Ornithine Aminotransferase Activity Assay
2.11. Transaldolase Activity Assay
2.12. Measurement of Human Bleomycin Hydrolase
3. Results
3.1. Characterization of MVs
3.2. Proteomics of Cell Line-Derived MVs
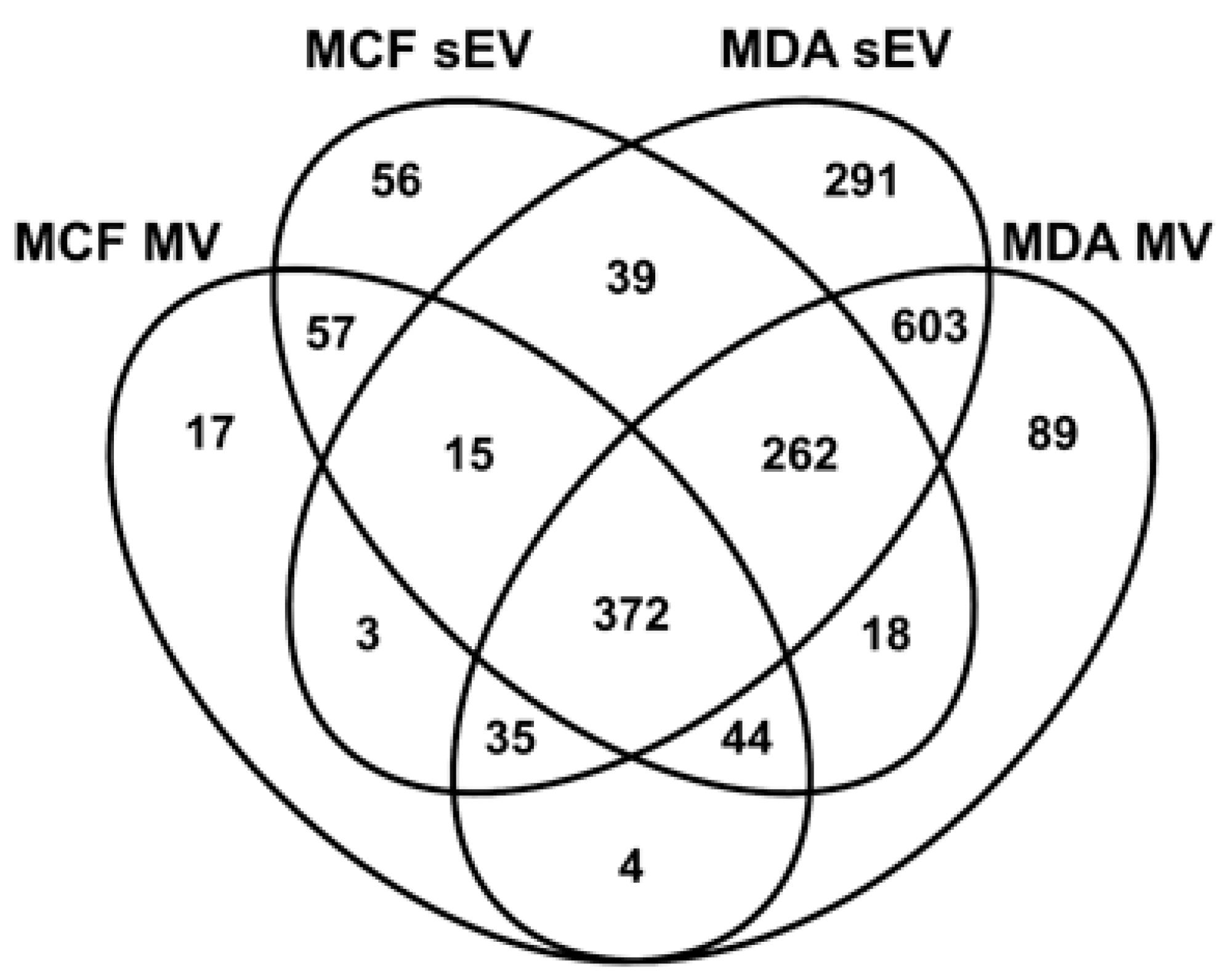
3.3. Comparative Proteomic Analysis of sEVs and MVs
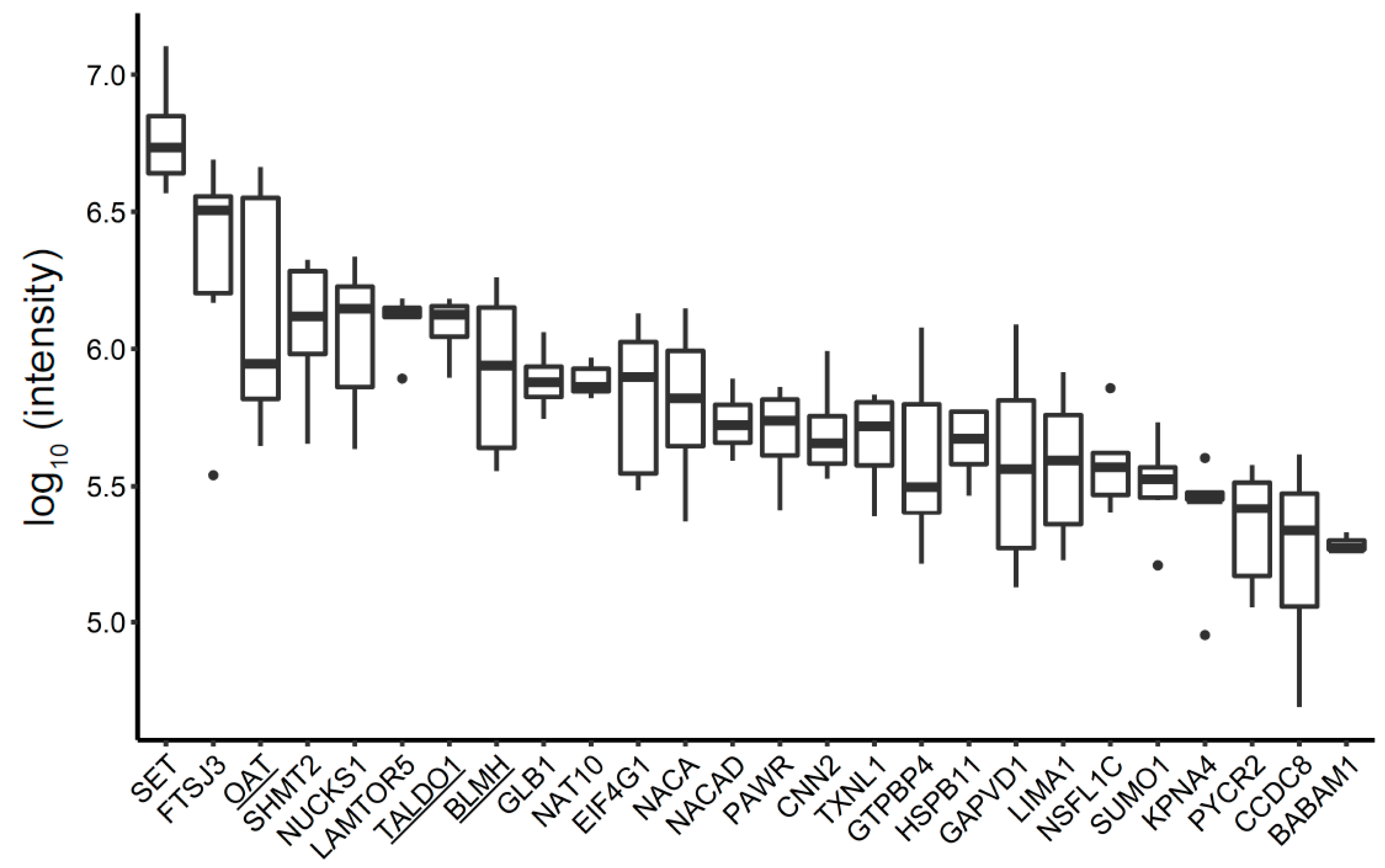
3.4. Unique BC MV Proteins
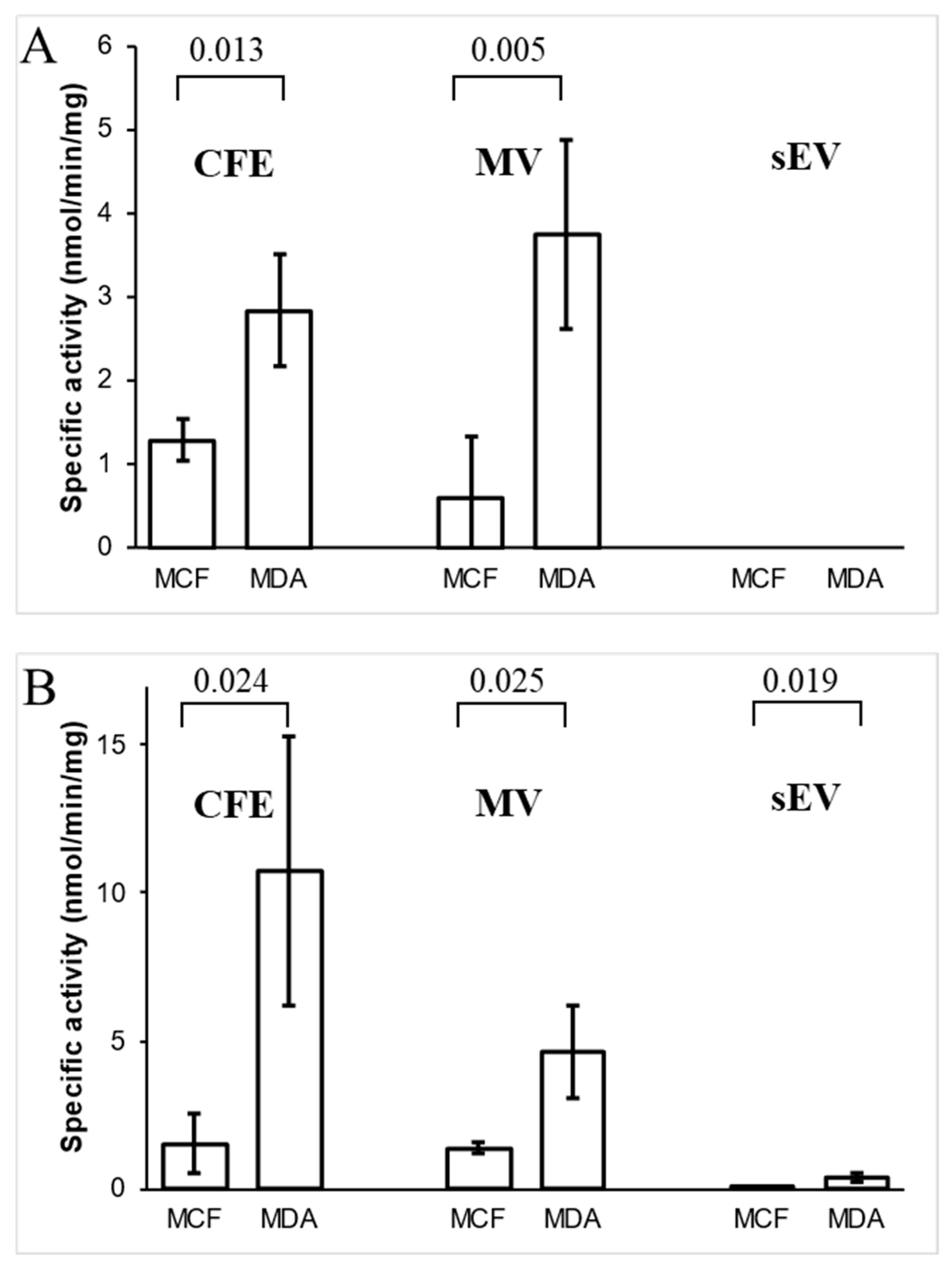
3.5. Analysis of Ornithine Aminotransferase and Transaldolase in Cells and Their EVs
3.6. Concentrations of Bleomycin Hydrolase in Cells and Their EVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Global Cancer Observatory (GCO), Cancer Today. IARC, Lyon. 2018. Available online: https://gco.iarc.fr (accessed on 11 October 2019).
- Hendrick, R.E.; Baker, J.A.; Helvie, M.A. Breast Cancer Deaths Averted over 3 Decades. Cancer 2019, 125, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Carioli, G.; Malvezzi, M.; Rodriguez, T.; Bertuccio, P.; Negri, E.; Vecchia, C.L. Trends and Predictions to 2020 in Breast Cancer Mortality in Europe. Breast 2017, 36, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Mariotto, A.B.; Etzioni, R.; Hurlbert, M.; Penberthy, L.; Mayer, M. Estimation of the Number of Women Living with Metastatic Breast Cancer in the United States. Cancer Epidemiol. Biomark. Prev. 2017, 26, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Calvo, F.; Sahai, E. Cell Communication Networks in Cancer Invasion. Curr. Opin. Cell Biol. 2011, 23, 621–629. [Google Scholar] [CrossRef]
- Kapoor, P.; Saunders, M.M.; Li, Z.; Zhou, Z.; Sheaffer, N.; Kunze, E.L.; Samant, R.S.; Welch, D.R.; Donahue, H.J. Breast Cancer Metastatic Potential: Correlation with Increased Heterotypic Gap Junctional Intercellular Communication between Breast Cancer Cells and Osteoblastic Cells. Int. J. Cancer 2004, 111, 693–697. [Google Scholar] [CrossRef]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Henrique Baeta, H.; Moraes, M.C.S.; Beck, H.C.; Rodriguez, M.S.; Saraswat, M.; Pandey, A.; Matthiesen, R. Extra-Cellular Vesicles Carry Proteome of Cancer Hallmarks. Front. Biosci. 2020, 25, 398–436. [Google Scholar]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; Mccague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic Profiling of Extracellular Vesicles Derived from Prostate and Prostate Cancer Cell Lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular Vesicle Isolation and Characterization: Toward Clinical Application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Vizio, D.D. Extracellular Vesicles in Cancer: Exosomes, Microvesicles and the Emerging Role of Large Oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Vizio, D.D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large Oncosomes in Human Prostate Cancer Tissues and in the Circulation of Mice with Metastatic Disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Li, I.; Nabet, B.Y. Exosomes in the Tumor Microenvironment as Mediators of Cancer Therapy Resistance. Mol. Cancer 2019, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jayaseelan, V.P. Emerging Role of Exosomes as Promising Diagnostic Tool for Cancer. Cancer Gene Ther. 2019, 27, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Du, S.-L.; Zhang, J.; Liang, A.-L.; Liu, Y.-J. Exosomes and Breast Cancer: A Comprehensive Review of Novel Therapeutic Strategies from Diagnosis to Treatment. Cancer Gene Ther. 2016, 24, 6–12. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Wang, Q.; Liu, Y.; Bao, W.; Wu, S. Exosomes in Cancer: Small Transporters with Big Functions. Cancer Lett. 2018, 435, 55–65. [Google Scholar] [CrossRef]
- An, M.; Zhu, J.; Wu, J.; Cuneo, K.C.; Lubman, D.M. Circulating Microvesicles from Pancreatic Cancer Accelerate the Migration and Proliferation of PANC-1 Cells. J. Proteome Res. 2018, 17, 1690–1699. [Google Scholar] [CrossRef]
- Huaitong, X.; Yuanyong, F.; Yueqin, T.; Peng, Z.; Wei, S.; Kai, S. Microvesicles Releasing by Oral Cancer Cells Enhance Endothelial Cell Angiogenesis via Shh/RhoA Signaling Pathway. Cancer Biol. Ther. 2017, 18, 783–791. [Google Scholar] [CrossRef]
- Menck, K.; Bleckmann, A.; Wachter, A.; Hennies, B.; Ries, L.; Schulz, M.; Balkenhol, M.; Pukrop, T.; Schatlo, B.; Rost, U.; et al. Characterisation of Tumour-Derived Microvesicles in Cancer Patients’ Blood and Correlation with Clinical Outcome. J. Extracell. Vesicles 2017, 6, 1340745. [Google Scholar] [CrossRef]
- Das, K.; Paul, S.; Singh, A.; Ghosh, A.; Roy, A.; Ansari, S.A.; Prasad, R.; Mukherjee, A.; Sen, P. Triple-Negative Breast Cancer-Derived Microvesicles Transfer microRNA221 to the Recipient Cells and Thereby Promote Epithelial-to-Mesenchymal Transition. J. Biol. Chem. 2019, 294, 13681–13696. [Google Scholar] [CrossRef]
- Pan, Y.; Li, J.; Zhang, Y.; Wang, N.; Liang, H.; Liu, Y.; Zhang, C.-Y.; Zen, K.; Gu, H. Slug-Upregulated MiR-221 Promotes Breast Cancer Progression through Suppressing E-Cadherin Expression. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C.; et al. Tumor-Derived Microvesicles Mediate Human Breast Cancer Invasion through Differentially Glycosylated EMMPRIN. J. Mol. Cell Biol. 2014, 7, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.; Johnson, M.S.; Pokharel, D.; Krishnan, S.R.; Bebawy, M. Microparticles Shed from Multidrug Resistant Breast Cancer Cells Provide a Parallel Survival Pathway through Immune Evasion. BMC Cancer 2017, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Maes, E.; Mertens, I.; Valkenborg, D.; Pauwels, P.; Rolfo, C.; Baggerman, G. Proteomics in Cancer Research: Are We Ready for Clinical Practice? Crit. Rev. Oncol. Hematol. 2015, 96, 437–448. [Google Scholar] [CrossRef]
- Moya-Alvarado, G.; Gershoni-Emek, N.; Perlson, E.; Bronfman, F.C. Neurodegeneration and Alzheimer’s Disease (AD). What Can Proteomics Tell Us About the Alzheimer’s Brain? Mol. Cell. Proteom. 2015, 15, 409–425. [Google Scholar] [CrossRef]
- Lindsey, M.L.; Mayr, M.; Gomes, A.V.; Delles, C.; Arrell, D.K.; Murphy, A.M.; Lange, R.A.; Costello, C.E.; Jin, Y.-F.; Laskowitz, D.T.; et al. Transformative Impact of Proteomics on Cardiovascular Health and Disease. Circulation 2015, 132, 852–872. [Google Scholar] [CrossRef]
- Barnabas, G.D.; Bahar-Shany, K.; Sapoznik, S.; Helpman, L.; Kadan, Y.; Beiner, M.; Weitzner, O.; Arbib, N.; Korach, J.; Perri, T.; et al. Microvesicle Proteomic Profiling of Uterine Liquid Biopsy for Ovarian Cancer Early Detection. Mol. Cell. Proteom. 2019, 18, 865–875. [Google Scholar] [CrossRef]
- Pokharel, D.; Padula, M.P.; Lu, J.F.; Tacchi, J.L.; Luk, F.; Djordjevic, S.P.; Bebawy, M. Proteome Analysis of Multidrug-Resistant, Breast Cancer–Derived Microparticles. J. Extracell. Vesicles 2014, 3, 24384. [Google Scholar] [CrossRef]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The Proteomic Analysis of Breast Cell Line Exosomes Reveals Disease Patterns and Potential Biomarkers. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized P.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 15 December 2020).
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-Associated Genes and Variants. Nucleic Acids Res. 2016, 45, D1. [Google Scholar] [CrossRef]
- Bradford, N.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kumar, H.R.; Ananda, S.; Devaraju, K.S.; Prakash, B.M.; Kumar, S.S.; Babu, S.V.S.; Ramachandraswamy, N.; Puttaraju, H.P. A Sensitive Assay for Ornithine Amino Transferase in Rat Brain Mitochondria by Ninhydrin Method. Indian J. Clin. Biochem. 2009, 24, 275–279. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsolas, O.; Joris, L. Transaldolase. Carbohydr. Metab. Part C Methods Enzymol. 1975, 42, 290–297. [Google Scholar]
- Sun, Y.; Huo, C.; Qiao, Z.; Shang, Z.; Uzzaman, A.; Liu, S.; Jiang, X.; Fan, L.-Y.; Ji, L.; Guan, X.; et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J. Proteome Res. 2018, 17, 1101–1107. [Google Scholar] [CrossRef]
- Kang, M.; Kim, S.; Ko, J. Roles of CD133 in Microvesicle Formation and Oncoprotein Trafficking in Colon Cancer. Faseb J. 2018, 33, 4248–4260. [Google Scholar] [CrossRef]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’souza-Schorey, C. ARF6-Regulated Shedding of Tumor Cell-Derived Plasma Membrane Microvesicles. Curr. Biol. 2009, 19, 1875–1885. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Didiot, M.-C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; Difiglia, M.; Kiebish, M.A.; et al. High-Resolution Proteomic and Lipidomic Analysis of Exosomes and Microvesicles from Different Cell Sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Asadi, S.; Weng, Z.; Sismanopoulos, N.; Theoharides, T.C. Stimulated Human Mast Cells Secrete Mitochondrial Components That Have Autocrine and Paracrine Inflammatory Actions. PLoS ONE 2012, 7, e49767. [Google Scholar] [CrossRef] [PubMed]
- Puhm, F.; Afonyushkin, T.; Resch, U.; Obermayer, G.; Rohde, M.; Penz, T.; Schuster, M.; Wagner, G.; Rendeiro, A.F.; Melki, I.; et al. Mitochondria Are a Subset of Extracellular Vesicles Released by Activated Monocytes and Induce Type I IFN and TNF Responses in Endothelial Cells. Circ. Res. 2019, 125, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Das, S.R.; Emin, M.T.; Wei, M.; Sun, L.; Westphalen, K.; Rowlands, D.J.; Quadri, S.K.; Bhattacharya, S.; Bhattacharya, J. Mitochondrial Transfer from Bone-Marrow–Derived Stromal Cells to Pulmonary Alveoli Protects against Acute Lung Injury. Nat. Med. 2012, 18, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.V.; Morrison, T.J.; Doherty, D.F.; Mcauley, D.F.; Matthay, M.A.; Kissenpfennig, A.; O’kane, C.M.; Krasnodembskaya, A.D. Mitochondrial Transfer via Tunneling Nanotubes Is an Important Mechanism by Which Mesenchymal Stem Cells Enhance Macrophage Phagocytosis in the In Vitro and In Vivo Models of ARDS. Stem Cells 2016, 34, 2210–2223. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Granata, S.; Santucci, L.; Candiano, G.; Fabris, A.; Antonucci, N.; Petretto, A.; Bartolucci, M.; Zotto, G.D.; Antonini, F.; et al. Proteomic Analysis of Urinary Microvesicles and Exosomes in Medullary Sponge Kidney Disease and Autosomal Dominant Polycystic Kidney Disease. Clin. J. Am. Soc. Nephrol. 2019, 14, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Tomsig, J.L.; Creutz, C.E. Copines: A Ubiquitous Family of Ca 2+ -Dependent Phospholipid-Binding Proteins. Cell. Mol. Life Sci. 2002, 59, 1467–1477. [Google Scholar] [CrossRef]
- Maitra, R.; Grigoryev, D.N.; Bera, T.K.; Pastan, I.H.; Lee, B. Cloning, Molecular Characterization, and Expression Analysis of Copine 8. Biochem. Biophys. Res. Commun. 2003, 303, 842–847. [Google Scholar] [CrossRef][Green Version]
- Ramsey, H.; Zhang, D.-E.; Richkind, K.; Burcoglu-O’ral, A.; Hromas, R. Fusion of AML1/Runx1 to Copine VIII, a Novel Member of the Copine Family, in an Aggressive Acute Myelogenous Leukemia with t(12;21) Translocation. Leukemia 2003, 17, 1665–1666. [Google Scholar] [CrossRef][Green Version]
- Ramsey, C.S.; Yeung, F.; Stoddard, P.B.; Li, D.; Creutz, C.E.; Mayo, M.W. Copine-I Represses NF-ΚB Transcription by Endoproteolysis of p65. Oncogene 2008, 27, 3516–3526. [Google Scholar] [CrossRef]
- Liu, S.; Tang, H.; Zhu, J.; Ding, H.; Zeng, Y.; Du, W.; Ding, Z.; Song, P.; Zhang, Y.; Liu, Z.; et al. High Expression of Copine 1 Promotes Cell Growth and Metastasis in Human Lung Adenocarcinoma. Int. J. Oncol. 2018, 53, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.K.B.A.; Shimizu, A.; Zankov, D.P.; Sato, A.; Kurita, S.; Ito, M.; Maeda, T.; Yoshida, T.; Sakaue, T.; Higashiyama, S.; et al. Epithelial Membrane Protein 1 Promotes Tumor Metastasis by Enhancing Cell Migration via Copine-III and Rac1. Oncogene 2018, 37, 5416–5434. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, C.; Keller, C.; Boulay, A.; Vecchi, M.; Bianchi, M.; Sack, R.; Lienhard, S.; Duss, S.; Hofsteenge, J.; Hynes, N.E. Copine-III Interacts with ErbB2 and Promotes Tumor Cell Migration. Oncogene 2009, 29, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Umeda, S.; Kanda, M.; Koike, M.; Tanaka, H.; Miwa, T.; Tanaka, C.; Kobayashi, D.; Suenaga, M.; Hayashi, M.; Yamada, S.; et al. Copine 5 Expression Predicts Prognosis Following Curative Resection of Esophageal Squamous Cell Carcinoma. Oncol. Rep. 2018, 40, 3772–3780. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, S.; Pallero, M.A.; Eggleton, P.; Michalak, M.; Murphy-Ullrich, J.E. The Anti-Adhesive Activity of Thrombospondin Is Mediated by the N-Terminal Domain of Cell Surface Calreticulin. J. Biol. Chem. 2002, 277, 37219–37228. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.I.; Eggleton, P.; Sweetwyne, M.T.; Duyn, L.B.V.; Greives, M.R.; Naylor, S.M.; Michalak, M.; Murphy-Ullrich, J.E. Calreticulin: Non-Endoplasmic Reticulum Functions in Physiology and Disease. Faseb J. 2009, 24, 665–683. [Google Scholar] [CrossRef]
- Chen, C.-N.; Chang, C.-C.; Su, T.-E.; Hsu, W.-M.; Jeng, Y.-M.; Ho, M.-C.; Hsieh, F.-J.; Lee, P.-H.; Kuo, M.-L.; Lee, H.; et al. Identification of Calreticulin as a Prognosis Marker and Angiogenic Regulator in Human Gastric Cancer. Ann. Surg. Oncol. 2008, 16, 524–533. [Google Scholar] [CrossRef]
- Du, X.-L.; Hu, H.; Lin, D.-C.; Xia, S.-H.; Shen, X.-M.; Zhang, Y.; Luo, M.-L.; Feng, Y.-B.; Cai, Y.; Xu, X.; et al. Proteomic Profiling of Proteins Dysregulted in Chinese Esophageal Squamous Cell Carcinoma. J. Mol. Med. 2007, 85, 863–875. [Google Scholar] [CrossRef]
- Chiang, W.-F.; Hwang, T.-Z.; Hour, T.-C.; Wang, L.-H.; Chiu, C.-C.; Chen, H.-R.; Wu, Y.-J.; Wang, C.-C.; Wang, L.-F.; Chien, C.-Y.; et al. Calreticulin, an Endoplasmic Reticulum-Resident Protein, Is Highly Expressed and Essential for Cell Proliferation and Migration in Oral Squamous Cell Carcinoma. Oral Oncol. 2013, 49, 534–541. [Google Scholar] [CrossRef]
- Chahed, K.; Kabbage, M.; Ehret-Sabatier, L.; Lemaitre-Guillier, C.; Remadi, S.; Hoebeke, J.; Chouchane, L. Expression of Fibrinogen E-Fragment and Fibrin E-Fragment Is Inhibited in the Human Infiltrating Ductal Carcinoma of the Breast: The Two-Dimensional Electrophoresis and MALDI-TOF-Mass Spectrometry Analyses. Int. J. Oncol. 2005, 27, 1425–1431. [Google Scholar] [CrossRef]
- Hellman, K.; Alaiya, A.A.; Schedvins, K.; Steinberg, W.; Hellström, A.-C.; Auer, G. Protein Expression Patterns in Primary Carcinoma of the Vagina. Br. J. Cancer 2004, 91, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Sivashanmugam, M.; Jaidev, J.; Umashankar, V.; Sulochana, K.N. Ornithine and Its Role in Metabolic Diseases: An Appraisal. Biomed. Pharmacother. 2017, 86, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tao, B.; Chong, Y.; Ma, S.; Wu, G.; Zhu, H.; Zhao, Y.; Zhao, S.; Niu, M.; Zhang, S.; et al. Ornithine and Breast Cancer: A Matched Case–Control Study. Sci. Rep. 2020, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zigmond, E.; Ya’Acov, A.B.; Lee, H.; Lichtenstein, Y.; Shalev, Z.; Smith, Y.; Zolotarov, L.; Ziv, E.; Kalman, R.; Le, H.V.; et al. Suppression of Hepatocellular Carcinoma by Inhibition of Overexpressed Ornithine Aminotransferase. Acs Med. Chem. Lett. 2015, 6, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Miyasaka, Y.; Enomoto, N.; Nagayama, K.; Izumi, N.; Marumo, F.; Watanabe, M.; Sato, C. Analysis of Differentially Expressed Genes in Human Hepatocellular Carcinoma Using Suppression Subtractive Hybridization. Br. J. Cancer 2001, 85, 228–234. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Klose, P.; Weise, C.; Bondzio, A.; Multhaup, G.; Einspanier, R.; Gruber, A.D. Proteome of Metastatic Canine Mammary Carcinomas: Similarities to and Differences from Human Breast Cancer. J. Proteome Res. 2010, 9, 6380–6391. [Google Scholar] [CrossRef]
- Li, W.; Zhang, W.; Deng, W.; Zhong, Y.; Zhang, Y.; Peng, Z.; Chen, H.; Sun, R.; Zhang, X.; Yang, S. Quantitative Proteomic Analysis of Mitochondrial Proteins Differentially Expressed between Small Cell Lung Cancer Cells and Normal Human Bronchial Epithelial Cells. Thorac. Cancer 2018, 9, 1366–1375. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, L.; Li, K.; Liu, F.; Wang, L.; Zhang, D.; Zhou, J.; Ma, X.; Wang, S.; Yang, S. Ornithine Aminotransferase Promoted the Proliferation and Metastasis of Non-Small Cell Lung Cancer via Upregulation of MiR-21. J. Cell. Physiol. 2018, 234, 12828–12838. [Google Scholar] [CrossRef]
- Perl, A.; Hanczko, R.; Telarico, T.; Oaks, Z.; Landas, S. Oxidative Stress, Inflammation and Carcinogenesis Are Controlled through the Pentose Phosphate Pathway by Transaldolase. Trends Mol. Med. 2011, 17, 395–403. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The Pentose Phosphate Pathway and Cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Silvers, C.R.; Miyamoto, H.; Messing, E.M.; Netto, G.J.; Lee, Y.-F. Characterization of Urinary Extracellular Vesicle Proteins in Muscle-Invasive Bladder Cancer. Oncotarget 2017, 8, 91199–91208. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, P.C.; Morris, H.P.; Weber, G. Behavior of transaldolase (EC 2.2.1.2) and transketolase (EC 2.2.1.1) Activities in normal, neoplastic, differentiating, and regenerating liver. Cancer Res. 1976, 36, 3189–3197. [Google Scholar] [PubMed]
- Wang, C.; Guo, K.; Gao, D.; Kang, X.; Jiang, K.; Li, Y.; Sun, L.; Zhang, S.; Sun, C.; Liu, X.; et al. Identification of Transaldolase as a Novel Serum Biomarker for Hepatocellular Carcinoma Metastasis Using Xenografted Mouse Model and Clinic Samples. Cancer Lett. 2011, 313, 154–166. [Google Scholar] [CrossRef]
- Basta, P.V.; Bensen, J.T.; Tse, C.-K.; Perou, C.M.; Sullivan, P.F.; Olshan, A.F. Genetic Variation in Transaldolase 1 and Risk of Squamous Cell Carcinoma of the Head and Neck. Cancer Detect. Prev. 2008, 32, 200–208. [Google Scholar] [CrossRef]
- Ding, Y.; Gong, C.; Huang, D.; Chen, R.; Sui, P.; Lin, K.H.; Liang, G.; Yuan, L.; Xiang, H.; Chen, J.; et al. Synthetic Lethality between HER2 and Transaldolase in Intrinsically Resistant HER2-Positive Breast Cancers. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Travis, E.L.; Wang, M.; Allard, J.D.; Peltz, G. Bleomycin hydrolase protects mice from bleomycin induced death and is involved in bleomycin induced lung fibrosis. Cancer Res. 2004, 45, 5127. [Google Scholar]
- Schwartz, D.R.; Homanics, G.E.; Hoyt, D.G.; Klein, E.; Abernethy, J.; Lazo, J.S. The Neutral Cysteine Protease Bleomycin Hydrolase Is Essential for Epidermal Integrity and Bleomycin Resistance. Proc. Natl. Acad. Sci. USA 1999, 96, 4680–4685. [Google Scholar] [CrossRef] [PubMed]
- Ramotar, D.; Wang, H. Protective Mechanisms against the Antitumor Agent Bleomycin: Lessons from Saccharomyces Cerevisiae. Curr. Genet. 2003, 43, 213–224. [Google Scholar] [CrossRef]
- Nicolay, N.H.; Rühle, A.; Perez, R.L.; Trinh, T.; Sisombath, S.; Weber, K.-J.; Ho, A.D.; Debus, J.; Saffrich, R.; Huber, P.E. Mesenchymal Stem Cells Are Sensitive to Bleomycin Treatment. Sci. Rep. 2016, 6, 26645. [Google Scholar] [CrossRef]
- Chen, J.; Stubbe, J. Bleomycins: Towards Better Therapeutics. Nat. Rev. Cancer 2005, 5, 102–112. [Google Scholar] [CrossRef]
- Ferrando, A.A.; Pendás, A.M.; Llano, E.; Velasco, G.; Lidereau, R.; López-Otıín, C. Gene Characterization, Promoter Analysis, and Chromosomal Localization of Human Bleomycin Hydrolase. J. Biol. Chem. 1997, 272, 33298–33304. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, H.; He, Q. Involvement of Bleomycin Hydrolase and Poly(ADP-Ribose) Polymerase-1 in Ubc9-Mediated Resistance to Chemotherapy Agents. Int. J. Oncol. 2016, 50, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jeon, J.; Mejia, S.; Yao, C.Q.; Ignatchenko, V.; Nyalwidhe, J.O.; Gramolini, A.O.; Lance, R.S.; Troyer, D.A.; Drake, R.R.; et al. Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer. Nat. Commun. 2016, 7, 11906. [Google Scholar] [CrossRef] [PubMed]
- Galieva, L.R.; James, V.; Mukhamedshina, Y.O.; Rizvanov, A.A. Therapeutic Potential of Extracellular Vesicles for the Treatment of Nerve Disorders. Front. Neurosci. 2019, 13, 163. [Google Scholar] [CrossRef]
- Cvjetkovic, A.; Jang, S.C.; Konečná, B.; Höög, J.L.; Sihlbom, C.; Lässer, C.; Lötvall, J. Detailed Analysis of Protein Topology of Extracellular Vesicles-Evidence of Unconventional Membrane Protein Orientation. Sci. Rep. 2016, 6, 36338. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Risha, Y.; Susevski, V.; Hüttmann, N.; Poolsup, S.; Minic, Z.; Berezovski, M.V. Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy. Biomedicines 2021, 9, 107. https://doi.org/10.3390/biomedicines9020107
Risha Y, Susevski V, Hüttmann N, Poolsup S, Minic Z, Berezovski MV. Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy. Biomedicines. 2021; 9(2):107. https://doi.org/10.3390/biomedicines9020107
Chicago/Turabian StyleRisha, Yousef, Vanessa Susevski, Nico Hüttmann, Suttinee Poolsup, Zoran Minic, and Maxim V. Berezovski. 2021. "Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy" Biomedicines 9, no. 2: 107. https://doi.org/10.3390/biomedicines9020107
APA StyleRisha, Y., Susevski, V., Hüttmann, N., Poolsup, S., Minic, Z., & Berezovski, M. V. (2021). Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy. Biomedicines, 9(2), 107. https://doi.org/10.3390/biomedicines9020107







