Identification of Candidate Genes for Endometriosis in a Three-Generation Family with Multiple Affected Members Using Whole-Exome Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. The Multiplex Family
2.2. Whole-Exome Sequencing
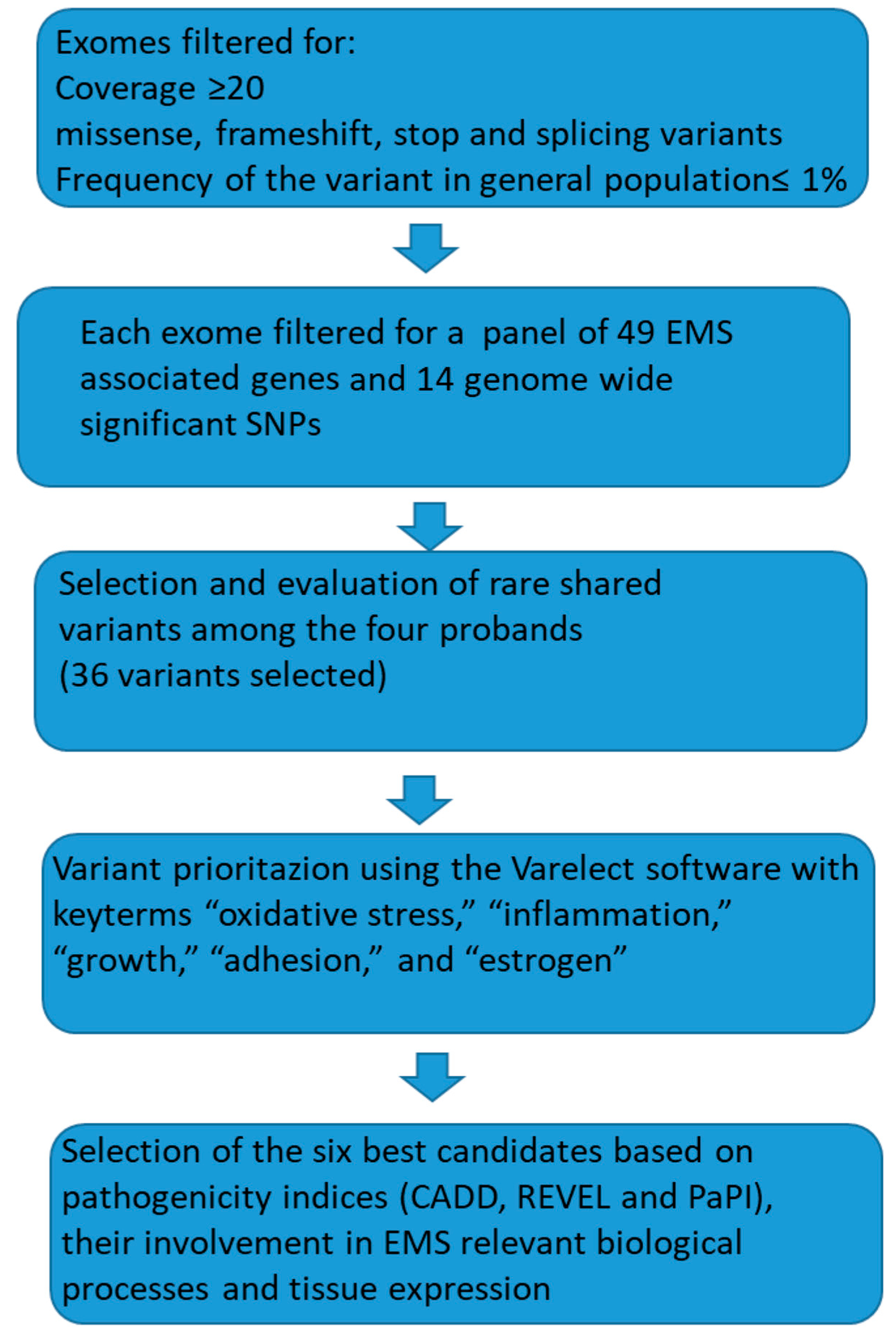
2.3. Bioinformatic Analysis
3. Results
- Minor allele frequency (MAF) and number of homozygotes, as per the GnomAD v2.1.1 database (https://gnomad.broadinstitute.org/ accessed on 22 September 2024).
- Predicted functional impact using in silico tools such as SIFT, MutationTaster, and PolyPhen, alongside integrated scores including CADD, REVEL, and PaPI indices.
- Biological relevance to processes like growth, inflammation, adhesion, and oxidative stress, assessed through a literature review.
- Tissue-specific expression profiles, utilizing data from the GTEx database (https://www.gtexportal.org/home/aboutAdultGtex V10 accessed on 22 September 2024).
| Gene | Gene Function | Variant | MAF | ClinVar | REVEL | CADD | PaPI | In Silico Tools: P/VUS/B+LB | CONS Score | Expression | Expression Levels in Uterine/Endometrial Tissues | Main Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAMB4 | Cell adhesion, growth, and migration (oncosuppressor) | NM_007356.3: c.3319G>A, Gly1107Arg | 3/251,316 | VUS (1) RCV004078131 | 0.715 | 24.6 | 1.0 | 10/9/8 | 4.512 | Skin, but also other tissues | low | [26] |
| EGFL6 | Growth factor (oncogene) | NM_015507.4: c.1414G>A, Gly472Arg | 5/183,514 | VUS (1) RCV004071274 | 0.367 | 23.5 | 1.0 | 10/9/8 | 5.076 | Cutaneous and visceral adipocytes, vagina, cervix, breast, and lung | moderate | [29] |
| NAV3 | Regulates microtubule dynamics, inhibits migration, and restrains dissemination of tumors (oncosuppressor) | NM_001024383.2: c.7154T>C, Leu2385Pro | 139/275,880 | / | 0.425 | 27.8 | 1.0 | 9/7/5 | 7.844 | Ovary, vagina, uterus, and cervix, but also other tissues | low | [30] |
| ADAMTS18 | Proliferation and migration (oncogene) | NM_199355.4: c.1474G>T, Gly492Trp | / | / | 0.088 | 24.1 | 0.92 | -/5/25 | 3.45 | Cervix, breast, cerebellum, and other tissues | moderate | [32] |
| SLIT1 | Growth (oncosuppressor) | NM_003061.3 c.967T>G; Ser323Ala | / | / | 0.132 | 23 | 0.98 | 2/6/20 | 5.514 | Mainly brain | low | [33] |
| MLH1 | Mismatch repair (oncosuppressor) | NM_000249.4: c.977T>C; Val326Ala | 131/282,782 | CIP (2VUS/25B-LB) RCV000035356 (most recent) | 0.846 | 24.5 | 1.0 | 8/11/1 | 7.661 | All tissues | high | [34] |
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- As-Sanie, S.; Mackenzie, S.C.; Morrison, L.; Schrepf, A.; Zondervan, K.T.; Horne, A.W.; Missmer, S.A. Endometriosis: A Review. JAMA 2025, 334, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Waseem, S.; Luo, L. Advances in the diagnosis and management of endometriosis: A com sprehensive review. Pathol. Res. Pract. 2025, 266, 155813. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Peng, H.; Huang, X.; Qi, X. The association between endometriosis and risk of endometrial cancer and breast cancer: A meta-analysis. BMC Womens Health 2022, 22, 455. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dentillo, D.B.; Meola, J.; Ferriani, R.A.; Rosa-E-Silva, J.C. Common Dysregulated Genes in Endometriosis and Malignancies. Rev. Bras. Ginecol. Obstet. 2016, 38, 253–262. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, R.; Zheng, Y.; Wang, J.; Xu, H.; Zhang, R.; Xie, Z.; Zhang, L.; Zhao, R. Identification of key genes associated with endometriosis and endometrial cancer by bioinformatics analysis. Front. Oncol. 2024, 14, 1387860. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Giudice, L.C.; Kao, L.C. Endometriosis. Lancet 2004, 364, 1789–1799. [Google Scholar] [CrossRef] [PubMed]
- Cousins, F.L.; McKinnon, B.D.; Mortlock, S.; Fitzgerald, H.C.; Zhang, C.; Montgomery, G.W.; Gargett, C.E. New concepts on the etiology of endometriosis. J. Obstet. Gynaecol. Res. 2023, 49, 1090–1105. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Polak, G.; Banaszewska, B.; Filip, M.; Radwan, M.; Wdowiak, A. Environmental Factors and Endometriosis. Int. J. Environ. Res. Public Health 2021, 18, 11025. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Simpson, J.L.; Elias, S.; Malinak, L.R.; Buttram, V.C., Jr. Heritable aspects of endometriosis. I. Genet. Stud. Am. J. Obstet. Gynecol. 1980, 137, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Parasar, P.; Ozcan, P.; Terry, K.L. Endometriosis: Epidemiology, Diagnosis and Clinical Management. Curr. Obstet. Gynecol. Rep. 2017, 6, 34–41. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Frey, G.H. The familial occurrence of endometriosis; report of five instances and review of the literature. Am. J. Obstet. Gynecol. 1957, 73, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, S.; Hadfield, R.; Mardon, H.; Barlow, D. Age of onset of pain symptoms in non-twin sisters concordant for endometriosis. Hum. Reprod. 1996, 11, 403–405. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sapkota, Y.; Steinthorsdottir, V.; Morris, A.P.; Fassbender, A.; Rahmioglu, N.; De Vivo, I.; Buring, J.E.; Zhang, F.; Edwards, T.L.; Jones, S.; et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017, 8, 15539. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tapmeier, T.T.; Rahmioglu, N.; Lin, J.; De Leo, B.; Obendorf, M.; Raveendran, M.; Fischer, O.M.; Bafligil, C.; Guo, M.; Harris, R.A.; et al. Neuropeptide S receptor 1 is a nonhormonal treatment target in endometriosis. Sci. Transl. Med. 2021, 13, eabd6469. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Malentacchi, F.; Fambrini, M.; Harrath, A.H.; Huang, H.; Petraglia, F. Epigenetics of Estrogen and Progesterone Receptors in Endometriosis. Reprod. Sci. 2020, 27, 1967–1974. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Seidita, I.; Vannuccini, S.; Prisinzano, M.; Donati, C.; Petraglia, F. Epigenetics, endometriosis and sex steroid receptors: An update on the epigenetic regulatory mechanisms of estrogen and progesterone receptors in patients with endometriosis. Vitam. Horm. 2023, 122, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Ochoa Bernal, M.A.; Fazleabas, A.T. The Known, the Unknown and the Future of the Pathophysiology of Endometriosis. Int. J. Mol. Sci. 2024, 25, 5815. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351, Erratum in Nucleic Acids Res. 2022, 50, 8999. https://doi.org/10.1093/nar/gkac610. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nicora, G.; Limongelli, I.; Gambelli, P.; Memmi, M.; Malovini, A.; Mazzanti, A.; Napolitano, C.; Priori, S.; Bellazzi, R. CardioVAI: An automatic implementation of ACMG-AMP variant interpretation guidelines in the diagnosis of cardiovascular diseases. Hum. Mutat. 2018, 39, 1835–1846. [Google Scholar] [CrossRef] [PubMed]
- Santin, A.; Spedicati, B.; Morgan, A.; Lenarduzzi, S.; Tesolin, P.; Nardone, G.G.; Mazzà, D.; Di Lorenzo, G.; Romano, F.; Buonomo, F.; et al. Puzzling Out the Genetic Architecture of Endometriosis: Whole-Exome Sequencing and Novel Candidate Gene Identification in a Deeply Clinically Characterised Cohort. Biomedicines 2023, 11, 2122. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stelzer, G.; Plaschkes, I.; Oz-Levi, D.; Alkelai, A.; Olender, T.; Zimmerman, S.; Twik, M.; Belinky, F.; Fishilevich, S.; Nudel, R.; et al. VarElect: The phenotype-based variation prioritizer of the GeneCards Suite. BMC Genom. 2016, 17 (Suppl. S2), 444. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Choi, M.R.; An, C.H.; Yoo, N.J.; Lee, S.H. Laminin gene LAMB4 is somatically mutated and expressionally altered in gastric and colorectal cancers. APMIS 2015, 123, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.G.; Naven, M.; Harris, R.; Colley, J.; West, H.; Li, N.; Liu, Y.; Adams, R.; Maughan, T.S.; Nichols, L.; et al. Exome resequencing identifies potential tumor-suppressor genes that predispose to colorectal cancer. Hum. Mutat. 2013, 34, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jiang, R.; Yao, Y.; Qian, L.; Zhao, Y.; Huang, X. Identification of endometriosis-associated genes and pathways based on bioinformatic analysis. Medicine 2021, 100, e26530. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bai, S.; Ingram, P.; Chen, Y.C.; Deng, N.; Pearson, A.; Niknafs, Y.S.; O’Hayer, P.; Wang, Y.; Zhang, Z.Y.; Boscolo, E.; et al. EGFL6 Regulates the Asymmetric Division, Maintenance, and Metastasis of ALDH+ Ovarian Cancer Cells. Cancer Res. 2016, 76, 6396–6409, Erratum in Cancer Res. 2017, 77, 2175. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garrett, A.A.; Bai, S.; Cascio, S.; Gupta, N.; Yang, D.; Buckanovich, R.J. EGFL6 promotes endometrial cancer cell migration and proliferation. Gynecol. Oncol. 2024, 185, 75–82. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cohen-Dvashi, H.; Ben-Chetrit, N.; Russell, R.; Carvalho, S.; Lauriola, M.; Nisani, S.; Mancini, M.; Nataraj, N.; Kedmi, M.; Roth, L.; et al. Navigator-3, a modulator of cell migration, may act as a suppressor of breast cancer progression. EMBO Mol. Med. 2015, 7, 299–314. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bugaeva, O.; Maliniemi, P.; Prestvik, W.S.; Leivo, E.; Kluger, N.; Salava, A.; Virtanen, S.; Jäntti, K.; Saksela, O.; Lehti, K.; et al. Tumour Suppressor Neuron Navigator 3 and Matrix Metalloproteinase 14 are Co-expressed in Most Melanomas but Downregulated in Thick Tumours. Acta Derm. Venereol. 2023, 103, adv00883. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wei, X.; Prickett, T.D.; Viloria, C.G.; Molinolo, A.; Lin, J.C.; Cardenas-Navia, I.; Cruz, P.; NISC Comparative Sequencing Program; Rosenberg, S.A.; Davies, M.A.; et al. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol. Cancer Res. 2010, 8, 1513–1525. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marlow, R.; Strickland, P.; Lee, J.S.; Wu, X.; Pebenito, M.; Binnewies, M.; Le, E.K.; Moran, A.; Macias, H.; Cardiff, R.D.; et al. SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res. 2008, 68, 7819–7827, Erratum in Cancer Res. 2010, 70, 3853. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Takahashi, M.; Shimodaira, H.; Andreutti-Zaugg, C.; Iggo, R.; Kolodner, R.D.; Ishioka, C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007, 67, 4595–4604. [Google Scholar] [CrossRef] [PubMed]
- Ducreux, B.; Patrat, C.; Firmin, J.; Ferreux, L.; Chapron, C.; Marcellin, L.; Parpex, G.; Bourdon, M.; Vaiman, D.; Santulli, P.; et al. Systematic review on the DNA methylation role in endometriosis: Current evidence and perspectives. Clin. Epigenetics 2025, 17, 32. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ioannidou, A.; Sakellariou, M.; Sarli, V.; Panagopoulos, P.; Machairiotis, N. New Evidence About Malignant Transformation of Endometriosis-A Systematic Review. J. Clin. Med. 2025, 14, 2975. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Anglesio, M.S.; Yong, P.J. Endometriosis-associated Ovarian Cancers. Clin. Obstet. Gynecol. 2017, 60, 711–727. [Google Scholar] [CrossRef] [PubMed]
- Lamlum, H.; Ilyas, M.; Rowan, A.; Clark, S.; Johnson, V.; Bell, J.; Frayling, I.; Efstathiou, J.; Pack, K.; Payne, S.; et al. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: A new facet to Knudson’s ‘two-hit’ hypothesis. Nat. Med. 1999, 5, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Nousiainen, S.; Kuismin, O.; Reinikka, S.; Manninen, R.; Khamaiseh, S.; Kuivalainen, M.; Terho, A.; Koivurova, S.; Niinimäki, M.; Salokas, K.; et al. Whole-exome sequencing reveals candidate high-risk susceptibility genes for endometriosis. Hum. Genom. 2023, 17, 88. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kina, B.G.; Topbas Selcuki, N.F.; Bahat, P.Y.; Usta, T.; Aydin, S.; Rahmioglu, N.; Tuncer, F.N.; Oral, E. Whole exome sequencing reveals novel candidate variants for endometriosis utilizing multiple affected members in a single family. Mol. Genet. Genom. Med. 2024, 12, e2312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, Y.; Pan, H.; Han, Y.; Li, T.; Liu, K.; Wang, B. Novel missense variant of CIITA contributing to endometriosis. Reprod. Biomed. Online 2022, 45, 544–551. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lintas, C.; Azzarà, A.; Panasiti, V.; Gurrieri, F. Identification of Candidate Genes for Endometriosis in a Three-Generation Family with Multiple Affected Members Using Whole-Exome Sequencing. Biomedicines 2025, 13, 1922. https://doi.org/10.3390/biomedicines13081922
Lintas C, Azzarà A, Panasiti V, Gurrieri F. Identification of Candidate Genes for Endometriosis in a Three-Generation Family with Multiple Affected Members Using Whole-Exome Sequencing. Biomedicines. 2025; 13(8):1922. https://doi.org/10.3390/biomedicines13081922
Chicago/Turabian StyleLintas, Carla, Alessia Azzarà, Vincenzo Panasiti, and Fiorella Gurrieri. 2025. "Identification of Candidate Genes for Endometriosis in a Three-Generation Family with Multiple Affected Members Using Whole-Exome Sequencing" Biomedicines 13, no. 8: 1922. https://doi.org/10.3390/biomedicines13081922
APA StyleLintas, C., Azzarà, A., Panasiti, V., & Gurrieri, F. (2025). Identification of Candidate Genes for Endometriosis in a Three-Generation Family with Multiple Affected Members Using Whole-Exome Sequencing. Biomedicines, 13(8), 1922. https://doi.org/10.3390/biomedicines13081922