Epigenetic Regulation of Cardiac Troponin Genes in Pediatric Patients with Heart Failure Supported by Ventricular Assist Device
Abstract
:1. Introduction
2. Materials and Methods
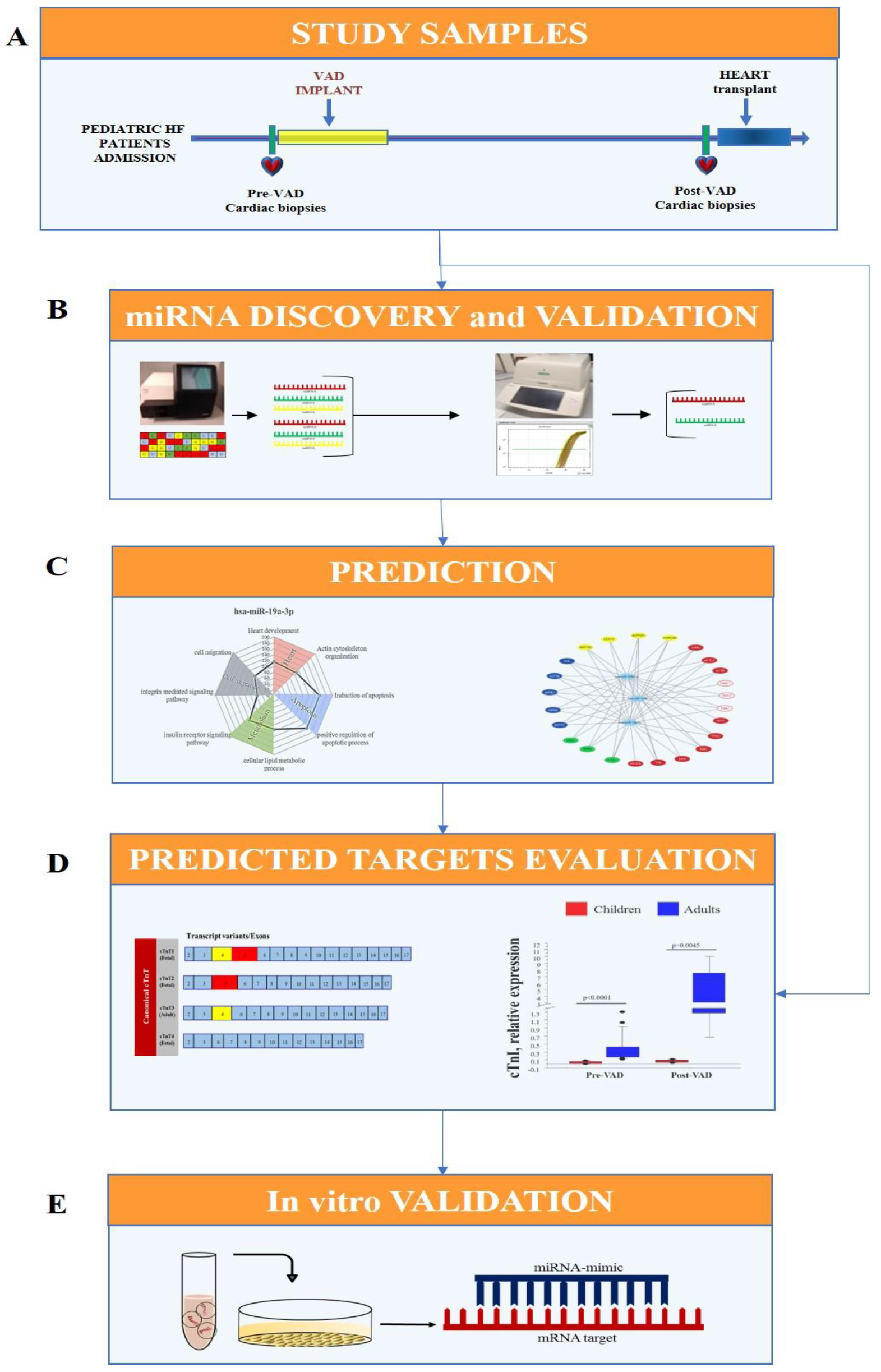
2.1. Experimental Design
- Cardiac samples were collected from pediatric HF patients at VAD implant (pre-VAD) and at heart transplant (post-VAD);
- Cardiac miRNA profile was performed by Next Generation Sequencing (NGS) in pre-VAD and post-VAD samples and sequencing results were confirmed by real-time PCR and the validated miRNA were selected for future analysis;
- Sarcomere components, including cardiac troponins, were identified as putative targets of selected miRNAs by an in-silico analysis;
- An in vitro transfection study using miRNA mimic in HL-1 cell line was carried out for testing the regulatory role of selected miRNA on predicted targets.
2.2. Study Sample
2.3. RNA Extraction
2.4. Libraries and NGS
2.5. Bioinformatic Analysis of Identified miRNAs
2.6. NGS Data Validation
2.7. Bioinformatic Prediction of miRNA Targets
2.8. Troponin Expression in Cardiac Tissue
2.9. In Vitro Study
2.10. Statistical Analysis
3. Results
3.1. Clinical Characteristics of HF Pediatric Patients
3.2. miRNA Profiling
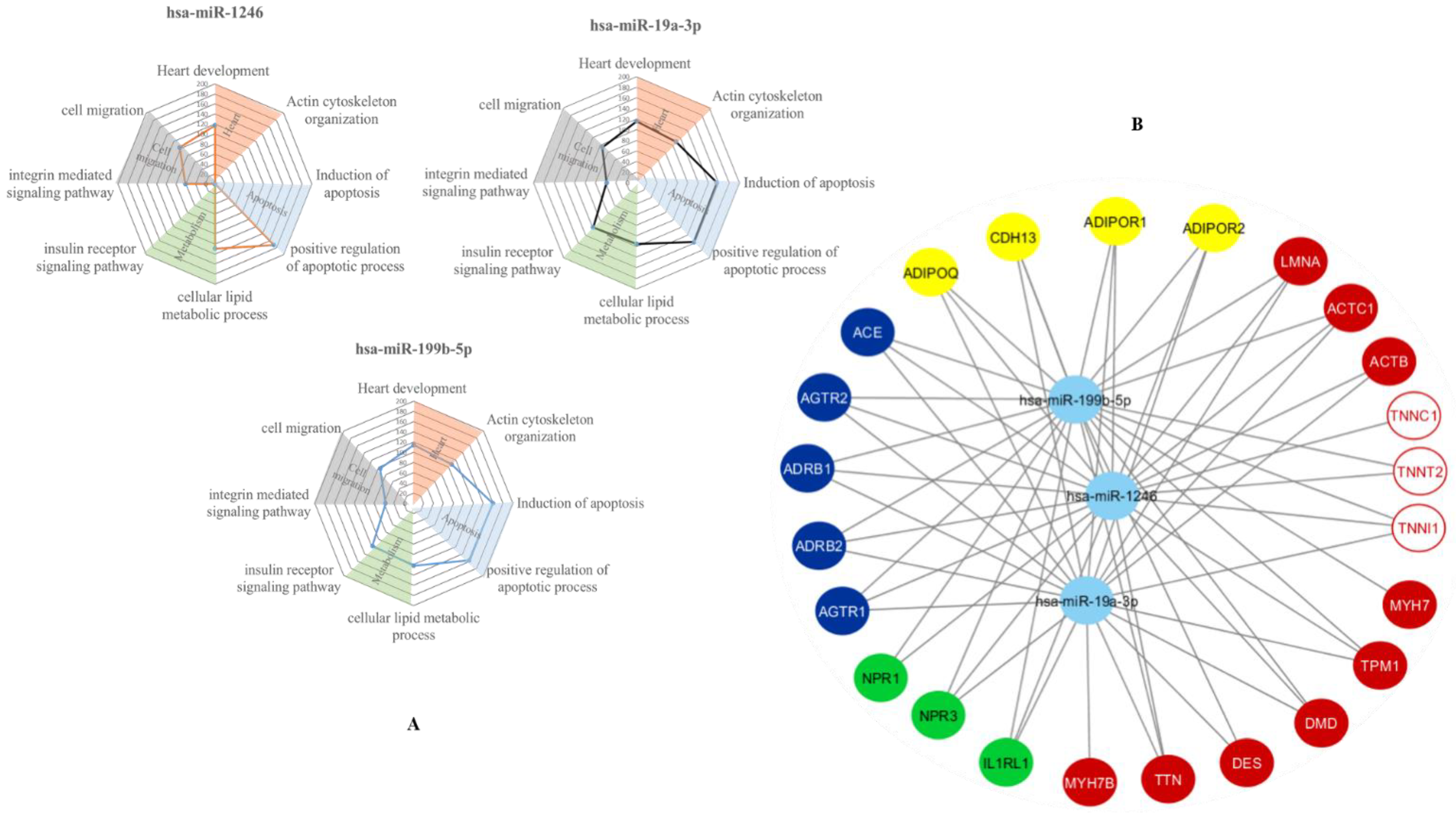
3.3. In-Silico Prediction of Putative miRNA Targets
3.4. Cardiac Troponin Complex
3.5. Comparison of Expression Levels of Validate Cardiac miRNAs and Cardiac Troponin Complex in HF Children Supported by VAD
3.6. In Vitro Validation of Putative Targets of Selected Cardiac miRNAs
4. Discussion
- -
- Cardiac miRNA profile was modified in HF children by VAD support, as assessed by NGS and real-time PCR;
- -
- The re-expression of fetal isoforms of cTnT and ssTnI can be observed in cardiac tissue from HF children;
- -
- The NGS-identified miRNAs were involved in the regulation of troponin complex, as confirmed by the in vitro study.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsu, D.T.; Pearson, G.D. Heart failure in children: Part I: History, etiology, and pathophysiology. Circulation. Heart Fail. 2009, 2, 63–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, D.T.; Pearson, G.D. Heart failure in children: Part II: Diagnosis, treatment, and future directions. Circulation. Heart Fail. 2009, 2, 490–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossano, J.W.; Kim, J.J.; Decker, J.A.; Price, J.F.; Zafar, F.; Graves, D.E.; Morales, D.L.; Heinle, J.S.; Bozkurt, B.; Towbin, J.A.; et al. Prevalence, morbidity, and mortality of heart failure-related hospitalizations in children in the United States: A population-based study. J. Card. Fail. 2012, 18, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Kirk, R.; Dipchand, A.I.; Rosenthal, D.N.; Addonizio, L.; Burch, M.; Chrisant, M.; Dubin, A.; Everitt, M.; Gajarski, R.; Mertens, L.; et al. The International Society for Heart and Lung Transplantation Guidelines for the management of pediatric heart failure: Executive summary. [Corrected]. J. Heart Lung Transplant. Off. Publ. Int. Soc. Heart Transplant. 2014, 33, 888–909. [Google Scholar] [CrossRef]
- Ragusa, R.; Di Molfetta, A.; Amodeo, A.; Trivella, M.G.; Caselli, C. Pathophysiology and molecular signalling in pediatric heart failure and VAD therapy. Clin. Chim. Acta 2020, 510, 751–759. [Google Scholar] [CrossRef]
- Zafar, F.; Castleberry, C.; Khan, M.S.; Mehta, V.; Bryant, R., 3rd; Lorts, A.; Wilmot, I.; Jefferies, J.L.; Chin, C.; Morales, D. Pediatric heart transplant waiting list mortality in the era of ventricular assist devices. J. Heart Lung Transplant. Off. Publ. Int. Soc. Heart Transplant. 2015, 34, 82–88. [Google Scholar] [CrossRef]
- Napoli, C.; Grimaldi, V.; De Pascale, M.R.; Sommese, L.; Infante, T.; Soricelli, A. Novel epigenetic-based therapies useful in cardiovascular medicine. World J. Cardiol. 2016, 8, 211–219. [Google Scholar] [CrossRef]
- Shah, P.; Bristow, M.R.; Port, J.D. MicroRNAs in Heart Failure, Cardiac Transplantation, and Myocardial Recovery: Biomarkers with Therapeutic Potential. Curr. Heart Fail. Rep. 2017, 14, 454–464. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, H.; Liu, F.; Chen, L.; Luo, W.; Su, P.; Li, W.; Yu, L.; Yang, X.; Cai, J. Identification of micro-RNA networks in end-stage heart failure because of dilated cardiomyopathy. J. Cell. Mol. Med. 2013, 17, 1173–1187. [Google Scholar] [CrossRef]
- Barsanti, C.; Trivella, M.G.; D’Aurizio, R.; El Baroudi, M.; Baumgart, M.; Groth, M.; Caruso, R.; Verde, A.; Botta, L.; Cozzi, L.; et al. Differential regulation of microRNAs in end-stage failing hearts is associated with left ventricular assist device unloading. BioMed Res. Int. 2015, 2015, 592512. [Google Scholar] [CrossRef]
- Jefferies, J.; Chang, A.; Rossano, J.; Shaddy, R.; Towbin, J. Heart Failure in the Child and Young Adult: From Bench to Bedside, 1st ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 1–824. [Google Scholar]
- Stauffer, B.L.; Russell, G.; Nunley, K.; Miyamoto, S.D.; Sucharov, C.C. miRNA expression in pediatric failing human heart. J. Mol. Cell. Cardiol. 2013, 57, 43–46. [Google Scholar] [CrossRef] [Green Version]
- Caselli, C.; D’Amico, A.; Ragusa, R.; Caruso, R.; Prescimone, T.; Cabiati, M.; Nonini, S.; Marraccini, P.; Del Ry, S.; Trivella, M.G.; et al. IL-33/ST2 pathway and classical cytokines in end-stage heart failure patients submitted to left ventricular assist device support: A paradoxic role for inflammatory mediators? Mediat. Inflamm. 2013, 2013, 498703. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, A.; Ragusa, R.; Caruso, R.; Prescimone, T.; Nonini, S.; Cabiati, M.; Del Ry, S.; Trivella, M.G.; Giannessi, D.; Caselli, C. Uncovering the cathepsin system in heart failure patients submitted to Left Ventricular Assist Device (LVAD) implantation. J. Transl. Med. 2014, 12, 350. [Google Scholar] [CrossRef] [Green Version]
- Bascetta, L.; Oliviero, A.; D’Aurizio, R.; Evangelista, M.; Mercatanti, A.; Pellegrini, M.; Marrocolo, F.; Bracarda, S.; Rizzo, M. The Prostate Cancer Cells Resistant to Docetaxel as in vitro Model for Discovering MicroRNAs Predictive of the Onset of Docetaxel Resistance. Int. J. Mol. Sci. 2017, 18, 1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragusa, R.; Di Molfetta, A.; D’Aurizio, R.; Del Turco, S.; Cabiati, M.; Del Ry, S.; Basta, G.; Pitto, L.; Amodeo, A.; Trivella, M.G.; et al. Variations of circulating miRNA in paediatric patients with Heart Failure supported with Ventricular Assist Device: A pilot study. Sci. Rep. 2020, 10, 5905. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, H. Myocardial Infarction: The Protective Role of MiRNAs in Myocardium Pathology. Front. Cardiovasc. Med. 2021, 8, 631817. [Google Scholar] [CrossRef] [PubMed]
- Vegter, E.L.; van der Meer, P.; de Windt, L.J.; Pinto, Y.M.; Voors, A.A. MicroRNAs in heart failure: From biomarker to target for therapy. Eur. J. Heart Fail. 2016, 18, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Matkovich, S.J.; Van Booven, D.J.; Youker, K.A.; Torre-Amione, G.; Diwan, A.; Eschenbacher, W.H.; Dorn, L.E.; Watson, M.A.; Margulies, K.B.; Dorn, G.W., 2nd. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation 2009, 119, 1263–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leptidis, S.; El Azzouzi, H.; Lok, S.I.; de Weger, R.; Olieslagers, S.; Kisters, N.; Silva, G.J.; Heymans, S.; Cuppen, E.; Berezikov, E.; et al. A deep sequencing approach to uncover the miRNOME in the human heart. PLoS ONE 2013, 8, e57800. [Google Scholar] [CrossRef]
- da Costa Martins, P.A.; Salic, K.; Gladka, M.M.; Armand, A.S.; Leptidis, S.; el Azzouzi, H.; Hansen, A.; Coenen-de Roo, C.J.; Bierhuizen, M.F.; van der Nagel, R.; et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat. Cell Biol. 2010, 12, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Duygu, B.; Poels, E.M.; Juni, R.; Bitsch, N.; Ottaviani, L.; Olieslagers, S.; de Windt, L.J.; da Costa Martins, P.A. miR-199b-5p is a regulator of left ventricular remodeling following myocardial infarction. Non-Coding RNA Res. 2017, 2, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Wang, F.; Gao, R.; Wu, J.; Ou, Y.; Chen, X.; Wang, T.; Zhou, X.; Zhu, W.; Li, P.; et al. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced fibrogenesis in human cardiac fibroblasts. Sci. Rep. 2016, 6, 24747. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Hao, Q.; Wei, J.; Li, G.H.; Wu, Y.; Zhao, Y.F. MicroRNA-19a/b-3p protect the heart from hypertension-induced pathological cardiac hypertrophy through PDE5A. J. Hypertens. 2018, 36, 1847–1857. [Google Scholar] [CrossRef]
- Mao, Z.J.; Zhang, Q.L.; Shang, J.; Gao, T.; Yuan, W.J.; Qin, L.P. Shenfu Injection attenuates rat myocardial hypertrophy by up-regulating miR-19a-3p expression. Sci. Rep. 2018, 8, 4660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sisakian, H. Cardiomyopathies: Evolution of pathogenesis concepts and potential for new therapies. World J. Cardiol. 2014, 6, 478–494. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.V.; Venkatraman, G.; Davis, J.P.; Tikunova, S.B.; Engel, P.; Solaro, R.J.; Potter, J.D. Cardiac troponin T isoforms affect the Ca(2+) sensitivity of force development in the presence of slow skeletal troponin I: Insights into the role of troponin T isoforms in the fetal heart. J. Biol. Chem. 2004, 279, 49579–49587. [Google Scholar] [CrossRef] [Green Version]
- Anderson, P.A.; Malouf, N.N.; Oakeley, A.E.; Pagani, E.D.; Allen, P.D. Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ. Res. 1991, 69, 1226–1233. [Google Scholar] [CrossRef] [Green Version]
- Li, M.X.; Hwang, P.M. Structure and function of cardiac troponin C (TNNC1): Implications for heart failure, cardiomyopathies, and troponin modulating drugs. Gene 2015, 571, 153–166. [Google Scholar] [CrossRef] [Green Version]



| HF Children Pre-VAD | HF Children Post-VAD | p-Value | |
|---|---|---|---|
| Age, months | 29 (5–123) | - | |
| Male gender (n) | 6 (13) | - | |
| Etiology, n (%) | |||
| DCM | 69.2% | - | |
| LV non-compaction | 15.4% | - | |
| RCM | 15.4% | - | |
| Weight, Kg | 9 (4.8–26) | - | |
| LVEF, % | 19 (13.75–20.75) | 37 (26–45) | p = 0.0273 |
| LVEDV, mL | 57 (37.5–83) | 24 (15.25–30.75) | p = 0.0087 |
| LVESV, mL | 37 (28.7–71) | 14 (8.72–20) | p = 0.0201 |
| LVEDD, mm | 47 (42.5–56.5) | 41 (30.5–49.5) | ns |
| LVESD, mm | 44.5 (39–57) | 37 (23–45) | ns |
| TAPSE, mm | 0.985 (0.7–1.25) | 0.72 (0.63–1.015) | ns |
| RVFAC, % | 33 (23.5–41) | 45 (40.5–56.25) | p = 0.049 |
| White blood cells, × 109/L | 9.2 (7.76–12.7) | 8.77 (8.07–9.65) | ns |
| Hb, g/dL | 12.9 (12.4–14.2) | 11.7 (10.3–12.67) | ns |
| Platelets, × 109/L | 272.5 (204–289) | 327 (252–403) | ns |
| INR | 1.2 (1.14–1.2) | 1.1 (1–2.55) | ns |
| Urea nitrogen, mg/dL | 15.5 (12.7–32.5) | 21 (10–31.25) | ns |
| Creatinine, mg/dL | 0.43 (0.275–0.615) | 0.37 (0.2–0.5) | ns |
| Albumin, g/dL | 4.2 (3.6–5.25) | 4.1 (3.9–4.4) | ns |
| Glucose, mg/dL | 113 (71–128) | 91 (87.75–120.75) | ns |
| C-reactive Protein, mg/dL | 0.41 (0.11–1.5) | 0.8 (0.2–1.7) | ns |
| Bilirubin tot, mg/dL | 1.2 (0.51–1.95) | 0.36 (0.25–0.72) | p = 0.0161 |
| Lactate Dehydrogenase, U/L | 812 (405–1011) | 742 (634–1088) | ns |
| Pre-VAD | Post-VAD | |||||
|---|---|---|---|---|---|---|
| Children | Adult | p-Value | Children | Adult | p-Value | |
| cTnI | 0.03 ± 0.009 | 0.38 ± 0.08 | p < 0.0001 | 0.07 ± 0.012 | 5.06 ± 1.75 * | p = 0.0045 |
| ssTnI | 0.3 ± 0.096 | 0.58 ± 0.38 | ns | 0.77 ± 0.49 | --- | --- |
| cTnC | 0.58 ± 0.176 | 8.87 ± 1.57 | p < 0.0001 | 1.179 ± 0.328 | 16.79 ± 3.8 * | p = 0.0045 |
| cTnT 1 | 0.48 ± 0.13 | 1 ± 0.13 | p = 0.01 | 0.89 ± 0.27 | 4.27 ± 1.17 * | p = 0.006 |
| cTnT 2 | 0.86 ± 0.22 | 1.08 ± 0.18 | ns | 1.04 ± 0.29 | 8.06 ± 2.79 * | p = 0.008 |
| cTnT 3 | 0.16 ± 0.03 | 0.88 ± 0.9 | p < 0.0001 | 0.54 ± 0.12 | 2.8 ± 0.6 * | p = 0.0074 |
| cTnT 4 | 0.15 ± 0.032 | 0.866 ± 0.09 | p < 0.0001 | 0.6 ± 0.14 | 1.66 ± 0.24 * | p = 0.007 |
| cTnT 12 | 0.11 ± 0.03 | 0.79 ± 0.09 | p < 0.0001 | 0.22 ± 0.04 | 0.82 ± 0.27 | p = 0.0045 |
| cTnT 10,11,12 | 0.37 ± 0.066 | 1 ± 0.129 | p = 0.0002 | 0.84 ± 0.13 | 5.08 ± 1.84 * | p = 0.0017 |
| hsa-miR-1246 | hsa-miR-19a-3p | hsa-miR-199b-5p | |
|---|---|---|---|
| cTnI | Rho = −0.744 p = 0.004 | Rho = −0.853 p = 0.001 | Rho = −0.591 p = 0.022 |
| ssTnI | ns | Rho = −0.808 p = 0.0051 | ns |
| cTnC | Rho = −0.686 p = 0.01 | Rho = −0.625 p = 0.0194 | ns |
| cTnT1 | ns | ns | ns |
| cTnT2 | ns | ns | ns |
| cTnT3 | Rho = −0.771 p = 0.0028 | Rho = −0.706 p = 0.0063 | Rho = −0.562 p = 0.0296 |
| cTnT4 | Rho = −0.791 p = 0.0022 | Rho = −0.732 p = 0.0046 | Rho = −0.624 p = 0.0157 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragusa, R.; Di Molfetta, A.; Del Turco, S.; Cabiati, M.; Del Ry, S.; Basta, G.; Mercatanti, A.; Pitto, L.; Amodeo, A.; Trivella, M.G.; et al. Epigenetic Regulation of Cardiac Troponin Genes in Pediatric Patients with Heart Failure Supported by Ventricular Assist Device. Biomedicines 2021, 9, 1409. https://doi.org/10.3390/biomedicines9101409
Ragusa R, Di Molfetta A, Del Turco S, Cabiati M, Del Ry S, Basta G, Mercatanti A, Pitto L, Amodeo A, Trivella MG, et al. Epigenetic Regulation of Cardiac Troponin Genes in Pediatric Patients with Heart Failure Supported by Ventricular Assist Device. Biomedicines. 2021; 9(10):1409. https://doi.org/10.3390/biomedicines9101409
Chicago/Turabian StyleRagusa, Rosetta, Arianna Di Molfetta, Serena Del Turco, Manuela Cabiati, Silvia Del Ry, Giuseppina Basta, Alberto Mercatanti, Letizia Pitto, Antonio Amodeo, Maria Giovanna Trivella, and et al. 2021. "Epigenetic Regulation of Cardiac Troponin Genes in Pediatric Patients with Heart Failure Supported by Ventricular Assist Device" Biomedicines 9, no. 10: 1409. https://doi.org/10.3390/biomedicines9101409
APA StyleRagusa, R., Di Molfetta, A., Del Turco, S., Cabiati, M., Del Ry, S., Basta, G., Mercatanti, A., Pitto, L., Amodeo, A., Trivella, M. G., Rizzo, M., & Caselli, C. (2021). Epigenetic Regulation of Cardiac Troponin Genes in Pediatric Patients with Heart Failure Supported by Ventricular Assist Device. Biomedicines, 9(10), 1409. https://doi.org/10.3390/biomedicines9101409











