Deciphering the Role of Pyrvinium Pamoate in the Generation of Integrated Stress Response and Modulation of Mitochondrial Function in Myeloid Leukemia Cells through Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Chemicals
2.2. Cell Viability
2.3. Cell Cycle Analysis
2.4. Immunoblotting
2.5. Transcriptomic Analysis of Pyrvinium-Treated Molm13 Cells
2.6. Confocal Microscopy Observation
2.7. Cellular Bioenergetic Analysis
2.8. Mitochondrial Parameters (Mitochondrial Mass and Reactive Oxygen Species (ROS) Levels)
2.9. Mitochondrial Complex I Activity
2.10. Subcutaneous Mouse Xenografts
2.11. Statistical Analysis
3. Results
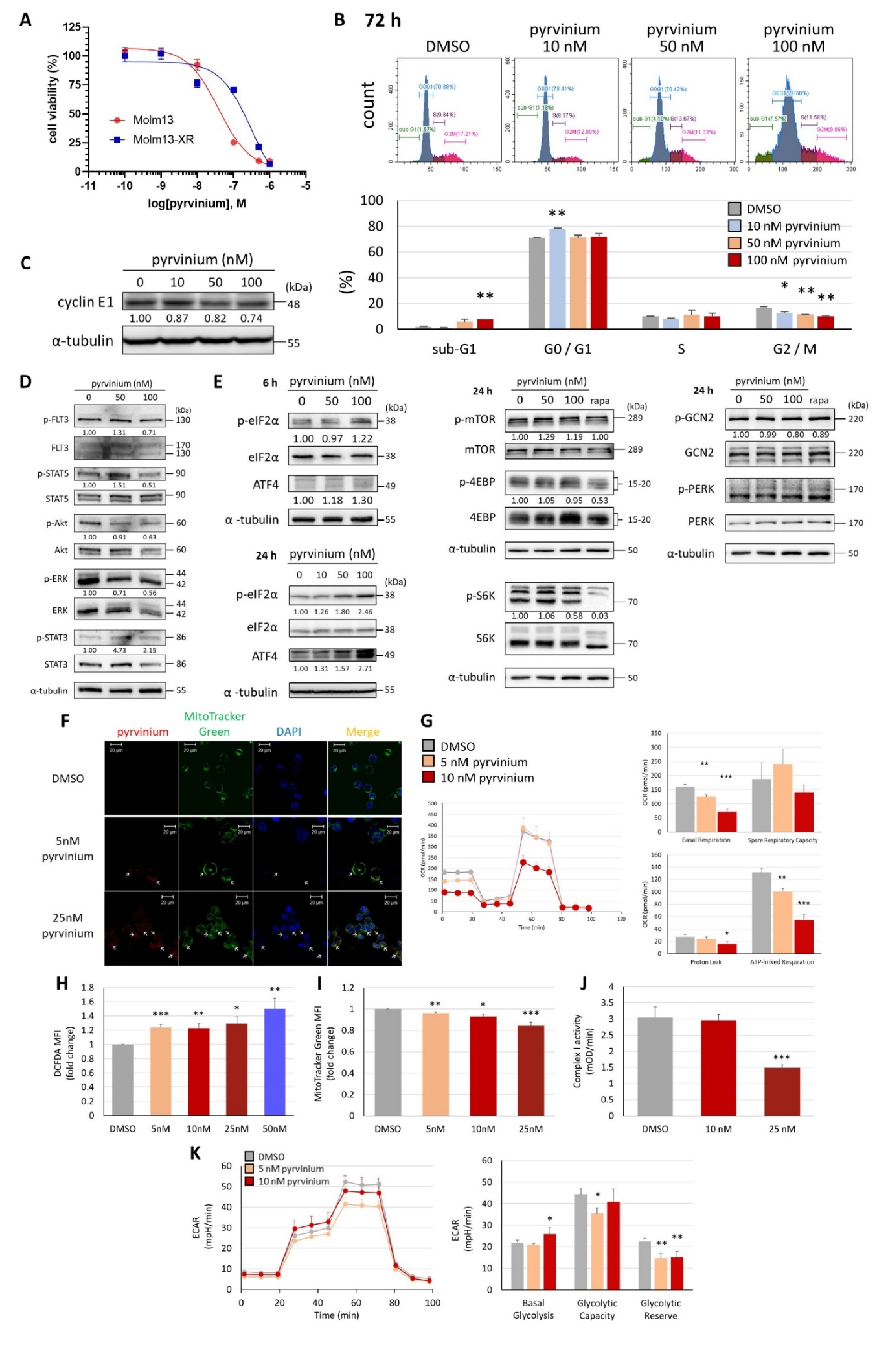
3.1. Pyrvinium Pamoate Can Inhibit Cell Proliferation and Induce Cell Death in Molm13 Cells
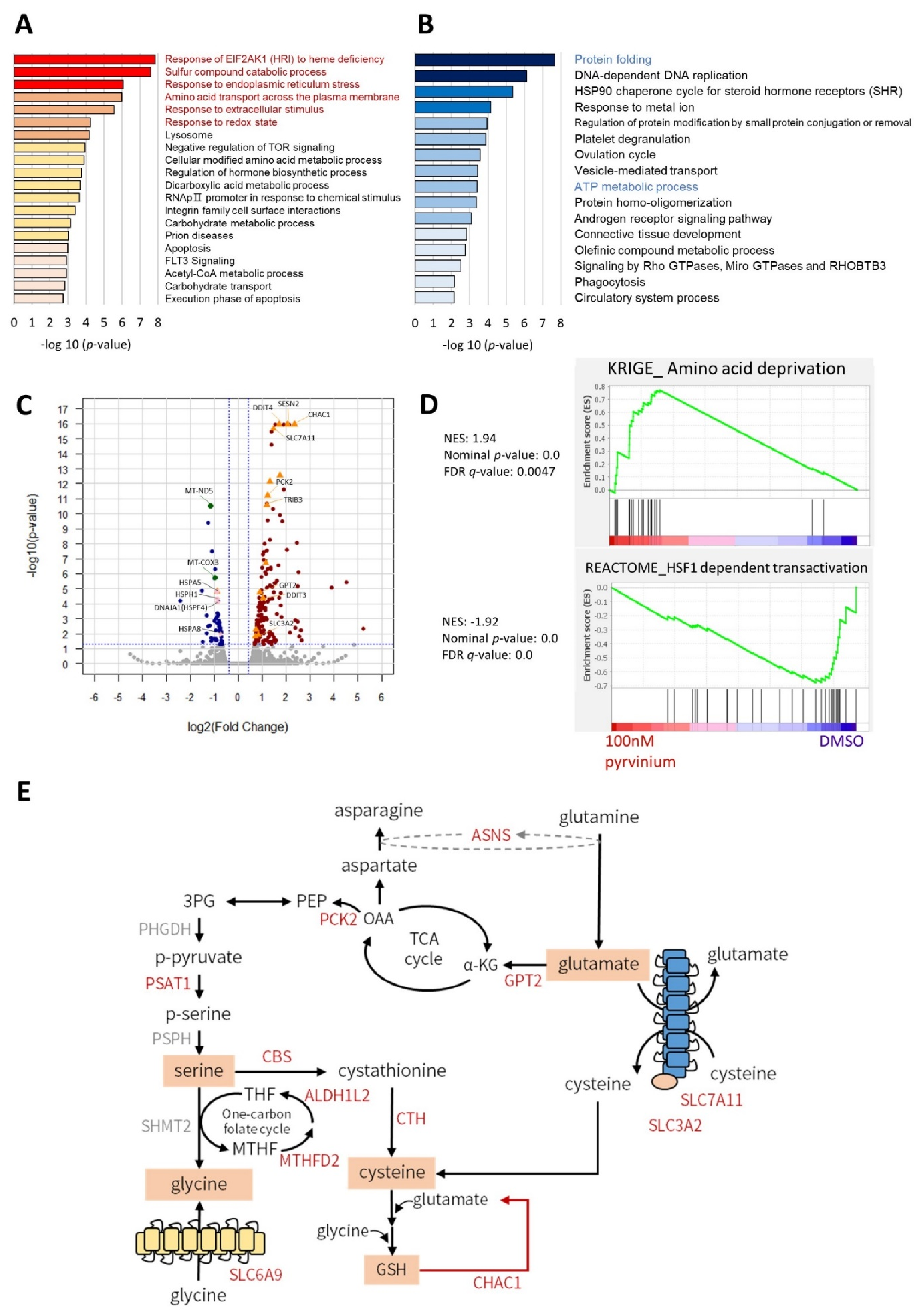
3.2. Transcriptome Analysis Revealed That Pyrvinium Pamoate Triggers an UPR-like Pathway and Redox Balance Modulation, but Blunts Protein Folding Function and ATP Synthesis in Molm13 Cells
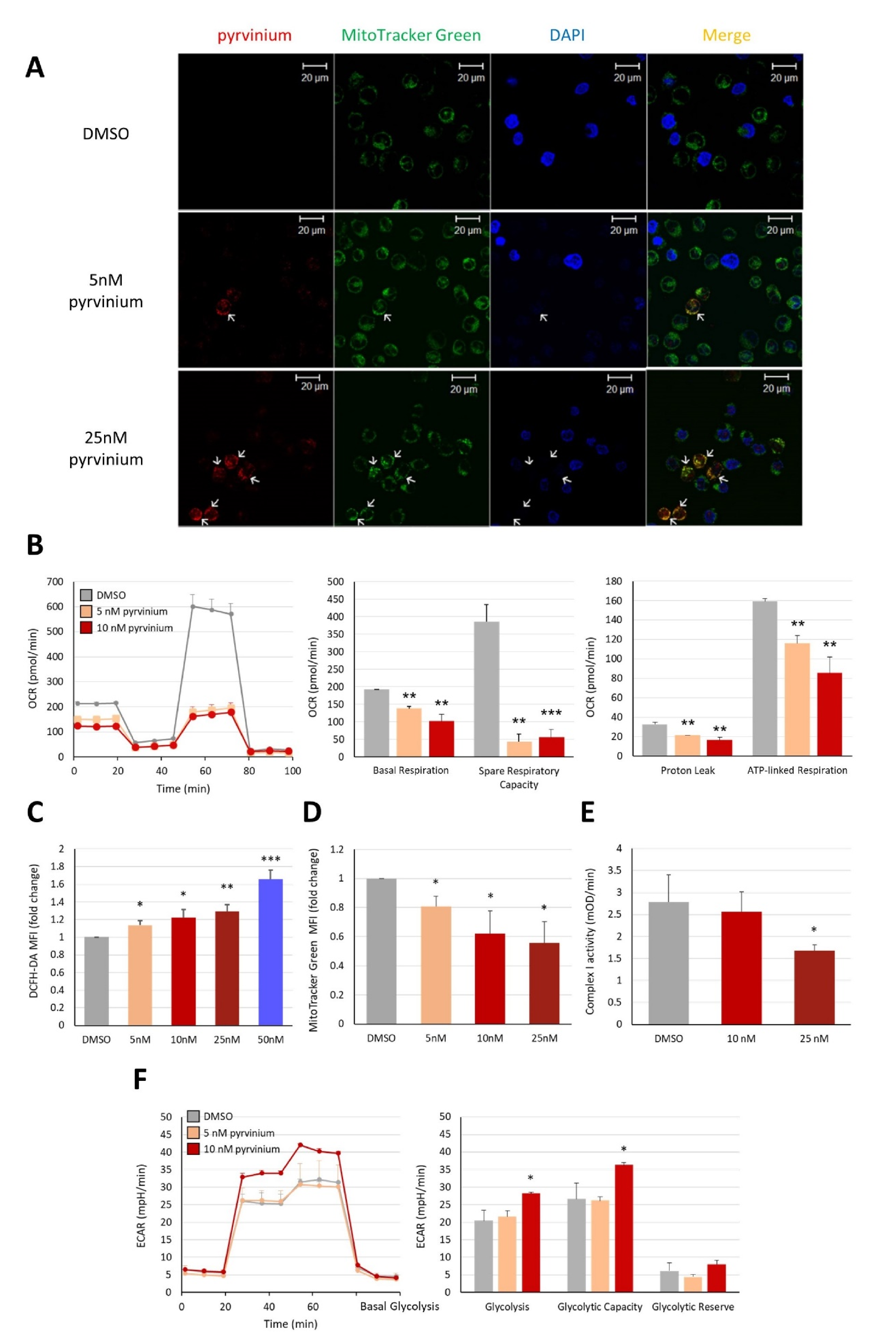
3.3. Pyrvinium Pamoate Targets Mitochondria and Has an Inhibitory Effect on Mitochondrial Respiration
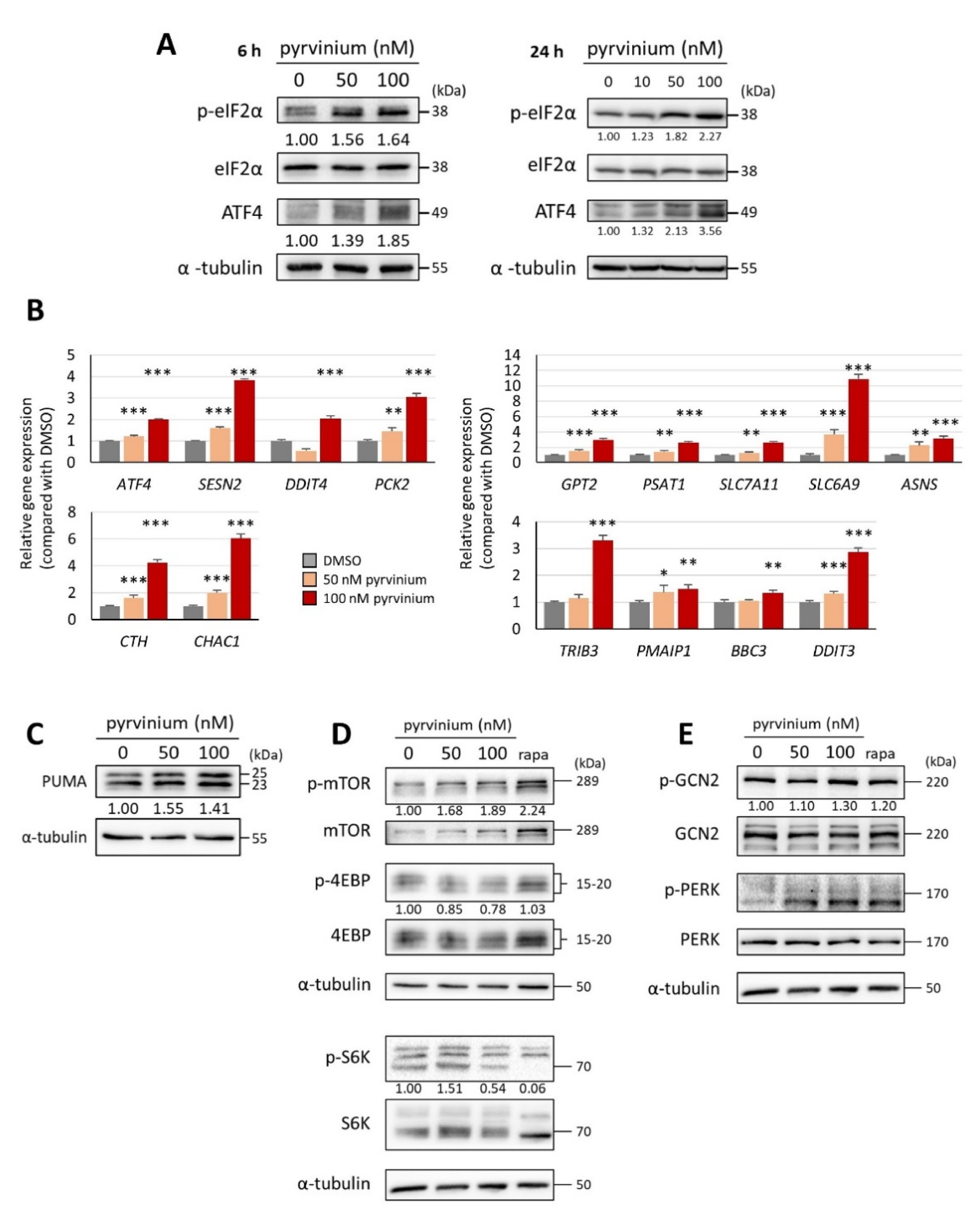
3.4. Pyrvinium Pamoate Triggers the Activation of the eIF2α-ATF4 Pathway and Inhibits mTORC1 Signaling
3.5. Pyrvinium Pamoate Exhibited Similar Activity in Molm13 Cells and Cabozantinib-Resistant Molm13-XR Cells
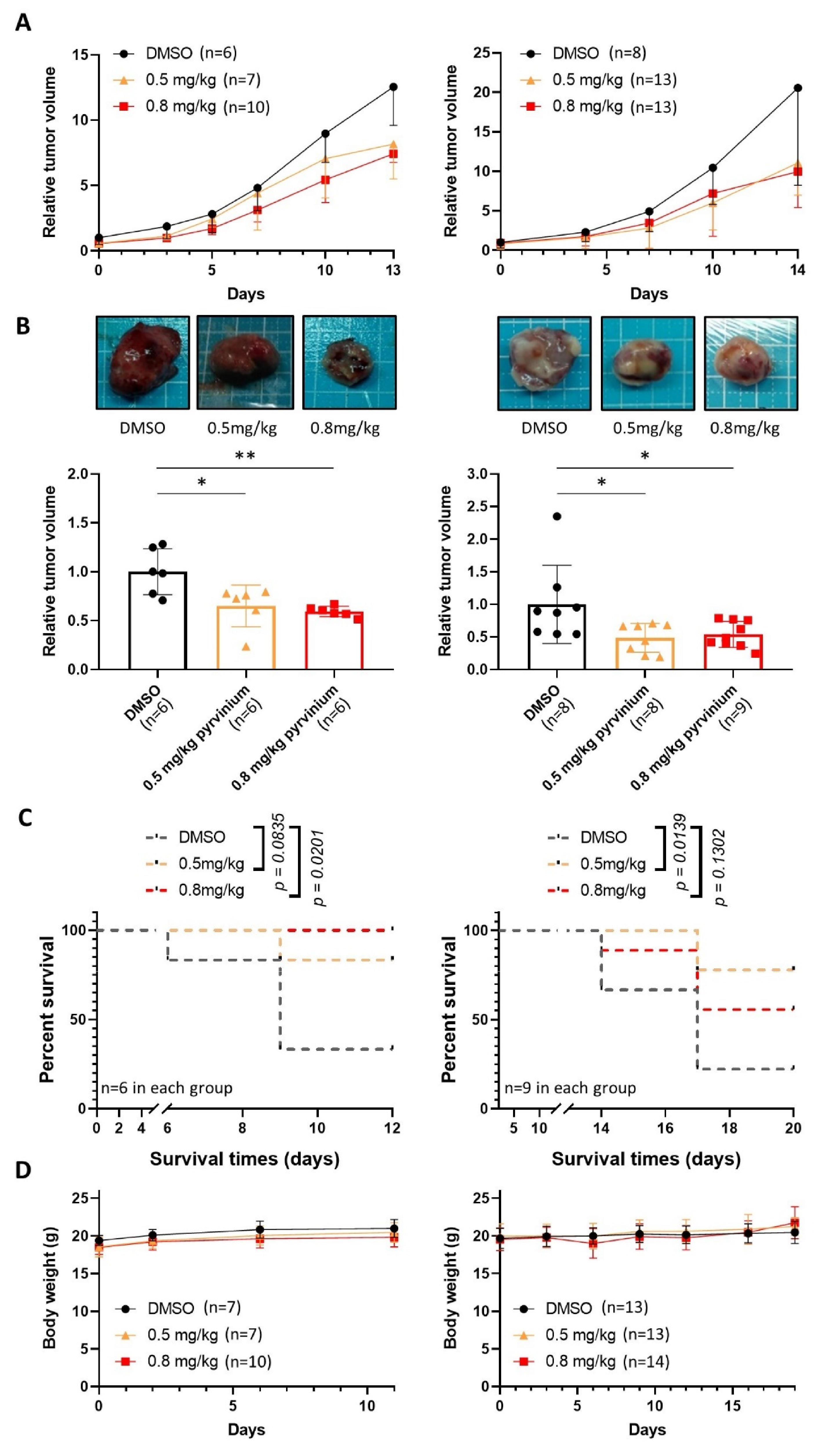
3.6. Pyrvinium Pamoate Retards the Growth of Subcutaneous Molm13 and Molm13-XR Xenograft Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dohner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medinger, M.; Passweg, J.R. Acute myeloid leukaemia genomics. Br. J. Haematol. 2017, 179, 530–542. [Google Scholar] [CrossRef]
- Short, N.J.; Konopleva, M.; Kadia, T.M.; Borthakur, G.; Ravandi, F.; DiNardo, C.D.; Daver, N. Advances in the Treatment of Acute Myeloid Leukemia: New Drugs and New Challenges. Cancer Discov. 2020, 10, 506–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cucchi, D.G.J.; Polak, T.B.; Ossenkoppele, G.J.; Uyl-De Groot, C.A.; Cloos, J.; Zweegman, S.; Janssen, J. Two decades of targeted therapies in acute myeloid leukemia. Leukemia 2021, 35, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Dohner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Kang, C.; Blair, H.A. Gilteritinib: A Review in Relapsed or Refractory FLT3-Mutated Acute Myeloid Leukaemia. Target. Oncol. 2020, 15, 681–689. [Google Scholar] [CrossRef]
- Pollyea, D.A. New drugs for acute myeloid leukemia inspired by genomics and when to use them. Hematol. Am. Soc. Hematol. Educ. Program. 2018, 2018, 45–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [PubMed] [Green Version]
- Fozard, G. Major helminthic diseases of North America: A review. J. Fam. Pract. 1978, 6, 1195–1203. [Google Scholar]
- Beck, J.W.; Saavedra, D.; Antell, G.J.; Tejeiro, B. The treatment of pinworm infections in humans (enterobiasis) with pyrvinium chloride and pyrvinium pamoate. Am. J. Trop. Med. Hyg. 1959, 8, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Esumi, H.; Lu, J.; Kurashima, Y.; Hanaoka, T. Antitumor activity of pyrvinium pamoate, 6-(dimethylamino)-2-[2-(2,5-dimethyl-1-phenyl-1H-pyrrol-3-yl)ethenyl]-1-methyl-qu inolinium pamoate salt, showing preferential cytotoxicity during glucose starvation. Cancer Sci. 2004, 95, 685–690. [Google Scholar] [CrossRef]
- Harada, Y.; Ishii, I.; Hatake, K.; Kasahara, T. Pyrvinium pamoate inhibits proliferation of myeloma/erythroleukemia cells by suppressing mitochondrial respiratory complex I and STAT3. Cancer Lett. 2012, 319, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Momtazi-Borojeni, A.A.; Abdollahi, E.; Ghasemi, F.; Caraglia, M.; Sahebkar, A. The novel role of pyrvinium in cancer therapy. J. Cell. Physiol. 2018, 233, 2871–2881. [Google Scholar] [CrossRef] [PubMed]
- Thorne, C.A.; Hanson, A.J.; Schneider, J.; Tahinci, E.; Orton, D.; Cselenyi, C.S.; Jernigan, K.K.; Meyers, K.C.; Hang, B.I.; Waterson, A.G.; et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat. Chem. Biol. 2010, 6, 829–836. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Zhu, Y.; Lu, Y.; Yu, Z.; Zhong, J.; Li, Y.; Pan, J. Anthelmintic pyrvinium pamoate blocks Wnt/β-catenin and induces apoptosis in multiple myeloma cells. Oncol. Lett. 2018, 15, 5871–5878. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fei, D.L.; Flaveny, C.A.; Dahmane, N.; Baubet, V.; Wang, Z.; Bai, F.; Pei, X.H.; Rodriguez-Blanco, J.; Hang, B.; et al. Pyrvinium attenuates Hedgehog signaling downstream of smoothened. Cancer Res. 2014, 74, 4811–4821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrella, D.; Manni, I.; Tumaini, B.; Dattilo, R.; Papaccio, F.; Mutarelli, M.; Sirci, F.; Amoreo, C.A.; Mottolese, M.; Iezzi, M.; et al. Computational drugs repositioning identifies inhibitors of oncogenic PI3K/AKT/P70S6K-dependent pathways among FDA-approved compounds. Oncotarget 2016, 7, 58743–58758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomitsuka, E.; Kita, K.; Esumi, H. An anticancer agent, pyrvinium pamoate inhibits the NADH-fumarate reductase system--a unique mitochondrial energy metabolism in tumour microenvironments. J. Biochem. 2012, 152, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, W.; Cheong, J.K.; Ang, S.H.; Teo, B.; Xu, P.; Asari, K.; Sun, W.T.; Than, H.; Bunte, R.M.; Virshup, D.M.; et al. Pyrvinium selectively targets blast phase-chronic myeloid leukemia through inhibition of mitochondrial respiration. Oncotarget 2015, 6, 33769–33780. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.H.; Macdonald, J.; Liu, G.; Lee, A.S.; Ly, M.; Davis, T.; Ke, N.; Zhou, D.; Wong-Staal, F.; Li, Q.X. Pyrvinium targets the unfolded protein response to hypoglycemia and its anti-tumor activity is enhanced by combination therapy. PLoS ONE 2008, 3, e3951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Lei, Y.; Liu, R.; Li, J.; Yuan, K.; Li, Y.; Chen, Y.; Liu, Y.; Lu, Y.; Edwards Iii, C.K.; et al. Pyrvinium targets autophagy addiction to promote cancer cell death. Cell Death Dis. 2013, 4, e614. [Google Scholar] [CrossRef]
- Jones, J.O.; Bolton, E.C.; Huang, Y.; Feau, C.; Guy, R.K.; Yamamoto, K.R.; Hann, B.; Diamond, M.I. Non-competitive androgen receptor inhibition in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2009, 106, 7233–7238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, S.K.; Tew, B.Y.; Lim, M.; Stankavich, B.; He, M.; Pufall, M.; Hu, W.; Chen, Y.; Jones, J.O. Mechanistic Investigation of the Androgen Receptor DNA-Binding Domain Inhibitor Pyrvinium. ACS Omega 2019, 4, 2472–2481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Ma, L.; Paik, H.; Sirota, M.; Wei, W.; Chua, M.S.; So, S.; Butte, A.J. Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nat. Commun. 2017, 8, 16022. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-T.; Hsieh, C.-H.; Oyang, Y.-J.; Huang, H.-C.; Juan, H.-F. A Large-Scale Gene Expression Intensity-Based Similarity Metric for Drug Repositioning. iScience 2018, 7, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, S.; Sears, T.; Cortopassi, G.; Woolard, K.; Angelastro, J.M. Repurposing FDA approved drugs inhibiting mitochondrial function for targeting glioma-stem like cells. Biomed. Pharmacother. 2021, 133, 111058. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, C.; Hallett, R.; Vora, P.; Manoranjan, B.; Mahendram, S.; Qazi, M.A.; McFarlane, N.; Subapanditha, M.; Nolte, S.M.; Singh, M.; et al. Pyrvinium Targets CD133 in Human Glioblastoma Brain Tumor-Initiating Cells. Clin. Cancer Res. 2015, 21, 5324–5337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Liu, S.; Jin, R.; Xu, H.; Li, Y.; Chen, Y.; Zhao, G. Pyrvinium pamoate regulates MGMT expression through suppressing the Wnt/β-catenin signaling pathway to enhance the glioblastoma sensitivity to temozolomide. Cell Death Discov. 2021, 7, 288. [Google Scholar] [CrossRef] [PubMed]
- Su, K.W.; Ou, D.L.; Fu, Y.H.; Tien, H.F.; Hou, H.A.; Lin, L.I. Repurposing cabozantinib with therapeutic potential in KIT-driven t(8;21) acute myeloid leukaemias. Cancer Gene Ther. 2021. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [Green Version]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C.; Trigoso, C.I.; Llorente, A.R.; Del Castillo, P. DNA fluorescence induced by polymethine cation pyrvinium binding. Histochem. J. 1991, 23, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Krige, D.; Needham, L.A.; Bawden, L.J.; Flores, N.; Farmer, H.; Miles, L.E.; Stone, E.; Callaghan, J.; Chandler, S.; Clark, V.L.; et al. CHR-2797: An antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 2008, 68, 6669–6679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohoka, N.; Yoshii, S.; Hattori, T.; Onozaki, K.; Hayashi, H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005, 24, 1243–1255. [Google Scholar] [CrossRef]
- Quiros, P.M.; Prado, M.A.; Zamboni, N.; D’Amico, D.; Williams, R.W.; Finley, D.; Gygi, S.P.; Auwerx, J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017, 216, 2027–2045. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Wang, A.N.; Liao, H.A.; Chen, C.Y.; Hou, H.A.; Hu, C.Y.; Tien, H.F.; Ou, D.L.; Lin, L.I. Cabozantinib is selectively cytotoxic in acute myeloid leukemia cells with FLT3-internal tandem duplication (FLT3-ITD). Cancer Lett. 2016, 376, 218–225. [Google Scholar] [CrossRef]
- Wander, P.; Arentsen-Peters, S.; Pinhanҫos, S.S.; Koopmans, B.; Dolman, M.E.M.; Ariese, R.; Bos, F.L.; Castro, P.G.; Jones, L.; Schneider, P.; et al. High-throughput drug screening reveals Pyrvinium pamoate as effective candidate against pediatric MLL-rearranged acute myeloid leukemia. Transl. Oncol. 2021, 14, 101048. [Google Scholar] [CrossRef]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Mungrue, I.N.; Pagnon, J.; Kohannim, O.; Gargalovic, P.S.; Lusis, A.J. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J. Immunol. 2009, 182, 466–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Galen, P.; Mbong, N.; Kreso, A.; Schoof, E.M.; Wagenblast, E.; Ng, S.W.K.; Krivdova, G.; Jin, L.; Nakauchi, H.; Dick, J.E. Integrated Stress Response Activity Marks Stem Cells in Normal Hematopoiesis and Leukemia. Cell Rep. 2018, 25, 1109–1117.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nii, T.; Prabhu, V.V.; Ruvolo, V.; Madhukar, N.; Zhao, R.; Mu, H.; Heese, L.; Nishida, Y.; Kojima, K.; Garnett, M.J.; et al. Imipridone ONC212 activates orphan G protein-coupled receptor GPR132 and integrated stress response in acute myeloid leukemia. Leukemia 2019, 33, 2805–2816. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, V.V.; Talekar, M.K.; Lulla, A.R.; Kline, C.L.B.; Zhou, L.; Hall, J.; Van den Heuvel, A.P.J.; Dicker, D.T.; Babar, J.; Grupp, S.A.; et al. Single agent and synergistic combinatorial efficacy of first-in-class small molecule imipridone ONC201 in hematological malignancies. Cell Cycle 2018, 17, 468–478. [Google Scholar] [CrossRef] [Green Version]
- Kline, C.L.; Van den Heuvel, A.P.; Allen, J.E.; Prabhu, V.V.; Dicker, D.T.; El-Deiry, W.S. ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2alpha kinases. Sci. Signal. 2016, 9, ra18. [Google Scholar] [CrossRef] [Green Version]
- Ishizawa, J.; Kojima, K.; Chachad, D.; Ruvolo, P.; Ruvolo, V.; Jacamo, R.O.; Borthakur, G.; Mu, H.; Zeng, Z.; Tabe, Y.; et al. ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci. Signal. 2016, 9, ra17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharon, D.; Cathelin, S.; Mirali, S.; Di Trani, J.M.; Yanofsky, D.J.; Keon, K.A.; Rubinstein, J.L.; Schimmer, A.D.; Ketela, T.; Chan, S.M. Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci. Transl. Med. 2019, 11, eaax2863. [Google Scholar] [CrossRef]
- Hlozkova, K.; Pecinova, A.; Alquezar-Artieda, N.; Pajuelo-Reguera, D.; Simcikova, M.; Hovorkova, L.; Rejlova, K.; Zaliova, M.; Mracek, T.; Kolenova, A.; et al. Metabolic profile of leukemia cells influences treatment efficacy of L-asparaginase. BMC Cancer 2020, 20, 526. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.M.; Xiang, M.; Heppler, L.N.; Tosic, I.; Jiang, K.; Munoz, J.O.; Gaikwad, A.S.; Horton, T.M.; Long, X.; Narayanan, P.; et al. Atovaquone is active against AML by upregulating the integrated stress pathway and suppressing oxidative phosphorylation. Blood Adv. 2019, 3, 4215–4227. [Google Scholar] [CrossRef]
- Feng, J.; Jiang, W.; Liu, Y.; Huang, W.; Hu, K.; Li, K.; Chen, J.; Ma, C.; Sun, Z.; Pang, X. Blocking STAT3 by Pyrvinium Pamoate Causes Metabolic Lethality in KRAS-mutant lung cancer. Biochem. Pharmacol. 2020, 177, 113960. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, L.; Hernandez-Garcia, D.; Schnabel, D.; Salas-Vidal, E.; Castro-Obregon, S. Function of reactive oxygen species during animal development: Passive or active? Dev. Biol. 2008, 320, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Wise, D.R.; Diehl, J.A.; Simon, M.C. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J. Biol. Chem. 2008, 283, 31153–31162. [Google Scholar] [CrossRef] [Green Version]
- Cantor, J.R.; Sabatini, D.M. Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2012, 2, 881–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabe, Y.; Lorenzi, P.L.; Konopleva, M. Amino acid metabolism in hematologic malignancies and the era of targeted therapy. Blood 2019, 134, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Fung, M.K.L.; Chan, G.C. Drug-induced amino acid deprivation as strategy for cancer therapy. J. Hematol. Oncol. 2017, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Gallipoli, P.; Giotopoulos, G.; Tzelepis, K.; Costa, A.S.H.; Vohra, S.; Medina-Perez, P.; Basheer, F.; Marando, L.; Di Lisio, L.; Dias, J.M.L.; et al. Glutaminolysis is a metabolic dependency in FLT3(ITD) acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood 2018, 131, 1639–1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matre, P.; Velez, J.; Jacamo, R.; Qi, Y.; Su, X.; Cai, T.; Chan, S.M.; Lodi, A.; Sweeney, S.R.; Ma, H.; et al. Inhibiting glutaminase in acute myeloid leukemia: Metabolic dependency of selected AML subtypes. Oncotarget 2016, 7, 79722–79735. [Google Scholar] [CrossRef] [Green Version]
- Zavorka Thomas, M.E.; Lu, X.; Talebi, Z.; Jeon, J.Y.; Buelow, D.R.; Gibson, A.A.; Uddin, M.E.; Brinton, L.T.; Nguyen, J.; Collins, M.; et al. Gilteritinib Inhibits Glutamine Uptake and Utilization in FLT3-ITD-Positive AML. Mol. Cancer Ther. 2021, 20, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chow, J.P.H.; Leung, Y.O.; Lu, X.; Yuen, C.H.; Lee, W.L.; Chau, K.C.; Yang, L.L.; Wong, R.M.H.; Lam, J.Y.T.; et al. NEI-01-Induced Arginine Deprivation Has Potent Activity Against Acute Myeloid Leukemia Cells Both In Vitro and In Vivo. Mol. Cancer Ther. 2021, 20, 2218–2227. [Google Scholar] [CrossRef] [PubMed]
- Feun, L.; You, M.; Wu, C.J.; Kuo, M.T.; Wangpaichitr, M.; Spector, S.; Savaraj, N. Arginine deprivation as a targeted therapy for cancer. Curr. Pharm. Des. 2008, 14, 1049–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miraki-Moud, F.; Ghazaly, E.; Ariza-McNaughton, L.; Hodby, K.A.; Clear, A.; Anjos-Afonso, F.; Liapis, K.; Grantham, M.; Sohrabi, F.; Cavenagh, J.; et al. Arginine deprivation using pegylated arginine deiminase has activity against primary acute myeloid leukemia cells in vivo. Blood 2015, 125, 4060–4068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.J.; Hsiao, H.H.; Hsu, Y.T.; Liu, Y.C.; Kao, H.W.; Liu, T.C.; Cho, S.F.; Feng, X.; Johnston, A.; Bomalaski, J.S.; et al. Phase I study of ADI-PEG20 plus low-dose cytarabine for the treatment of acute myeloid leukemia. Cancer Med. 2021, 10, 2946–2955. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryoo, I.J.; Park, H.R.; Choo, S.J.; Hwang, J.H.; Park, Y.M.; Bae, K.H.; Shin-Ya, K.; Yoo, I.D. Selective cytotoxic activity of valinomycin against HT-29 Human colon carcinoma cells via down-regulation of GRP78. Biol. Pharm. Bull. 2006, 29, 817–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephanou, A.; Latchman, D.S. Transcriptional modulation of heat-shock protein gene expression. Biochem. Res. Int. 2011, 2011, 238601. [Google Scholar] [CrossRef] [Green Version]
- Hensen, S.M.; Heldens, L.; van Enckevort, C.M.; van Genesen, S.T.; Pruijn, G.J.; Lubsen, N.H. Heat shock factor 1 is inactivated by amino acid deprivation. Cell Stress Chaperones 2012, 17, 743–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Home, T.; Jensen, R.A.; Rao, R. Heat shock factor 1 in protein homeostasis and oncogenic signal integration. Cancer Res. 2015, 75, 907–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.-H.; Tseng, C.-Y.; Lu, J.-W.; Lu, W.-H.; Lan, P.-Q.; Chen, C.-Y.; Ou, D.-L.; Lin, L.-I. Deciphering the Role of Pyrvinium Pamoate in the Generation of Integrated Stress Response and Modulation of Mitochondrial Function in Myeloid Leukemia Cells through Transcriptome Analysis. Biomedicines 2021, 9, 1869. https://doi.org/10.3390/biomedicines9121869
Fu Y-H, Tseng C-Y, Lu J-W, Lu W-H, Lan P-Q, Chen C-Y, Ou D-L, Lin L-I. Deciphering the Role of Pyrvinium Pamoate in the Generation of Integrated Stress Response and Modulation of Mitochondrial Function in Myeloid Leukemia Cells through Transcriptome Analysis. Biomedicines. 2021; 9(12):1869. https://doi.org/10.3390/biomedicines9121869
Chicago/Turabian StyleFu, Yu-Hsuan, Chi-Yang Tseng, Jeng-Wei Lu, Wen-Hui Lu, Pei-Qi Lan, Chien-Yuan Chen, Da-Liang Ou, and Liang-In Lin. 2021. "Deciphering the Role of Pyrvinium Pamoate in the Generation of Integrated Stress Response and Modulation of Mitochondrial Function in Myeloid Leukemia Cells through Transcriptome Analysis" Biomedicines 9, no. 12: 1869. https://doi.org/10.3390/biomedicines9121869
APA StyleFu, Y.-H., Tseng, C.-Y., Lu, J.-W., Lu, W.-H., Lan, P.-Q., Chen, C.-Y., Ou, D.-L., & Lin, L.-I. (2021). Deciphering the Role of Pyrvinium Pamoate in the Generation of Integrated Stress Response and Modulation of Mitochondrial Function in Myeloid Leukemia Cells through Transcriptome Analysis. Biomedicines, 9(12), 1869. https://doi.org/10.3390/biomedicines9121869





