Dissecting the Transcriptomes of Multiple Metronidazole-Resistant and Sensitive Trichomonas vaginalis Strains Identified Distinct Genes and Pathways Associated with Drug Resistance and Cell Death
Abstract
:1. Introduction
2. Materials and Methods
2.1. T. vaginalis Strains and Cell Culture
2.2. MTZ Susceptibility Assay of Various T. vaginalis Strains
2.3. RNA Extraction and Quality Assessment
2.4. RNA-Seq and Bioinformatics Analysis
2.5. Homology Model
2.6. Protein-Ligand Docking Analysis
3. Results
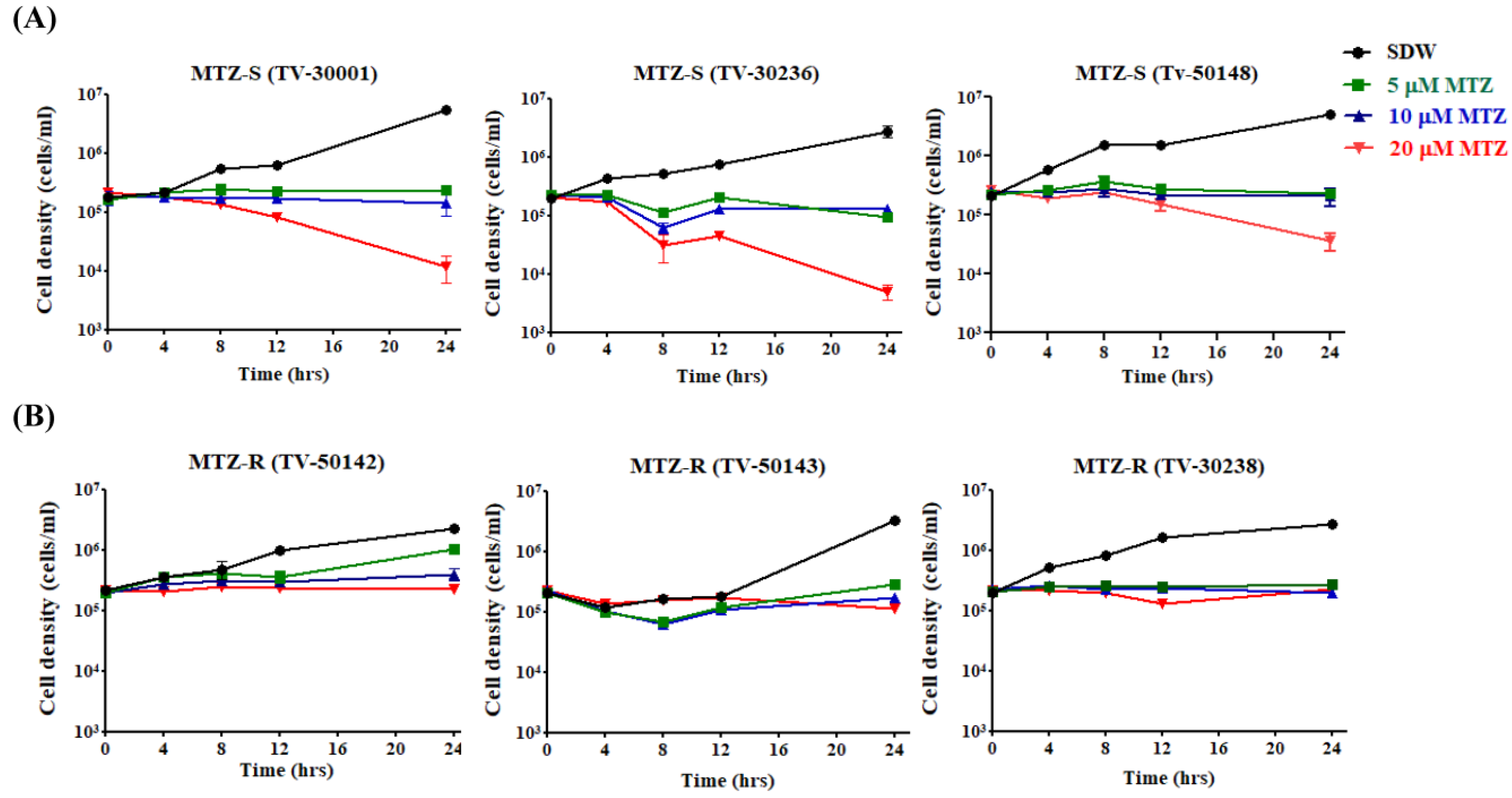
3.1. MTZ Susceptibility of Six MTZ-S and MTZ-R T. vaginalis Strains
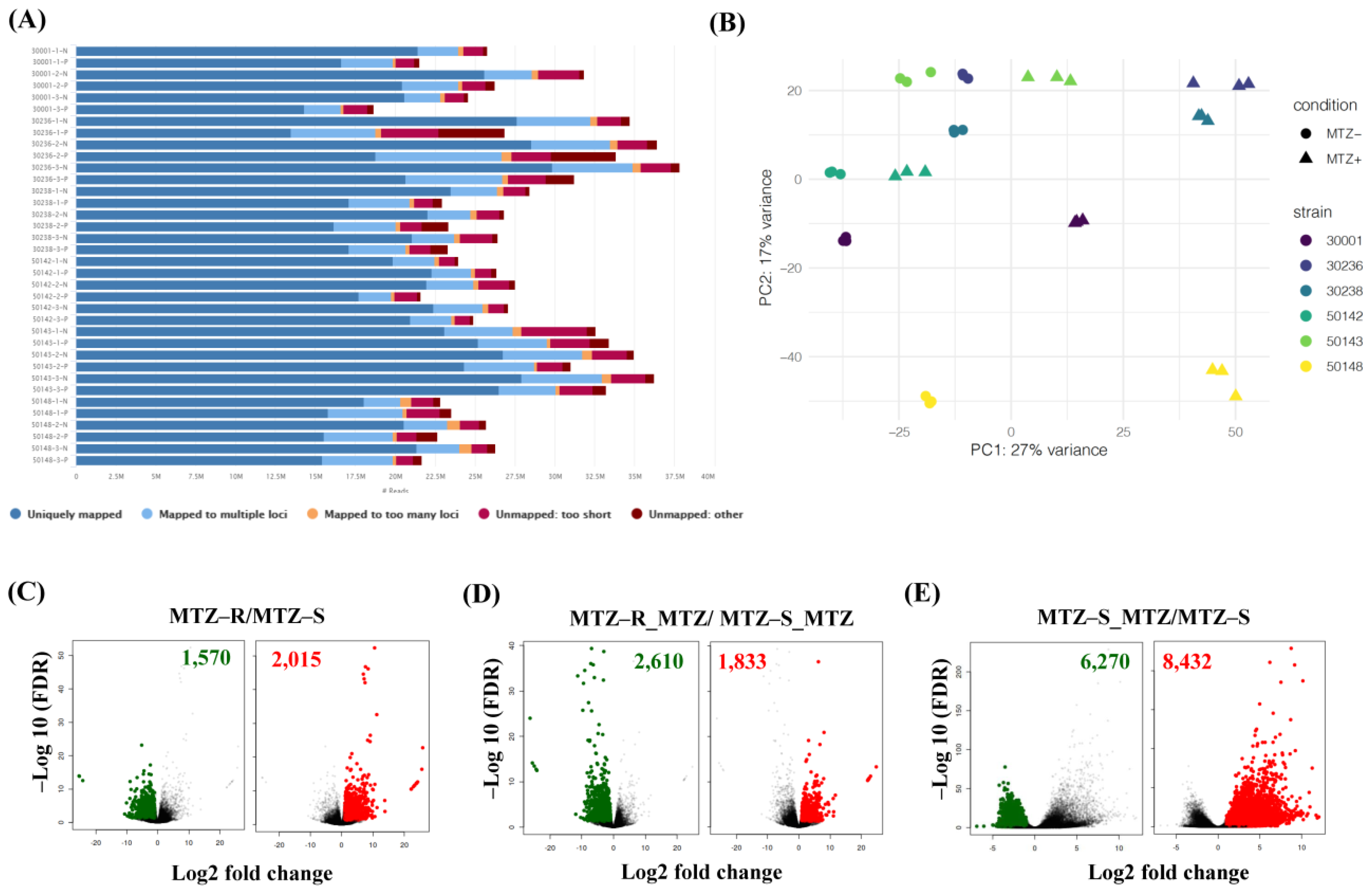
3.2. RNA-Seq of Six MTZ-S and MTZ-R T. vaginalis Strains in the Presence or Absence of MTZ
3.3. Differential Expression of the Previously Identified Genes Involved in Drug Resistance in the MTZ-R Transcriptomes Compared with Those of MTZ-S
3.3.1. Hydrogenosomal Metabolism
3.3.2. Oxygen Stress Response
3.3.3. Iron-Regulated Genes
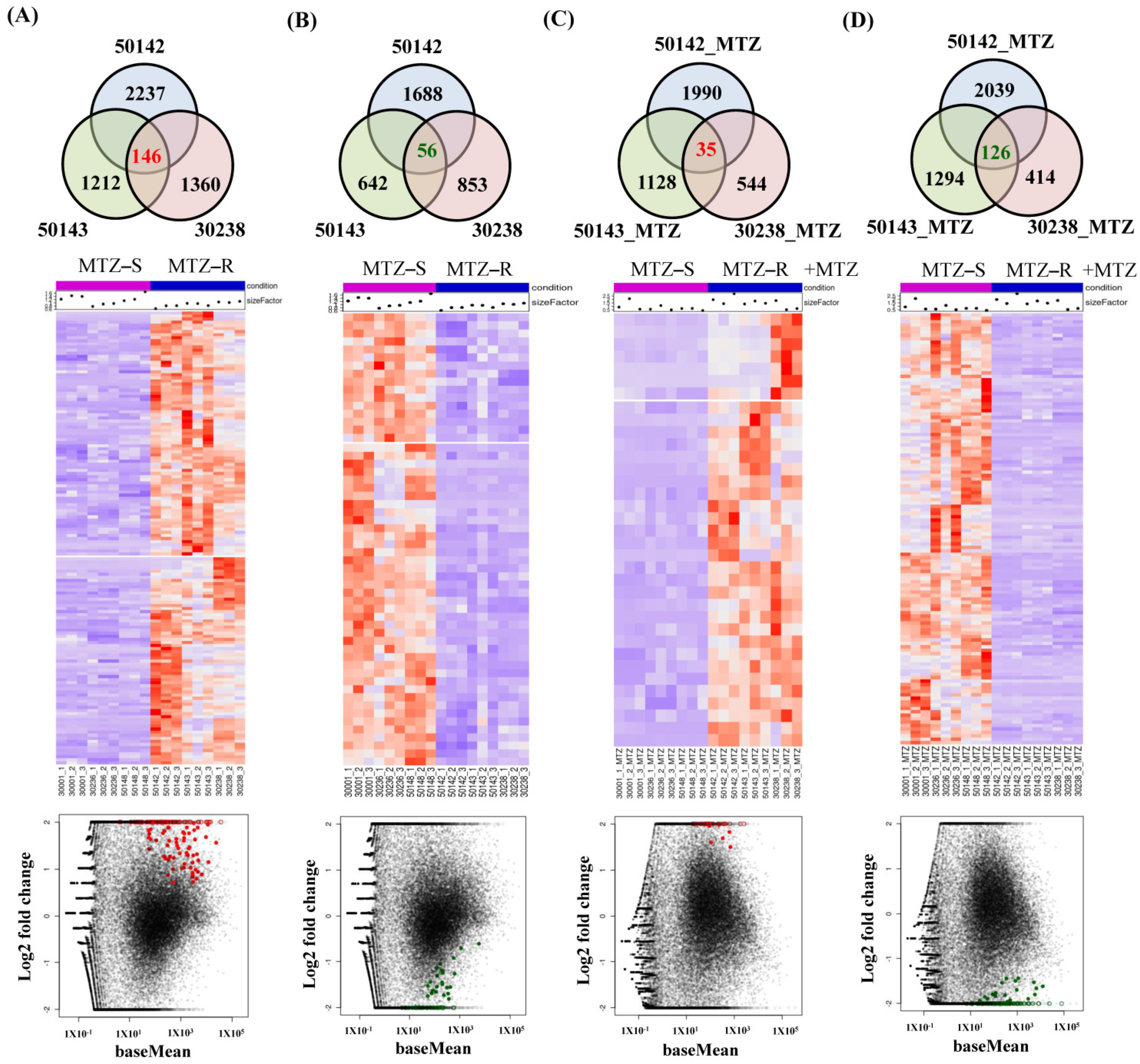
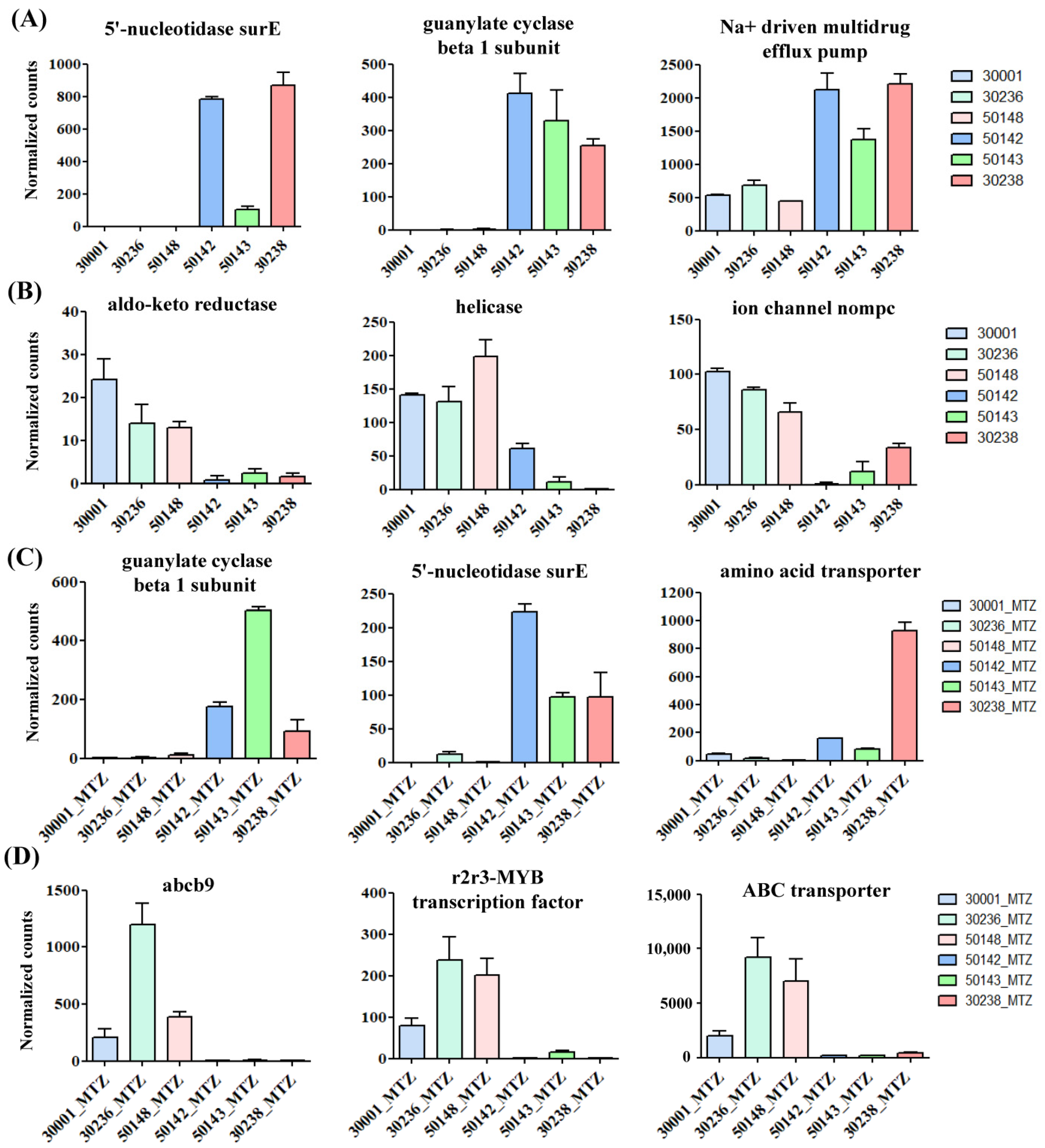
3.4. Identification of Shared Upregulated or Downregulated Genes in All MTZ-R Strains
3.5. Identification of Uniquely Upregulated Genes in Each MTZ-R and MTZ-S Strain
3.6. Functional Enrichment Analysis of MTZ-S Parasites Treated with MTZ Identifies Novel Genes and Pathways Associated with Drug-Induced Stress Responses
3.7. Molecular Docking Studies for the Identification of Novel MTZ-Binding Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MTZ | metronidazole |
| MTZ-S | MTZ-sensitive |
| MTZ-R | MTZ-resistant |
| ABC | ATP-binding cassette |
| MATE | multidrug and toxic compound extrusion |
| ERAD | endoplasmic reticulum-associated degradation |
| MdaB | modulator of drug activity B |
| HIV | human immunodeficiency virus |
| TNZ | tinidazole |
| PFOR | pyruvate: ferredoxin oxidoreductase |
| Fd | ferredoxin |
| ZIP | zinc regulated transporter, iron regulated transporter-like gene |
| Isc | Fe–S cluster |
| FeSOD | Fe-containing superoxide dismutase |
| SNPs | single nucleotide polymorphisms |
| SDW | sterile distilled water |
| RIN | RNA integrity number |
| NCBI | National Center for Biotechnology Information |
| PDB | Protein Data Bank |
| RMSD | root-mean-square deviation |
| PCA | principle component analysis |
| DEGs | differentially expressed genes |
| STK | succinate thiokinase |
| ID | iron-deficient |
| IR | iron-rich |
| MDR | multidrug resistance |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| feoAB | ferrous iron transporter |
| ABCB9 | ABC subfamily B member 9 |
| EhBspA1 | E. histolytica BspA |
| UPR | unfolded protein response |
| UPS | ubiquitin–proteasome system |
References
- World Health Organization. Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infection; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Menezes, C.B.; Frasson, A.P.; Tasca, T. Trichomoniasis—Are we giving the deserved attention to the most common non-viral sexually transmitted disease worldwide? Microb. Cell 2016, 3, 404–419. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Zhao, W.; Wang, H.; Wang, Y.; Li, J.; Wu, X. Trichomonas vaginalis infection-associated risk of cervical cancer: A meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 228, 166–173. [Google Scholar] [CrossRef]
- Silver, B.J.; Guy, R.J.; Kaldor, J.M.; Jamil, M.S.; Rumbold, A.R. Trichomonas vaginalis as a cause of perinatal morbidity: A systematic review and meta-analysis. Sex. Transm. Dis. 2014, 41, 369–376. [Google Scholar] [CrossRef]
- El-Shazly, A.M.; El-Naggar, H.M.; Soliman, M.; El-Negeri, M.; El-Nemr, H.E.; Handousa, A.E.; Morsy, T.A. A study on Trichomoniasis vaginalis and female infertility. J. Egypt Soc. Parasitol. 2001, 31, 545–553. [Google Scholar]
- McClelland, R.S.; Sangare, L.; Hassan, W.M.; Lavreys, L.; Mandaliya, K.; Kiarie, J.; Ndinya-Achola, J.; Jaoko, W.; Baeten, J.M. Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J. Infect. Dis. 2007, 195, 698–702. [Google Scholar] [CrossRef] [Green Version]
- Sutcliffe, S.; Alderete, J.F.; Till, C.; Goodman, P.J.; Hsing, A.W.; Zenilman, J.M.; De Marzo, A.M.; Platz, E.A. Trichomonosis and subsequent risk of prostate cancer in the Prostate Cancer Prevention Trial. Int. J. Cancer 2009, 124, 2082–2087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, G.; Narcisi, E.; Mosure, D.; Secor, W.E.; Higgins, J.; Moreno, H. Prevalence of metronidazole-resistant Trichomonas vaginalis in a gynecology clinic. Int. J. Reprod. Med. 2001, 46, 545–549. [Google Scholar] [CrossRef]
- Crowell, A.L.; Sanders-Lewis, K.A.; Secor, W.E. In vitro metronidazole and tinidazole activities against metronidazole-resistant strains of Trichomonas vaginalis. Antimicrob. Agents Chemother. 2003, 47, 1407–1409. [Google Scholar] [CrossRef] [Green Version]
- Kirkcaldy, R.D.; Augostini, P.; Asbel, L.E.; Bernstein, K.T.; Kerani, R.P.; Mettenbrink, C.J.; Pathela, P.; Schwebke, J.R.; Secor, W.E.; Workowski, K.A.; et al. Trichomonas vaginalis antimicrobial drug resistance in 6 US cities, STD Surveillance Network, 2009–2010. Emerg. Infect. Dis. 2012, 18, 939–943. [Google Scholar] [CrossRef] [Green Version]
- Sobel, J.D.; Nyirjesy, P.; Brown, W. Tinidazole therapy for metronidazole-resistant vaginal trichomoniasis. Clin. Infect. Dis. 2001, 33, 1341–1346. [Google Scholar] [CrossRef]
- Mead, J.R.; Fernadez, M.; Romagnoli, P.A.; Secor, W.E. Use of Trichomonas vaginalis clinical isolates to evaluate correlation of gene expression and metronidazole resistance. J. Parasitol. 2006, 92, 196–199. [Google Scholar] [CrossRef]
- Wright, J.M.; Webb, R.I.; O’Donoghue, P.; Upcroft, P.; Upcroft, J.A. Hydrogenosomes of laboratory-induced metronidazole-resistant Trichomonas vaginalis lines are downsized while those from clinically metronidazole-resistant isolates are not. J. Eukaryot. Microbiol. 2010, 57, 171–176. [Google Scholar] [CrossRef]
- Rasoloson, D.; Vanacova, S.; Tomkova, E.; Razga, J.; Hrdy, I.; Tachezy, J.; Kulda, J. Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis. Microbiology 2002, 148, 2467–2477. [Google Scholar] [CrossRef] [Green Version]
- Yarlett, N.; Yarlett, N.C.; Lloyd, D. Ferredoxin-dependent reduction of nitroimidazole derivatives in drug-resistant and susceptible strains of Trichomonas vaginalis. Biochem. Pharmacol. 1986, 35, 1703–1708. [Google Scholar] [CrossRef]
- Leitsch, D.; Janssen, B.D.; Kolarich, D.; Johnson, P.J.; Duchene, M. Trichomonas vaginalis flavin reductase 1 and its role in metronidazole resistance. Mol. Microbiol. 2014, 91, 198–208. [Google Scholar] [CrossRef] [Green Version]
- Leitsch, D.; Drinic, M.; Kolarich, D.; Duchene, M. Down-regulation of flavin reductase and alcohol dehydrogenase-1 (ADH1) in metronidazole-resistant isolates of Trichomonas vaginalis. Mol. Biochem. Parasitol. 2012, 183, 177–183. [Google Scholar] [CrossRef]
- Argaez-Correa, W.; Alvarez-Sanchez, M.E.; Arana-Argaez, V.E.; Ramirez-Camacho, M.A.; Novelo-Castilla, J.S.; Coral-Martinez, T.I.; Torres-Romero, J.C. The Role of Iron Status in the Early Progression of Metronidazole Resistance in Trichomonas vaginalis Under Microaerophilic Conditions. J. Eukaryot. Microbiol. 2019, 66, 309–315. [Google Scholar] [CrossRef]
- Paulish-Miller, T.E.; Augostini, P.; Schuyler, J.A.; Smith, W.L.; Mordechai, E.; Adelson, M.E.; Gygax, S.E.; Secor, W.E.; Hilbert, D.W. Trichomonas vaginalis metronidazole resistance is associated with single nucleotide polymorphisms in the nitroreductase genes ntr4Tv and ntr6Tv. Antimicrob. Agents Chemother. 2014, 58, 2938–2943. [Google Scholar] [CrossRef] [Green Version]
- Carlton, J.M.; Hirt, R.P.; Silva, J.C.; Delcher, A.L.; Schatz, M.; Zhao, Q.; Wortman, J.R.; Bidwell, S.L.; Alsmark, U.C.; Besteiro, S.; et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 2007, 315, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Bradic, M.; Warring, S.D.; Tooley, G.E.; Scheid, P.; Secor, W.E.; Land, K.M.; Huang, P.J.; Chen, T.W.; Lee, C.C.; Tang, P.; et al. Genetic Indicators of Drug Resistance in the Highly Repetitive Genome of Trichomonas vaginalis. Genome Biol. Evol. 2017, 9, 1658–1672. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.C.; Chu, L.J.; Huang, P.J.; Cheng, W.H.; Zheng, Y.H.; Huang, C.Y.; Hong, S.W.; Chen, L.C.; Lin, H.A.; Wang, J.Y.; et al. Proteomic signatures of metronidazole-resistant Trichomonas vaginalis reveal novel proteins associated with drug resistance. Parasites Vectors 2020, 13, 274. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Macarthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Kaplan, W.; Littlejohn, T.G. Swiss-PDB Viewer (Deep View). Brief. Bioinform. 2001, 2, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Horvathova, L.; Safarikova, L.; Basler, M.; Hrdy, I.; Campo, N.B.; Shin, J.W.; Huang, K.Y.; Huang, P.J.; Lin, R.; Tang, P.; et al. Transcriptomic identification of iron-regulated and iron-independent gene copies within the heavily duplicated Trichomonas vaginalis genome. Genome Biol. Evol. 2012, 4, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Ansell, B.R.; Baker, L.; Emery, S.J.; McConville, M.J.; Svard, S.G.; Gasser, R.B.; Jex, A.R. Transcriptomics Indicates Active and Passive Metronidazole Resistance Mechanisms in Three Seminal Giardia Lines. Front. Microbiol. 2017, 8, 398. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.M.; Upcroft, J.A.; Dodd, H.N.; Chen, N.; Upcroft, P. Alternative 2-keto acid oxidoreductase activities in Trichomonas vaginalis. Mol. Biochem. Parasitol. 1999, 98, 203–214. [Google Scholar] [CrossRef]
- Land, K.M.; Clemens, D.L.; Johnson, P.J. Loss of multiple hydrogenosomal proteins associated with organelle metabolism and high-level drug resistance in trichomonads. Exp. Parasitol. 2001, 97, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Kulda, J.; Tachezy, J.; Cerkasovova, A. In vitro induced anaerobic resistance to metronidazole in Trichomonas vaginalis. J. Eukaryot. Microbiol. 1993, 40, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Tazreiter, M.; Leitsch, D.; Hatzenbichler, E.; Mair-Scorpio, G.E.; Steinborn, R.; Schreiber, M.; Duchene, M. Entamoeba histolytica: Response of the parasite to metronidazole challenge on the levels of mRNA and protein expression. Exp. Parasitol. 2008, 120, 403–410. [Google Scholar] [CrossRef]
- Penuliar, G.M.; Nakada-Tsukui, K.; Nozaki, T. Phenotypic and transcriptional profiling in Entamoeba histolytica reveal costs to fitness and adaptive responses associated with metronidazole resistance. Front. Microbiol. 2015, 6, 354. [Google Scholar] [CrossRef]
- Emery, S.J.; Baker, L.; Ansell, B.R.E.; Mirzaei, M.; Haynes, P.A.; McConville, M.J.; Svard, S.G.; Jex, A.R. Differential protein expression and post-translational modifications in metronidazole-resistant Giardia duodenalis. GigaScience 2018, 7, giy024. [Google Scholar] [CrossRef]
- Tsugawa, H.; Suzuki, H.; Satoh, K.; Hirata, K.; Matsuzaki, J.; Saito, Y.; Suematsu, M.; Hibi, T. Two amino acids mutation of ferric uptake regulator determines Helicobacter pylori resistance to metronidazole. Antioxid. Redox Signal. 2011, 14, 15–23. [Google Scholar] [CrossRef]
- Veeranagouda, Y.; Husain, F.; Boente, R.; Moore, J.; Smith, C.J.; Rocha, E.R.; Patrick, S.; Wexler, H.M. Deficiency of the ferrous iron transporter FeoAB is linked with metronidazole resistance in Bacteroides fragilis. J. Antimicrob. Chemother. 2014, 69, 2634–2643. [Google Scholar] [CrossRef] [Green Version]
- Pramanik, P.K.; Alam, M.N.; Roy Chowdhury, D.; Chakraborti, T. Drug Resistance in Protozoan Parasites: An Incessant Wrestle for Survival. J. Glob. Antimicrob. Resist. 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Duraisingh, M.T.; Cowman, A.F. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005, 94, 181–190. [Google Scholar] [CrossRef]
- Doliwa, C.; Escotte-Binet, S.; Aubert, D.; Sauvage, V.; Velard, F.; Schmid, A.; Villena, I. Sulfadiazine resistance in Toxoplasma gondii: No involvement of overexpression or polymorphisms in genes of therapeutic targets and ABC transporters. Parasite 2013, 20, 19. [Google Scholar] [CrossRef] [Green Version]
- Messaritakis, I.; Christodoulou, V.; Mazeris, A.; Koutala, E.; Vlahou, A.; Papadogiorgaki, S.; Antoniou, M. Drug resistance in natural isolates of Leishmania donovani s.l. promastigotes is dependent of Pgp170 expression. PLoS ONE 2013, 8, e65467. [Google Scholar] [CrossRef] [Green Version]
- Shahi, S.K.; Krauth-Siegel, R.L.; Clayton, C.E. Overexpression of the putative thiol conjugate transporter TbMRPA causes melarsoprol resistance in Trypanosoma brucei. Mol. Microbiol. 2002, 43, 1129–1138. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Lohia, A.; Kumar, A.; Samuelson, J. Overexpression of P-glycoprotein gene 1 by transfected Entamoeba histolytica confers emetine-resistance. Mol. Biochem. Parasitol. 1996, 82, 257–260. [Google Scholar] [CrossRef]
- Kay, C.; Woodward, K.D.; Lawler, K.; Self, T.J.; Dyall, S.D.; Kerr, I.D. The ATP-binding cassette proteins of the deep-branching protozoan parasite Trichomonas vaginalis. PLoS Negl. Trop. Dis. 2012, 6, e1693. [Google Scholar] [CrossRef] [Green Version]
- de Miguel, N.; Lustig, G.; Twu, O.; Chattopadhyay, A.; Wohlschlegel, J.A.; Johnson, P.J. Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteom. 2010, 9, 1554–1566. [Google Scholar] [CrossRef] [Green Version]
- Araujo-Santos, J.M.; Parodi-Talice, A.; Castanys, S.; Gamarro, F. The overexpression of an intracellular ABCA-like transporter alters phospholipid trafficking in Leishmania. Biochem. Biophys. Res. Commun. 2005, 330, 349–355. [Google Scholar] [CrossRef]
- Hlavata, I.; Mohelnikova-Duchonova, B.; Vaclavikova, R.; Liska, V.; Pitule, P.; Novak, P.; Bruha, J.; Vycital, O.; Holubec, L.; Treska, V.; et al. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis 2012, 27, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Hou, L.; Zhang, X.; Jiao, Y.; Li, Y.; Zhao, Y.; Guan, Y.; Liu, Z. ATP binding cassette subfamily B member 9 (ABCB9) is a prognostic indicator of overall survival in ovarian cancer. Medicine 2019, 98, e15698. [Google Scholar] [CrossRef]
- Ohashi-Kobayashi, A.; Ohashi, K.; Du, W.B.; Omote, H.; Nakamoto, R.; Al-Shawi, M.; Maeda, M. Examination of drug resistance activity of human TAP-like (ABCB9) expressed in yeast. Biochem. Biophys. Res. Commun. 2006, 343, 597–601. [Google Scholar] [CrossRef]
- Handrich, M.R.; Garg, S.G.; Sommerville, E.W.; Hirt, R.P.; Gould, S.B. Characterization of the BspA and Pmp protein family of trichomonads. Parasites Vectors 2019, 12, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penuliar, G.M.; Furukawa, A.; Nakada-Tsukui, K.; Husain, A.; Sato, D.; Nozaki, T. Transcriptional and functional analysis of trifluoromethionine resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 2012, 67, 375–386. [Google Scholar] [CrossRef]
- Li, C.; Wu, P.Y.; Hsieh, M. Growth-phase-dependent transcriptional regulation of the pcm and surE genes required for stationary-phase survival of Escherichia coli. Microbiology 1997, 143, 3513–3520. [Google Scholar] [CrossRef] [Green Version]
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 1778–1782. [Google Scholar] [CrossRef] [Green Version]
- Kaatz, G.W.; McAleese, F.; Seo, S.M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005, 49, 1857–1864. [Google Scholar] [CrossRef] [Green Version]
- Plant, C.W.; Edwards, D.I. Effect of tinidazole, metronidazole and nitrofurazone on nucleic acid synthesis in Clostridium bifermentans. J. Antimicrob. Chemother. 1976, 2, 203–209. [Google Scholar] [CrossRef]
- Sigeti, J.S.; Guiney, D.G., Jr.; Davis, C.E. Mechanism of action of metronidazole on Bacteroides fragilis. J. Infect. Dis. 1983, 148, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Wixon, J.; Kell, D. The Kyoto encyclopedia of genes and genomes—KEGG. Yeast 2000, 17, 48–55. [Google Scholar] [CrossRef]
- Tiengwe, C.; Muratore, K.A.; Bangs, J.D. Surface proteins, ERAD and antigenic variation in Trypanosoma brucei. Cell Microbiol. 2016, 18, 1673–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Shared Upregulated Genes in the MTZ-R Transcriptomes | |||
|---|---|---|---|
| Gene ID | MTZ-R/MTZ-S (log2FoldChange) | Padj | TrichDB Annotation |
| TVAG_195930 | 11.214 | 0 | 5′-nucleotidase surE |
| TVAG_154640 | 9.198 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_407240 | 9.146 | 0 | 60S ribosomal protein L10 |
| TVAG_355510 | 8.626 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_219680 | 8.463 | 0 | guanylate cyclase beta 1 subunit |
| TVAG_177180 | 7.166 | 0 | WD-repeat protein |
| TVAG_195940 | 7.125 | 0 | acyltransferase |
| TVAG_387850 | 6.553 | 0 | ankyrin repeat-containing protein |
| TVAG_113870 | 5.905 | 0 | acetyl-CoA hydrolase |
| TVAG_233960 | 5.346 | 0 | vps9-ankyrin repeat-containing protein |
| TVAG_043070 | 5.156 | 0 | fructose-bisphosphate aldolase |
| TVAG_333180 | 4.864 | 0 | ankyrin repeat-containing protein |
| TVAG_011440 | 4.644 | 0 | 2-5A-dependent ribonuclease |
| TVAG_241690 | 4.58 | 0 | ankyrin repeat domain protein |
| TVAG_156760 | 4.125 | 0 | clan MA, family M8, leishmanolysin-like metallopeptidase |
| TVAG_410370 | 3.616 | 0 | MYB81 |
| TVAG_446260 | 3.599 | 0 | guanylate cyclase |
| TVAG_277640 | 3.498 | 0 | 2-5A-dependent ribonuclease |
| TVAG_338210 | 3.305 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_005050 | 3.217 | 0 | ankyrin repeat domain protein |
| TVAG_103510 | 3.207 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_316160 | 3.19 | 0 | protein phosphatase-5 |
| TVAG_108400 | 3.072 | 0.003 | histidinol-phosphate aminotransferase |
| TVAG_114330 | 3.045 | 0 | golgin IMH1 |
| TVAG_396470 | 2.97 | 0.003 | leucine-rich repeat protein, BspA family |
| TVAG_214470 | 2.868 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_005060 | 2.828 | 0 | ankyrin repeat domain protein |
| TVAG_031910 | 2.788 | 0 | calcium/calmodulin-dependent protein kinase |
| TVAG_271790 | 2.735 | 0 | clan MA, family M8, leishmanolysin-like metallopeptidase |
| TVAG_315580 | 2.657 | 0.011 | CAMK family protein kinase |
| TVAG_247080 | 2.572 | 0 | adenylate cyclase |
| TVAG_157870 | 2.325 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_387720 | 2.281 | 0 | ankyrin repeat-rich membrane-spanning protein |
| TVAG_262530 | 2.268 | 0 | cortactin-binding protein |
| TVAG_161100 | 2.169 | 0 | heat shock protein 70kD |
| TVAG_040090 | 2.163 | 0 | cysteine synthase |
| TVAG_361490 | 2.143 | 0.025 | CAMK family protein kinase |
| TVAG_397320 | 2.116 | 0 | adenosine diphosphatase |
| TVAG_062550 | 2.106 | 0 | clan SB, family S8, subtilisin-like serine peptidase |
| TVAG_426540 | 2.105 | 0 | AGC family protein kinase |
| TVAG_185150 | 2.041 | 0 | CMGC family protein kinase |
| TVAG_109730 | 2.04 | 0.012 | 40S ribosomal protein S23 |
| TVAG_151620 | 2.008 | 0 | heat shock protein 70 (HSP70)-4 |
| TVAG_422640 | 2.006 | 0 | 26S proteasome non-ATPase regulatory subunit |
| TVAG_465230 | 1.974 | 0 | dihydrodiol dehydrogenase |
| TVAG_099320 | 1.884 | 0.002 | fibroblast growth factor, BASic, antisense |
| TVAG_339630 | 1.883 | 0 | thioredoxin |
| TVAG_072440 | 1.882 | 0 | coiled-coil domain-containing protein |
| TVAG_475160 | 1.827 | 0 | HSP90 co-chaperone |
| TVAG_475150 | 1.821 | 0 | AGC family protein kinase |
| TVAG_483040 | 1.784 | 0 | Na+-driven multidrug efflux pump |
| TVAG_336370 | 1.76 | 0 | pc-MYB2 |
| TVAG_099020 | 1.754 | 0.156 | eukaryotic translation initiation factor 4g |
| TVAG_383560 | 1.644 | 0 | chaperone protein DNAj |
| TVAG_057020 | 1.615 | 0 | 60S ribosomal protein L22 |
| TVAG_440390 | 1.604 | 0 | CAMK family protein kinase |
| TVAG_284390 | 1.507 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_457350 | 1.498 | 0.016 | r2r3-MYB transcription factor |
| TVAG_391100 | 1.439 | 0 | clan MA, family M8, leishmanolysin-like metallopeptidase |
| TVAG_494730 | 1.347 | 0 | lipid A export ATP-binding/permease protein msba |
| TVAG_359170 | 1.332 | 0 | serine/threonine protein kinase |
| TVAG_201990 | 1.316 | 0 | ubiquitin protein ligase E3a |
| TVAG_052020 | 1.289 | 0 | PIKK family atypical protein kinase |
| TVAG_031010 | 1.289 | 0 | triadin |
| TVAG_150550 | 1.236 | 0 | DNAj/HSP40 |
| TVAG_212040 | 1.137 | 0 | sua5 protein |
| TVAG_143490 | 1.11 | 0 | glycogen debranching enzyme |
| TVAG_368970 | 1.027 | 0 | peptidylprolyl isomerase |
| Shared Downregulated Genes in the MTZ-R Transcriptomes | |||
| TVAG_462960 | −4.027 | 0 | ankyrin |
| TVAG_602270 | −3.429 | 0 | aldo-keto reductase |
| TVAG_TEG_DS113597_1_3 | −3.086 | 0 | maverick transposable element conserved hypothetical protein |
| TVAG_357190 | −2.748 | 0 | calcium-dependent protein kinase |
| TVAG_524510 | −2.735 | 0.005 | helicase |
| TVAG_355170 | −2.575 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_248020 | −2.501 | 0 | ankyrin repeat-containing protein |
| TVAG_197120 | −2.461 | 0.002 | ion channel nompc |
| TVAG_409820 | −2.407 | 0 | riboflavin kinase/fmn adenylyltransferase |
| TVAG_410410 | −2.212 | 0 | DNA double-strand break repair Rad50 ATPase |
| TVAG_473500 | −2.175 | 0 | inversin |
| TVAG_249030 | −1.989 | 0 | DNA double-strand break repair Rad50 ATPase |
| TVAG_002620 | −1.73 | 0.097 | ankyrin repeat-containing protein |
| TVAG_212110 | −1.697 | 0 | dynein light chain Tctex-type |
| TVAG_136630 | −1.629 | 0 | ankyrin repeat-containing protein |
| TVAG_455660 | −1.564 | 0 | clan CA, family C12, ubiquitin hydrolase-like cysteine peptidase |
| TVAG_290850 | −1.451 | 0 | casein kinase II beta chain |
| TVAG_095200 | −1.25 | 0.001 | RNA polymerase II ctd phosphatase |
| Shared Upregulated Genes in All MTZ-R Transcriptomes Treated with MTZ | |||
|---|---|---|---|
| Gene ID | MTZ-R_MTZ/ MTZ-S_MTZ (log2FoldChange) | Padj | TrichDB Annotation |
| TVAG_177180 | 6.268 | 0 | WD-repeat protein |
| TVAG_219680 | 5.775 | 0 | guanylate cyclase beta 1 subunit |
| TVAG_195930 | 5.149 | 0 | 5′-nucleotidase surE |
| TVAG_019720 | 4.133 | 0 | amino acid transporter |
| TVAG_396470 | 3.167 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_316160 | 3.112 | 0 | protein phosphatase-5 |
| TVAG_214840 | 2.522 | 0 | CMGC family protein kinase |
| TVAG_038060 | 2.287 | 0 | ubiquitin-conjugating enzyme E2-25kD |
| TVAG_254920 | 2.087 | 0.011 | Na+-driven multidrug efflux pump |
| TVAG_397320 | 1.975 | 0 | adenosine diphosphatase |
| TVAG_145340 | 1.823 | 0 | alpha-galactosidase/alpha-N-acetylgalactosaminidase |
| TVAG_360900 | 1.499 | 0 | bsu-protein phosphatase |
| Shared Downregulated Genes in All MTZ-R Transcriptomes Treated with MTZ | |||
| TVAG_073330 | −7.547 | 0 | CAMK family protein kinase |
| TVAG_078520 | −7.152 | 0 | abcb9 |
| TVAG_262830 | −6.299 | 0 | hydroxyacyl dehydrogenase |
| TVAG_262850 | −5.986 | 0 | short-chain dehydrogenases/reductase |
| TVAG_394080 | −5.978 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_524510 | −5.46 | 0 | helicase |
| TVAG_273350 | −4.943 | 0 | r2r3-MYB transcription factor |
| TVAG_208780 | −4.935 | 0 | ankyrin repeat-containing protein |
| TVAG_RG_DS113355_8 | −4.864 | 0 | metallophosphoesterase domain-containing protein |
| TVAG_431960 | −4.565 | 0 | ABC transporter |
| TVAG_462960 | −4.472 | 0.01 | ankyrin |
| TVAG_309070 | −3.952 | 0 | ankyrin repeat-containing protein |
| TVAG_248020 | −3.602 | 0 | ankyrin repeat-containing protein |
| TVAG_048690 | −3.558 | 0 | aldo-keto reductase |
| TVAG_456680 | −3.513 | 0 | tankyrase |
| TVAG_060380 | −3.503 | 0 | 5′->3′ exoribonuclease |
| TVAG_360070 | −3.458 | 0 | TKL family protein kinase |
| TVAG_477880 | −3.449 | 0 | CAMK family protein kinase |
| TVAG_343700 | −3.408 | 0 | ankyrin repeat-containing protein |
| TVAG_373800 | −3.212 | 0 | CAMK family protein kinase |
| TVAG_080130 | −3.174 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_371570 | −3.061 | 0 | 4-alpha-glucanotransferase |
| TVAG_248040 | −2.897 | 0 | actin |
| TVAG_262740 | −2.857 | 0 | ankyrin 2,3/unc44 |
| TVAG_078510 | −2.832 | 0 | synaptonemal complex protein |
| TVAG_152410 | −2.828 | 0 | sugar transporter |
| TVAG_328000 | −2.719 | 0.122 | MYB-1 |
| TVAG_078450 | −2.615 | 0 | leucine-rich repeat protein, BspA family |
| TVAG_329660 | −2.585 | 0 | alcohol dehydrogenase |
| TVAG_108070 | −2.561 | 0 | alpha-amylase |
| TVAG_318200 | −2.544 | 0.001 | r2r3-MYB transcription factor |
| TVAG_171590 | −2.473 | 0.009 | histone-lysine N-methyltransferase, bat/ehmt |
| TVAG_415420 | −2.453 | 0 | zinc-iron transporter |
| TVAG_473500 | −2.365 | 0 | inversin |
| TVAG_455660 | −2.252 | 0 | clan CA, family C12, ubiquitin hydrolase-like cysteine peptidase |
| TVAG_174020 | −2.189 | 0.006 | CAMK family protein kinase |
| TVAG_480660 | −2.168 | 0 | aromatic amino acid transporter |
| TVAG_050900 | −2.131 | 0 | myotrophin |
| TVAG_437170 | −2.094 | 0 | rolling pebbles |
| TVAG_425530 | −2.091 | 0.019 | CK1 family protein kinase |
| TVAG_457450 | −2.087 | 0.001 | KS1 protein precursor |
| TVAG_020440 | −2.045 | 0 | tankyrase |
| TVAG_096770 | −2.032 | 0 | ankyrin repeat-containing protein |
| TVAG_198600 | −1.896 | 0 | DNA polymerase alpha catalytic subunit |
| TVAG_479220 | −1.8 | 0.003 | heat shock protein |
| TVAG_198590 | −1.698 | 0 | homo sapiens cgi-128 protein |
| TVAG_254890 | −1.441 | 0 | pyruvate–flavodoxin oxidoreductase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.-J.; Huang, C.-Y.; Li, Y.-X.; Liu, Y.-C.; Chu, L.-J.; Yeh, Y.-M.; Cheng, W.-H.; Chen, R.-M.; Lee, C.-C.; Chen, L.-C.; et al. Dissecting the Transcriptomes of Multiple Metronidazole-Resistant and Sensitive Trichomonas vaginalis Strains Identified Distinct Genes and Pathways Associated with Drug Resistance and Cell Death. Biomedicines 2021, 9, 1817. https://doi.org/10.3390/biomedicines9121817
Huang P-J, Huang C-Y, Li Y-X, Liu Y-C, Chu L-J, Yeh Y-M, Cheng W-H, Chen R-M, Lee C-C, Chen L-C, et al. Dissecting the Transcriptomes of Multiple Metronidazole-Resistant and Sensitive Trichomonas vaginalis Strains Identified Distinct Genes and Pathways Associated with Drug Resistance and Cell Death. Biomedicines. 2021; 9(12):1817. https://doi.org/10.3390/biomedicines9121817
Chicago/Turabian StyleHuang, Po-Jung, Ching-Yun Huang, Yu-Xuan Li, Yi-Chung Liu, Lichieh-Julie Chu, Yuan-Ming Yeh, Wei-Hung Cheng, Ruei-Ming Chen, Chi-Ching Lee, Lih-Chyang Chen, and et al. 2021. "Dissecting the Transcriptomes of Multiple Metronidazole-Resistant and Sensitive Trichomonas vaginalis Strains Identified Distinct Genes and Pathways Associated with Drug Resistance and Cell Death" Biomedicines 9, no. 12: 1817. https://doi.org/10.3390/biomedicines9121817
APA StyleHuang, P.-J., Huang, C.-Y., Li, Y.-X., Liu, Y.-C., Chu, L.-J., Yeh, Y.-M., Cheng, W.-H., Chen, R.-M., Lee, C.-C., Chen, L.-C., Lin, H.-C., Chiu, S.-F., Lin, W.-N., Lyu, P.-C., Tang, P., & Huang, K.-Y. (2021). Dissecting the Transcriptomes of Multiple Metronidazole-Resistant and Sensitive Trichomonas vaginalis Strains Identified Distinct Genes and Pathways Associated with Drug Resistance and Cell Death. Biomedicines, 9(12), 1817. https://doi.org/10.3390/biomedicines9121817







