Non-Coding RNAs in Neurodevelopmental Disorders—From Diagnostic Biomarkers to Therapeutic Targets: A Systematic Review
Abstract
1. Introduction
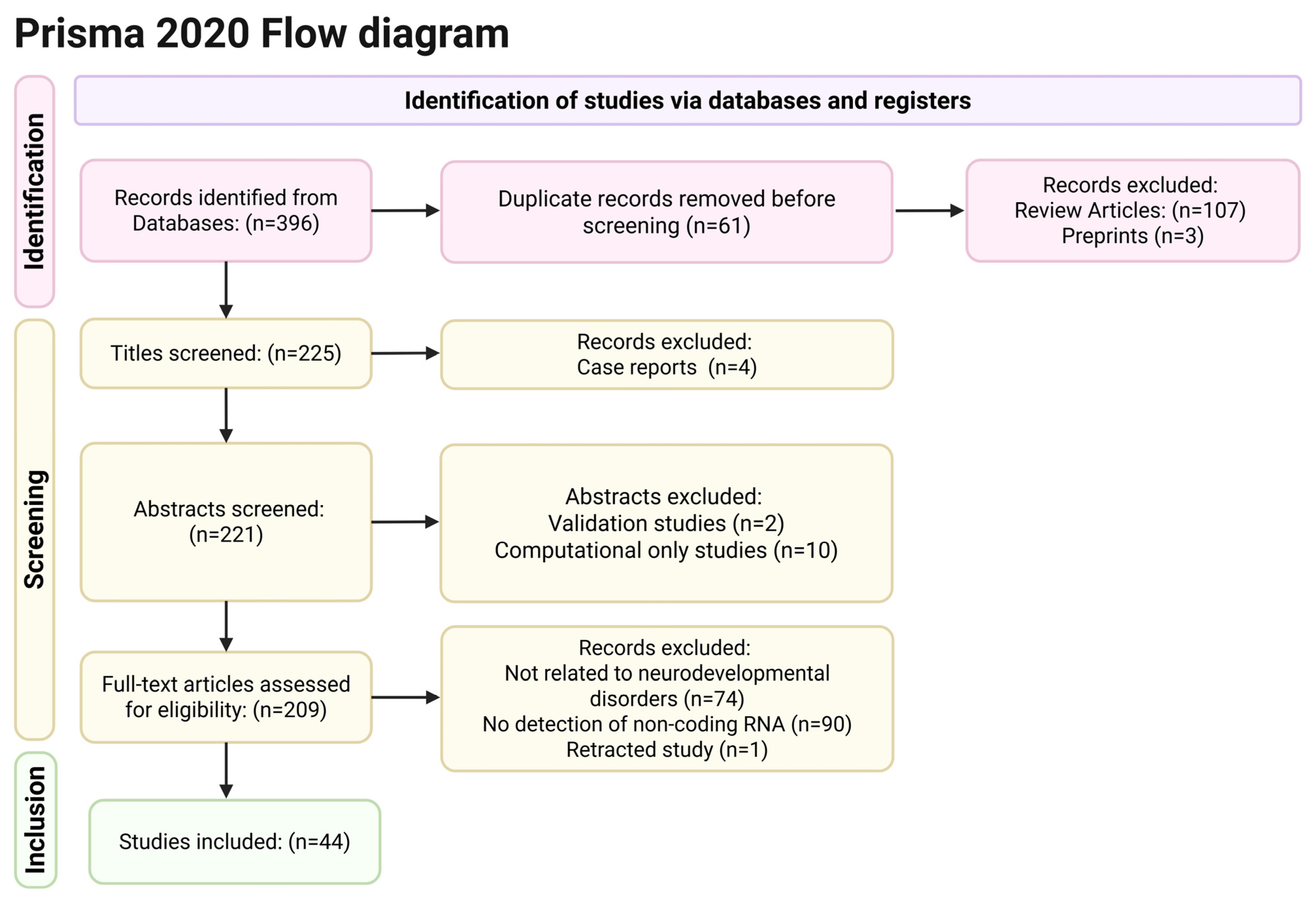
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Study Selection and Data Extraction
3. Results
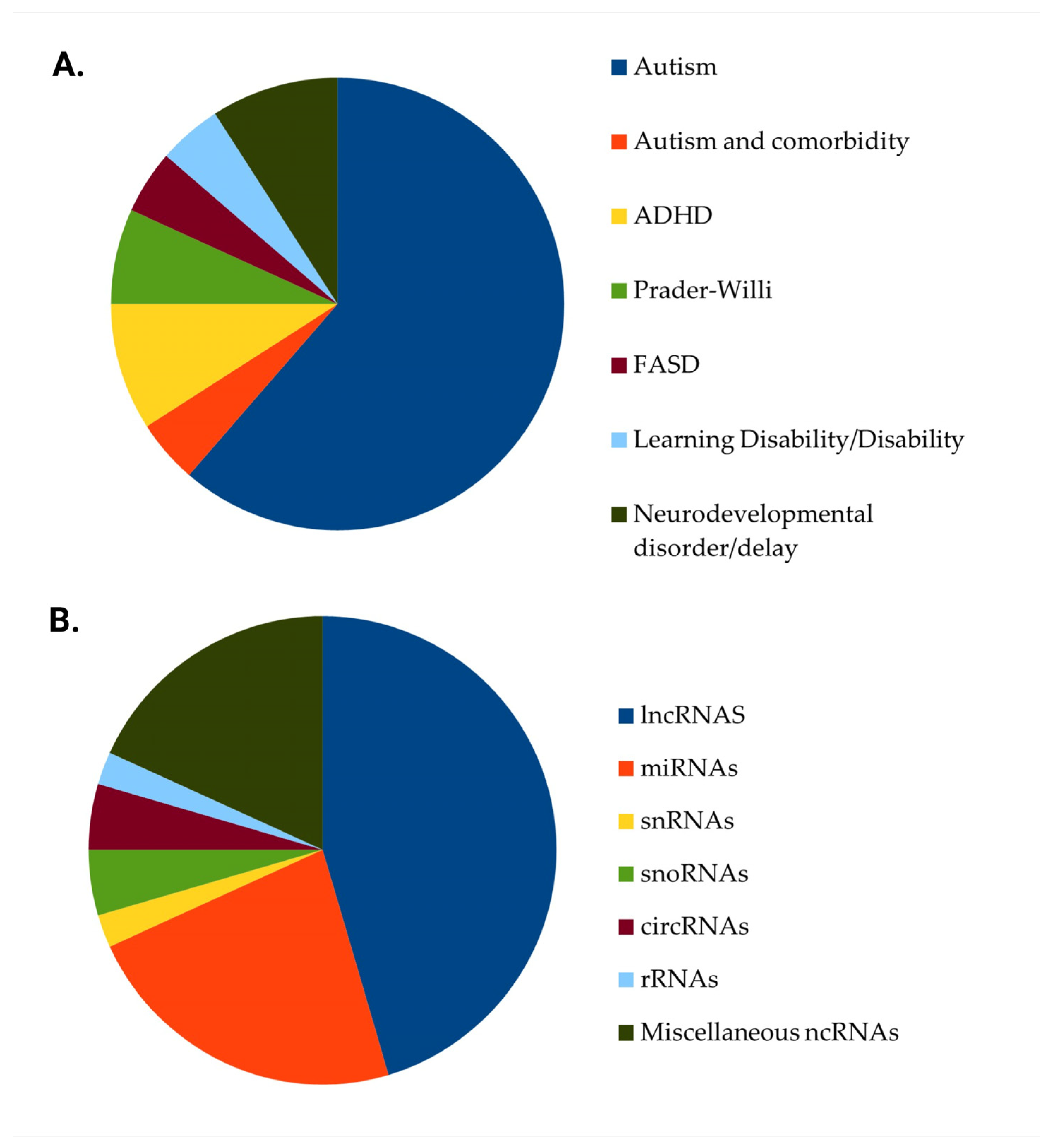
3.1. Studies’ Characteristics
3.2. Long Non-Coding RNAs
3.3. MicroRNAs
3.4. Small Nucleolar RNAs
3.5. Circular RNAs
3.6. Miscellaneous Classes of ncRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, S.; Shan, C.; Zhang, R.; Wang, T. Genetic advances in neurodevelopmental disorders. Med. Rev. 2024, 5, 139–151. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Liu, X.; Guo, R.; Xu, W.; Guo, Q.; Hao, C.; Ni, X.; Li, W. Biological implications of genetic variations in autism spectrum disorders from genomics studies. Biosci. Rep. 2021, 41, BSR20210593. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dias, C.M.; Walsh, C.A. Recent Advances in Understanding the Genetic Architecture of Autism. Annu. Rev. Genom. Hum. Genet. 2020, 21, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, X.; Zhong, L.; Zeng, L.; Li, L.; Yao, P. Understanding autism: Causes, diagnosis, and advancing therapies. Brain Res. Bull. 2025, 227, 111411. [Google Scholar] [CrossRef] [PubMed]
- Weiner, D.J.; Wigdor, E.M.; Ripke, S.; Walters, R.K.; Kosmicki, J.A.; Grove, J.; Samocha, K.E.; Goldstein, J.I.; Okbay, A.; Bybjerg-Grauholm, J.; et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 2017, 49, 978–985. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cirnigliaro, M.; Chang, T.S.; Arteaga, S.A.; Pérez-Cano, L.; Ruzzo, E.K.; Gordon, A.; Bicks, L.K.; Jung, J.Y.; Lowe, J.K.; Wall, D.P.; et al. The contributions of rare inherited and polygenic risk to ASD in multiplex families. Proc. Natl. Acad. Sci. USA 2023, 120, e2215632120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tioleco, N.; Silberman, A.E.; Stratigos, K.; Banerjee-Basu, S.; Spann, M.N.; Whitaker, A.H.; Turner, J.B. Prenatal maternal infection and risk for autism in offspring: A meta-analysis. Autism Res. 2021, 14, 1296–1316. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Rahman, M.M.; Carter, S.A.; Lin, J.C.; Zhuang, Z.; Chow, T.; Lurmann, F.W.; Kleeman, M.J.; Martinez, M.P.; van Donkelaar, A.; et al. Prenatal air pollution, maternal immune activation, and autism spectrum disorder. Environ. Int. 2023, 179, 108148. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, H.; Ding, L.; Qu, G.; Guo, X.; Liang, M.; Ma, S.; Sun, Y. Particulate matter exposure during pregnancy and infancy and risks of autism spectrum disorder in children: A systematic review and meta-analysis. Sci. Total Environ. 2023, 855, 158830. [Google Scholar] [CrossRef] [PubMed]
- O’Sharkey, K.; Mitra, S.; Chow, T.; Paik, S.A.; Thompson, L.; Su, J.; Cockburn, M.; Ritz, B. Prenatal exposure to criteria air pollution and traffic-related air toxics and risk of autism spectrum disorder: A population-based cohort study of California births (1990-2018). Environ. Int. 2025, 201, 109562. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Mejía, D.; Rodas, J.A.; Leon-Rojas, J.E. From Womb to Mind: Prenatal Epigenetic Influences on Mental Health Disorders. Int. J. Mol. Sci. 2025, 26, 6096. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, Y.; Takenaka, K.; Xu, S.M.; Cheng, Y.; Janitz, M. Recent advances in investigation of circRNA/lncRNA-miRNA-mRNA networks through RNA sequencing data analysis. Brief. Funct. Genom. 2025, 24, elaf005. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khoodoruth, M.A.S.; Khoodoruth, W.N.C.; Uroos, M.; Al-Abdulla, M.; Khan, Y.S.; Mohammad, F. Diagnostic and mechanistic roles of MicroRNAs in neurodevelopmental & neurodegenerative disorders. Neurobiol. Dis. 2024, 202, 106717. [Google Scholar] [CrossRef]
- Ilieva, M.S. Non-Coding RNAs in Neurological and Neuropsychiatric Disorders: Unraveling the Hidden Players in Disease Pathogenesis. Cells 2024, 13, 1063. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gurer, D.C.; Akgül, B. Noncoding RNAs: A New Layer of Functional RNAs. Curr. Pharm. Biotechnol. 2023, 24, 856–871. [Google Scholar] [CrossRef] [PubMed]
- Saju, A.F.; Mukundan, A.; Divyashree, M.; Chandrashekhar, R.; Rao, A.M. RNA diagnostics and therapeutics: A comprehensive review. RNA Biol. 2025, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, S.; Walitza, S.; Grünblatt, E. Emerging role of miRNA in attention deficit hyperactivity disorder: A systematic review. Atten. Deficit Hyperact. Disord. 2018, 10, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.F.; Gao, J.; Liu, C.M. The Role of Non-Coding RNAs in Neurodevelopmental Disorders. Front. Genet. 2019, 10, 1033. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ghafouri-Fard, S.; Noroozi, R.; Brand, S.; Hussen, B.M.; Eghtedarian, R.; Taheri, M.; Ebrahimzadeh, K. Emerging Role of Non-coding RNAs in Autism Spectrum Disorder. J. Mol. Neurosci. 2022, 72, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Torres, N.; Guzmán-Torres, K.; García-Cerro, S.; Bermúdez, G.P.; Cruz-Baquero, C.; Ochoa, H.; García-González, D.; Canal-Rivero, M.; Crespo-Facorro, B.; Ruiz-Veguilla, M. miRNAs as biomarkers of autism spectrum disorder: A systematic review and meta-analysis. Eur. Child Adolesc. Psychiatry 2024, 33, 2957–2990. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stott, J.; Wright, T.; Holmes, J.; Wilson, J.; Griffiths-Jones, S.; Foster, D.; Wright, B. A systematic review of non-coding RNA genes with differential expression profiles associated with autism spectrum disorders. PLoS ONE 2023, 18, e0287131. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, Y.; Dawes, R.; Kim, H.C.; Ljungdahl, A.; Stenton, S.L.; Walker, S.; Lord, J.; Lemire, G.; Martin-Geary, A.C.; Ganesh, V.S.; et al. De novo variants in the RNU4-2 snRNA cause a frequent neurodevelopmental syndrome. Nature 2024, 632, 832–840. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheng, W.; Zhou, S.; Zhou, J.; Wang, X. Identification of a robust non-coding RNA signature in diagnosing autism spectrum disorder by cross-validation of microarray data from peripheral blood samples. Medicine 2020, 99, e19484. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chiappori, F.; Cupaioli, F.A.; Consiglio, A.; Di Nanni, N.; Mosca, E.; Licciulli, V.F.; Mezzelani, A. Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host-Gut Microbiota Cross-Talk. Nutrients 2022, 14, 1340. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Darbinian, N.; Gallia, G.L.; Darbinyan, A.; Vadachkoria, E.; Merabova, N.; Moore, A.; Goetzl, L.; Amini, S.; Selzer, M.E. Effects of In Utero EtOH Exposure on 18S Ribosomal RNA Processing: Contribution to Fetal Alcohol Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 13714. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Duan, K.; Chen, J.; Calhoun, V.D.; Jiang, W.; Rootes-Murdy, K.; Schoenmacker, G.; Silva, R.F.; Franke, B.; Buitelaar, J.K.; Hoogman, M.; et al. Genomic patterns linked to gray matter alterations underlying working memory deficits in adults and adolescents with attention-deficit/hyperactivity disorder. Transl. Psychiatry. 2023, 13, 50. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heimdörfer, D.; Vorleuter, A.; Eschlböck, A.; Spathopoulou, A.; Suarez-Cubero, M.; Farhan, H.; Reiterer, V.; Spanjaard, M.; Schaaf, C.P.; Huber, L.A.; et al. Truncated variants of MAGEL2 are involved in the etiologies of the Schaaf-Yang and Prader-Willi syndromes. Am. J. Hum. Genet. 2024, 111, 1383–1404. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hicks, S.D.; Carpenter, R.L.; Wagner, K.E.; Pauley, R.; Barros, M.; Tierney-Aves, C.; Barns, S.; Greene, C.D.; Middleton, F.A. Saliva microRNA Differentiates Children with Autism from Peers with Typical and Atypical Development. J. Am. Acad. Child Adolesc. Psychiatry 2020, 59, 296–308. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hicks, S.D.; Confair, A. Infant Saliva Levels of microRNA miR-151a-3p Are Associated with Risk for Neurodevelopmental Delay. Int. J. Mol. Sci. 2023, 24, 1476. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Isik, C.M.; Bayyurt, E.B.T.; Sahin, N.O. The MNK-SYNGAP1 axis in specific learning disorder: Gene expression pattern and new perspectives. Eur. J. Pediatr. 2025, 184, 260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kichukova, T.; Petrov, V.; Popov, N.; Minchev, D.; Naimov, S.; Minkov, I.; Vachev, T. Identification of serum microRNA signatures associated with autism spectrum disorder as promising candidate biomarkers. Heliyon 2021, 7, e07462. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, D.; Xu, J.; Yang, M.Q. Gene Regulation Analysis Reveals Perturbations of Autism Spectrum Disorder during Neural System Development. Genes 2021, 12, 1901. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Y.; Liu, C.; Jin, Q.; Yu, H.; Long, H. H19/miR-484 axis serves as a candidate biomarker correlated with autism spectrum disorder. Int. J. Dev. Neurosci. 2025, 85, e10403. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Rao, S.; Xu, Y.; Li, J.; Huang, H.; Zhang, X.; Fu, H.; Wang, Q.; Cao, H.; Baranova, A.; et al. Identifying common genome-wide risk genes for major psychiatric traits. Hum. Genet. 2020, 139, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.; Chen, D.Z.; Engchuan, W.; Leal, T.P.; Thiruvahindrapuram, B.; Trost, B.; Howe, J.L.; Pellecchia, G.; Nalpathamkalam, T.; Alexandrova, R.; et al. Chromosome X-wide common variant association study in autism spectrum disorder. Am. J. Hum. Genet. 2025, 112, 135–153. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Piras, I.S.; Manti, F.; Costa, A.; Carone, V.; Scalese, B.; Talboom, J.S.; Veronesi, C.; Tabolacci, C.; Persico, A.M.; Huentelman, M.J.; et al. Molecular biomarkers to track clinical improvement following an integrative treatment model in autistic toddlers. Acta Neuropsychiatr. 2021, 33, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, Z.; Rahmani, D.; Jazi, M.S.; Ghasemi, M.R.; Sadeghi, H.; Miryounesi, M.; Razjouyan, K.; Fayyazi Bordbar, M.R. Altered expression of Csnk1a1p in Autism Spectrum Disorder in Iranian population: Case-control study. Sci. Rep. 2024, 14, 28307. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Safari, M.; Noroozi, R.; Taheri, M.; Ghafouri-Fard, S. The rs12826786 in HOTAIR lncRNA Is Associated with Risk of Autism Spectrum Disorder. J. Mol. Neurosci. 2020, 70, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Salloum-Asfar, S.; Ltaief, S.M.; Taha, R.Z.; Nour-Eldine, W.; Abdulla, S.A.; Al-Shammari, A.R. MicroRNA Profiling Identifies Age-Associated MicroRNAs and Potential Biomarkers for Early Diagnosis of Autism. Int. J. Mol. Sci. 2025, 26, 2044. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Salloum-Asfar, S.; Elsayed, A.K.; Elhag, S.F.; Abdulla, S.A. Circulating Non-Coding RNAs as a Signature of Autism Spectrum Disorder Symptomatology. Int. J. Mol. Sci. 2021, 22, 6549. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sane, S.; Ebrahimi, V.; Farsani, Z.S.; Ghafouri-Fard, S. Assessment of Expression of lncRNAs in Autistic Patients. J. Mol. Neurosci. 2024, 74, 81. [Google Scholar] [CrossRef] [PubMed]
- Sayad, A.; Badrlou, E.; Ghafouri-Fard, S.; Taheri, M. Association Analysis Between the rs1899663 Polymorphism of HOTAIR and Risk of Psychiatric Conditions in an Iranian Population. J. Mol. Neurosci. 2020, 70, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Scheper, M.; Romagnolo, A.; Besharat, Z.M.; Iyer, A.M.; Moavero, R.; Hertzberg, C.; Weschke, B.; Riney, K.; Feucht, M.; Scholl, T.; et al. miRNAs and isomiRs: Serum-Based Biomarkers for the Development of Intellectual Disability and Autism Spectrum Disorder in Tuberous Sclerosis Complex. Biomedicines 2022, 10, 1838. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sehovic, E.; Spahic, L.; Smajlovic-Skenderagic, L.; Pistoljevic, N.; Dzanko, E.; Hajdarpasic, A. Identification of developmental disorders including autism spectrum disorder using salivary miRNAs in children from Bosnia and Herzegovina. PLoS ONE 2020, 15, e0232351. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sledziowska, M.; Winczura, K.; Jones, M.; Almaghrabi, R.; Mischo, H.; Hebenstreit, D.; Garcia, P.; Grzechnik, P. Non-coding RNAs associated with Prader-Willi syndrome regulate transcription of neurodevelopmental genes in human induced pluripotent stem cells. Hum. Mol. Genet. 2023, 32, 608–620. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, J.J.; Chen, B.; Yu, T. Construction of an immune-related ceRNA network to screen for potential diagnostic markers for autism spectrum disorder. Front. Genet. 2022, 13, 1025813. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taheri, M.; Younesi, Z.; Moradi, S.; Tamizkar, K.H.; Razjouyan, K.; Arsang-Jang, S.; Ghafouri-Fard, S.; Neishabouri, S.M. Altered expression of CCAT1 and CCAT2 lncRNAs in autism spectrum disorder. Gene Rep. 2021, 23, 101172. [Google Scholar] [CrossRef]
- Taheri, M.; Tamizkar, K.H.; Omrani, S.; Arsang-Jang, S.; Ghafouri-Fard, S.; Omrani, M.D. MEG3 lncRNA is over-expressed in autism spectrum disorder. Metab. Brain Dis. 2021, 36, 2235–2242. [Google Scholar] [CrossRef] [PubMed]
- Tamizkar, K.H.; Ghafouri-Fard, S.; Omrani, M.D.; Pouresmaeili, F.; Arsang-Jang, S.; Taheri, M. Altered expression of lncRNAs in autism spectrum disorder. Metab. Brain Dis. 2021, 36, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Potter, K.J.; Burnett, L.C.; Orsso, C.E.; Inman, M.; Ryman, D.C.; Haqq, A.M. Prader-Willi-Like Phenotype Caused by an Atypical 15q11.2 Microdeletion. Genes 2020, 11, 128. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, J.; Wang, L. Prediction and prioritization of autism-associated long non-coding RNAs using gene expression and sequence features. BMC Bioinform. 2020, 21, 505. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wong, A.; Zhou, A.; Cao, X.; Mahaganapathy, V.; Azaro, M.; Gwin, C.; Wilson, S.; Buyske, S.; Bartlett, C.W.; Flax, J.F.; et al. MicroRNA and MicroRNA-Target Variants Associated with Autism Spectrum Disorder and Related Disorders. Genes 2022, 13, 1329. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zarantonello, G.; Arnoldi, M.; Filosi, M.; Tebaldi, T.; Spirito, G.; Barbieri, A.; Gustincich, S.; Sanges, R.; Domenici, E.; Di Leva, F.; et al. Natural SINEUP RNAs in Autism Spectrum Disorders: RAB11B-AS1 Dysregulation in a Neuronal CHD8 Suppression Model Leads to RAB11B Protein Increase. Front. Genet. 2021, 12, 745229. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bai, M.; Ye, D.; Guo, X.; Xi, J.; Liu, N.; Wu, Y.; Jia, W.; Wang, G.; Chen, W.; Li, G.; et al. Critical regulation of a NDIME/MEF2C axis in embryonic stem cell neural differentiation and autism. EMBO Rep. 2020, 21, e50283. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Butler, M.C.; Long, C.N.; Kinkade, J.A.; Green, M.T.; Martin, R.E.; Marshall, B.L.; Willemse, T.E.; Schenk, A.K.; Mao, J.; Rosenfeld, C.S. Endocrine disruption of gene expression and microRNA profiles in hippocampus and hypothalamus of California mice: Association of gene expression changes with behavioural outcomes. J. Neuroendocr. 2020, 32, e12847. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fu, Y.; Zhang, Y.L.; Liu, R.Q.; Xu, M.M.; Xie, J.L.; Zhang, X.L.; Xie, G.M.; Han, Y.T.; Zhang, X.M.; Zhang, W.T.; et al. Exosome lncRNA IFNG-AS1 derived from mesenchymal stem cells of human adipose ameliorates neurogenesis and ASD-like behavior in BTBR mice. J. Nanobiotechnol. 2024, 22, 66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gasparini, S.; Del Vecchio, G.; Gioiosa, S.; Flati, T.; Castrignano, T.; Legnini, I.; Licursi, V.; Ricceri, L.; Scattoni, M.L.; Rinaldi, A.; et al. Differential Expression of Hippocampal Circular RNAs in the BTBR Mouse Model for Autism Spectrum Disorder. Mol. Neurobiol. 2020, 57, 2301–2313. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fu, Q.; Zhong, M.; Long, Y.; Zhao, F.; Huang, Y.; Zhang, Z.; Wen, M.; Chen, K.; Chen, R.; et al. Quantitative proteomics of the miR-301a/SOCS3/STAT3 axis reveals underlying autism and anxiety-like behavior. Mol. Ther. Nucleic Acids 2024, 35, 102136. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, H.; Wan, X.; Yao, L.; Zhao, Q.; Yang, Y.; Liu, H.; Shang, J.; Zeng, F.; Wang, X.; Huang, S. Differentially expressed long non-coding RNAs and mRNAs of cadmium exposure on learning disability of offspring rats. Eur. J. Med. Res. 2024, 29, 82. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mizuno, S.; Hirota, J.N.; Ishii, C.; Iwasaki, H.; Sano, Y.; Furuichi, T. Comprehensive Profiling of Gene Expression in the Cerebral Cortex and Striatum of BTBRTF/ArtRbrc Mice Compared to C57BL/6J Mice. Front. Cell. Neurosci. 2020, 14, 595607. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paudel, P.; Pierotti, C.; Lozano, E.; Amoah, S.K.; Gardiner, A.S.; Caldwell, K.K.; Allan, A.M.; Mellios, N. Prenatal Alcohol Exposure Results in Sex-Specific Alterations in Circular RNA Expression in the Developing Mouse Brain. Front. Neurosci. 2020, 14, 581895. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Su, Z.; Frost, E.L.; Lammert, C.R.; Przanowska, R.K.; Lukens, J.R.; Dutta, A. tRNA-derived fragments and microRNAs in the maternal-fetal interface of a mouse maternal-immune-activation autism model. RNA Biol. 2020, 17, 1183–1195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, S.; You, L.; Xu, Q.; Ou, J.; Wu, D.; Yuan, X.; Liu, Z.; Hong, Q.; Tong, M.; Yang, L.; et al. Distinct long non-coding RNA and mRNA expression profiles in the hippocampus of an attention deficit hyperactivity disorder model in spontaneously hypertensive rats and control wistar Kyoto rats. Brain Res. Bull. 2020, 161, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, F.; Arnoldi, M.; Santarelli, S.; Massonot, M.; Lemée, M.V.; Bon, C.; Pellegrini, M.; Castellini, M.E.; Zarantonello, G.; Messina, A.; et al. SINEUP RNA rescues molecular phenotypes associated with CHD8 suppression in autism spectrum disorder model systems. Mol. Ther. 2025, 33, 1180–1196. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, X.; Haugh, W.; Zhang, Z.; Huang, J. Emerging Role of Long, Non-Coding RNA Nuclear-Enriched Abundant Transcript 1 in Stress- and Immune-Related Diseases. Int. J. Mol. Sci. 2025, 26, 4413. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chodurska, B.; Kunej, T. Long non-coding RNAs in humans: Classification, genomic organization and function. Non-Coding RNA Res. 2025, 11, 313–327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luo, C.; Lin, K.; Hu, C.; Zhu, X.; Zhu, J.; Zhu, Z. LINC01094 promotes pancreatic cancer progression by sponging miR-577 to regulate LIN28B expression and the PI3K/AKT pathway. Mol. Ther. Nucleic Acids 2021, 26, 523–535. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- O’Brien, E.M.; Selfe, J.L.; Martins, A.S.; Walters, Z.S.; Shipley, J.M. The long non-coding RNA MYCNOS-01 regulates MYCN protein levels and affects growth of MYCN-amplified rhabdomyosarcoma and neuroblastoma cells. BMC Cancer 2018, 18, 217. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yi, Q.; Feng, J.; Lan, W.; Shi, H.; Sun, W.; Sun, W. CircRNA and lncRNA-encoded peptide in diseases, an update review. Mol. Cancer 2024, 23, 214. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Deguchi, S.; Katsushima, K.; Hatanaka, A.; Shinjo, K.; Ohka, F.; Wakabayashi, T.; Zong, H.; Natsume, A.; Kondo, Y. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene 2017, 36, 4629–4640. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.T.; Chen, L.; Du, Y.; Jiang, Z.Y.; Cheng, Y. MicroRNAs as Regulators, Biomarkers, and Therapeutic Targets in Autism Spectrum Disorder. Mol. Neurobiol. 2025, 62, 5039–5056. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F. microRNA biogenesis and function. An overview. Adv. Exp. Med. Biol. 2010, 700, 1–14. [Google Scholar] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Boivin, V.; Faucher-Giguère, L.; Scott, M.; Abou-Elela, S. The cellular landscape of mid-size noncoding RNA. Wiley Interdiscip. Rev. RNA 2019, 10, e1530. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fafard-Couture, É.; Labialle, S.; Scott, M.S. The regulatory roles of small nucleolar RNAs within their host locus. RNA Biol. 2024, 21, 496–506. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bergeron, D.; Fafard-Couture, É.; Scott, M.S. Small nucleolar RNAs: Continuing identification of novel members and increasing diversity of their molecular mechanisms of action. Biochem. Soc. Trans. 2020, 48, 645–656. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, L.; Chen, M.; Hu, X.; Deng, L. Graph Convolutional Network and Contrastive Learning Small Nucleolar RNA (snoRNA) Disease Associations (GCLSDA): Predicting snoRNA-Disease Associations via Graph Convolutional Network and Contrastive Learning. Int. J. Mol. Sci. 2023, 24, 14429. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Y.; Chen, X.; Xiao, S.; Wang, H.; Li, B.; Zhang, M.; Wang, K. Unlocking the life code: A review of SnoRNA functional diversity and disease relevance. Cell Commun. Signal. 2025, 23, 266. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bai, H.; Meng, F.; Ke, K.; Fang, L.; Xu, W.; Huang, H.; Liang, X.; Li, W.; Zeng, F.; Chen, C. The significance of small noncoding RNAs in the pathogenesis of cardiovascular diseases. Genes Dis. 2024, 12, 101342. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dudzik, J.; Czechowicz, P.; Więch-Walów, A.; Sławski, J.; Collawn, J.F.; Bartoszewski, R. PiRNAs, PiRNA-Like, and PIWI Proteins in Somatic Cells: From Genetic Regulation to Disease Mechanisms. Wiley Interdiscip. Rev. RNA 2025, 16, e70012. [Google Scholar] [CrossRef] [PubMed]
- Cherlin, T.; Magee, R.; Jing, Y.; Pliatsika, V.; Loher, P.; Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol. 2020, 18, 38. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khundrakpam, B.S.; Lewis, J.D.; Kostopoulos, P.; Carbonell, F.; Evans, A.C. Cortical Thickness Abnormalities in Autism Spectrum Disorders Through Late Childhood, Adolescence, and Adulthood: A Large-Scale MRI Study. Cereb. Cortex 2017, 27, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Pretzsch, C.M.; Ecker, C. Structural neuroimaging phenotypes and associated molecular and genomic underpinnings in autism: A review. Front. Neurosci. 2023, 17, 1172779. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tang, G.; Gudsnuk, K.; Kuo, S.H.; Cotrina, M.L.; Rosoklija, G.; Sosunov, A.; Sonders, M.S.; Kanter, E.; Castagna, C.; Yamamoto, A.; et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014, 83, 1131–1143, Erratum in: Neuron 2014, 83, 1482. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hegarty, J.P.; Pegoraro, L.F.L.; Lazzeroni, L.C.; Raman, M.M.; Hallmayer, J.F.; Monterrey, J.C.; Cleveland, S.C.; Wolke, O.N.; Phillips, J.M.; Reiss, A.L.; et al. Genetic and environmental influences on structural brain measures in twins with autism spectrum disorder. Mol. Psychiatry 2020, 25, 2556–2566. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- de Groot, D.M.G.; Linders, L.; Kayser, R.; Nederlof, R.; de Esch, C.; Slieker, R.C.; Kuper, C.F.; Wolterbeek, A.; de Groot, V.J.; Veltien, A.; et al. Perinatal exposure to the immune-suppressant di-n-octyltin dichloride affects brain development in rats. Toxicol. Mech. Methods 2024, 34, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.; Akhtar, M.W.; Escorihuela, R.M.; Amador-Arjona, A.; Swarup, V.; Parker, J.; Zaremba, J.D.; Holland, T.; Bansal, N.; Holohan, D.R.; et al. NitroSynapsin therapy for a mouse MEF2C haploinsufficiency model of human autism. Nat. Commun. 2017, 8, 1488. [Google Scholar] [CrossRef] [PubMed]
- Harrington, A.J.; Raissi, A.; Rajkovich, K.; Berto, S.; Kumar, J.; Molinaro, G.; Raduazzo, J.; Guo, Y.; Loerwald, K.; Konopka, G.; et al. MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. eLife 2016, 5, e20059. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Krainc, D.; Sherman, K.; Lipton, S.A. Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc. Natl. Acad. Sci. USA 2000, 97, 7561–7566. [Google Scholar] [CrossRef] [PubMed]
- Flavell, S.W.; Cowan, C.W.; Kim, T.K.; Greer, P.L.; Lin, Y.; Paradis, S.; Griffith, E.C.; Hu, L.S.; Chen, C.; Greenberg, M.E. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 2006, 311, 1008–1012. [Google Scholar] [CrossRef]
- Shalizi, A.; Gaudilliere, B.; Yuan, Z.; Stegmuller, J.; Shirogane, T.; Ge, Q.; Tan, Y.; Schulman, B.; Harper, J.W.; Bonni, A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 2006, 311, 1012–1017. [Google Scholar] [CrossRef]
- Li, H.; Radford, J.C.; Ragusa, M.J.; Shea, K.L.; McKercher, S.R.; Zaremba, J.D.; Soussou, W.; Nie, Z.; Kang, Y.J.; Nakanishi, N.; et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 9397–9402. [Google Scholar] [CrossRef]
- Woelfle, S.; Pedro, M.T.; Wagner, J.; Schön, M.; Boeckers, T.M. Expression profiles of the autism-related SHANK proteins in the human brain. BMC Biol. 2023, 21, 254. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Tang, R.; Hu, Z.M.; Wang, X.H.; Gao, X.; Wang, T.; Tang, M.X. Key Synaptic Pathology in Autism Spectrum Disorder: Genetic Mechanisms and Recent Advances. J. Integr. Neurosci. 2024, 23, 184. [Google Scholar] [CrossRef] [PubMed]
- Siddiqua, H.; Akter, Y.; Uddin, M.N.; Kumkum, M.; Hossain, M.A.; Aziz, M.A.; Ahmed, M.S.; Chowdhury, M.A.; Islam, M.S.; Marzan, L.W. SHANK3 genetic polymorphism and susceptibility to ASD: Evidence from molecular, in silico, and meta-analysis approaches. Mol. Biol. Rep. 2022, 49, 8449–8460. [Google Scholar] [CrossRef] [PubMed]
- Na, E.S.; Nelson, E.D.; Kavalali, E.T.; Monteggia, L.M. The impact of MeCP2 loss- or gain-of-function on synaptic plasticity. Neuropsychopharmacology 2013, 38, 212–219. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arias-Aragón, F.; Robles-Lanuza, E.; Sánchez-Gómez, Á.; Martinez-Mir, A.; Scholl, F.G. Analysis of neurexin-neuroligin complexes supports an isoform-specific role for beta-neurexin-1 dysfunction in a mouse model of autism. Mol. Brain. 2025, 18, 20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hauw, J.J.; Hausser-Hauw, C.; Barthélémy, C. Synapse and primary cilia dysfunctions in Autism Spectrum Disorders. Avenues to normalize these functions. Rev. Neurol. 2024, 180, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Autism Spectrum Disorder in Under 19s: Recognition, Referral and Diagnosis; National Institute for Health and Care Excellence (NICE): London, UK, 2017. [PubMed]
- Panni, S. The Relevance of the Accurate Annotation of Micro and Long Non-Coding RNA Interactions for the Development of Therapies. Genes 2025, 16, 262. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, G.T.; Geschwind, D.H. Challenges and opportunities for precision medicine in neurodevelopmental disorders. Adv. Drug Deliv. Rev. 2022, 191, 114564. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Author | Ref. No | Date | Country | Type of Study | Disorder | Type of ncRNAs | Dysregulated ncRNAs | Biological Function | Biomaterial |
|---|---|---|---|---|---|---|---|---|---|
| Bai M et al. | [55] | 2020 | China | Animal | Autism | lncRNAs | NDIME | Neural differentiation | Hippocampus |
| Butler MC et al. | [56] | 2020 | USA | Animal | Socio-communicative deficits | miRNAs | miR-153, miR-181a, miR-9, miR-7a | Memory, neurons´ proliferation and differentiation | Hippocampus, Hypothalamus |
| Chen Y et al. | [23] | 2024 | UK, USA, Australia | Human | Neurodevelopmental disorder | snRNAs | U4 snRNA | - | Blood |
| Cheng W et al. | [24] | 2020 | China | Human | Autism | lncRNAs, pseudogenes, miscRNAs, rRNAs, snoRNAs, snRNAs | AQP4-AS1, FTH1P2, FTH1P3, GMCL2, HMGN2P11, IGHV3-47, MUC20-OT1, MYCLP1, POLR2KP2, RN7SL132P, RNA5SP160, RNU105B, RNU1–16P, RNU6-258P, RNU6-485P, RNU6-549P, RNVU1-15 | - | Blood |
| Chiappori F et al. | [25] | 2022 | Italy | Human | Autism | miRNAS and piRNAs | Hsa-miR-657, hsa-miR-2110, hsa-piR-28059 | Cell-cell junction, metabolite signalling, bacterial invasion, inflammation | Faeces |
| Darbinian N et al. | [26] | 2023 | USA | Human | Fetal Alcohol Spectrum Disorders | small ribosomal RNAs | srRNA 10, srRNA 11, srRNA 14, srRNA 47 | Neural cell proliferation, migration and apoptosis | Fetal brain tissue |
| Duan K et al. | [27] | 2023 | USA, Netherlands | Human | ADHD | lncRNAs | RP11_6N13.1 | - | Blood |
| Fu Y et al. | [57] | 2024 | China | Animal | Autism | lncRNAs | IFNG-AS1 | Neurogenesis, inhibition of apoptosis | Prefrontal cortex |
| Gasparini S et al. | [58] | 2020 | Italy | Animal | Autism | circRNAs | CircCdh9, circCacna1a, circCacna1c, circHivep2, circCdc14b, circTrpc6, circCep112, circWdr49, circNcoa2, circZcchc11, circRmst, circMyrip | Neuron development, glutamate receptor signaling, synaptogenesis, synaptic plasticity | Hippocampus |
| Heimdorfer D et al. | [28] | 2024 | Austria, Germany | Human | Prader-Willi, Schaaf-Yang | snoRNAs | SNORD116 | Cell lines | |
| Hicks SD et al. | [29] | 2023 | USA | Human | Neurodevelopmental delay | miRNAs | miR-125a-5p, miR-148a-5p, miR-151a-3p, miR-28-3p | Inflammatory pathways, neuron apoptosis | Saliva |
| Hicks SD et al. | [30] | 2020 | USA | Human | Autism | miRNAs | miR-28-3p, miR-148a-5p, miR-151a-3p, miR-125b-2-3p, miR-7706, miR-665, miR-4705, miR-620, miR-1277-5p | Axonal guidance, neurotrophic signaling, GABAergic synapse, addiction pathways | Saliva |
| Isik CM et al. | [31] | 2025 | Turkey | Human | Specific learning disorder | lncRNA | SYNGAP1-AS1 | Synaptic plasticity, learning, long-term potentiation | Blood |
| Kichukova T et al. | [32] | 2021 | Bulgaria | Human | Autism | miRNAs | miR-500a-5p, miR-197-5p, miR-424-5p, and miR-664a-3p | Synaptic pathways, axon guidance, neuroactive ligand-receptor interaction | Serum |
| di Leva F et al. | [65] | 2025 | Italy, France, USA | Ιn vitro human and animal cells, in vivo animal model | Autism | antisense long ncRNA | SINEUP-CHD8 | Regulation of cell proliferation and migration | Cell lines |
| Li D et al. | [33] | 2021 | USA | Human | Autism | lncRNAs | DNM3OS, IGFBP7-AS1 and LINC01139 | Axonal sprouting | Neural progenitor cells, neurons |
| Li X et al. | [59] | 2024 | China | Animal | Autism | miRNA-301a | miR-301a | Cytokine pathways Synaptogenesis signaling pathway | Brain tissue |
| Li Y et al. | [34] | 2024 | China | Human | Autism | lncRNAs, miRNAs | H19, miR-484 | Postsynaptic density, presynaptic active zone, synaptic membrane, nucleobase-containing compound kinase activity, and regulation of RNA stability | Blood |
| Liu Y et al. | [35] | 2020 | USA | Human | ADHD | miRNAs, lncRNAs | miR137 Long ncRNA LINC00461 | Neuroactive ligand-receptor interaction pathway | Blood |
| Liu S et al. | [35] | 2020 | China | Animal | Autism and ADHD | lncRNAs | LINC00461 | Neural migration, neural differentiation | Hippocampus |
| Liu H et al. | [60] | 2024 | Animal | Learning disability | lncRNAs | gi|672032535, uc.447, gi|672048769, NON-RATT015986, NONRATT027328, NONRATT022126, gi|672015918, gi|672027113gi|672071577, NON-RATT016007, NONRATT028784, uc.447, NONRATT022126, gi|672015918, NONRATT016007 | Pancreatic secretion pathway | Hippocampus | |
| Mendes M et al. | [36] | 2025 | Canada | Human | Autism | lncRNA | DDX53/PTCHD1-AS | - | |
| Mizuno S et al. | [61] | 2020 | Japan | Animal | Autism | ncRNAs | LncRNAs Gm26917 and Gm37194 | Synaptic transmission, neuron signalling, immune signalling | Cerebral cortex, striatum |
| Paudel P et al. | [62] | 2020 | USA | Animal | Fetal Alcohol Spectrum Disorder | circRNAs | CircSatb2, circPtchd2 | GABA receptor signaling, semaphoring signaling, dopamine signaling, neuron development, neuritogenesis, axonal guidance | Brain |
| Piras IS et al. | [37] | 2021 | USA, Italy | Human | Autism | lncRNAs | MALAT-1 | Synaptogenesis, dendritic density | Blood |
| Rahmani Z et al. | [38] | 2024 | Iran | Human | Autism | lncRNAs | DISC2, Linc00945, Foxg1-as1, Csnk1a1p, Evf2 | Neuron differentiation, migration | Blood |
| Safari M et al. | [39] | 2020 | Iran | Human | Autism | lncRNA | HOTAIR | Immune-mediated pathways | Blood |
| Salloum-Asfar S et al. | [40] | 2025 | Qatar | Human | Autism | miRNAs | miR-4433b-5p, miR-151a-5p, miR-335-5p, and miR-1180-3p | Neural differentiation and apoptosis, synaptogenesis, immune-related pathways | Plasma |
| Salloum-Asfar S et al. | [41] | 2021 | Qatar | Human | Autism | miRNAs, piRNAs, snoRNAs, | hsa-miR-302b-3p, hsa-miR-302a-3p, hsa-miR-302d-3p, piR-hsa-1282, piR-hsa-12790, SNORD3C, SNORD69, SNORD51 | Neural development Synaptic plasticity Gut homeostasis | Plasma |
| Sane S et al. | [42] | 2024 | Iran | Human | Autism | lncRNAs | LincRNA-ROR, LINC-PINT, LincRNA-p21, PCAT-29, PCAT-1 l | Blood | |
| Sayad A et al. | [43] | 2020 | Iran | Human | ADHD | lncRNA | HOTAIR | Neuron apoptosis | Blood |
| Scheper M et al. | [44] | 2022 | Netherlands, Italy, Germany, Austria, Belgium, Australia, France, UK, USA, Poland, Czech Republic | Human | Autism, Intellectual Disability | miRNAs and isomiRNAs | hsa-miR-409-5p, hsa-miR-1301-3p, hsa-miR-145-5p, hsa-miR-412-5p, hsa-miR-423-3p, hsa-miR-154-5p, hsa-miR-214-5p, hsa-miR-376b-3p, hsa-miR-379-3p, hsa-miR-494-3p, hsa-miR-103a-3p, hsa-miR-410-3p_miRNA, hsa-miR-221-3p_trim3 | Neurotransmitter signaling, neuroplasticity | Serum |
| Sehovic E et al. | [45] | 2020 | Bosnia Herzegovina | Human | Neurodevelopmental delay/disorder | miRNAs | miR-7-5p, miR-23a-3p, miR-27a-3p, miR-32-5p, miR-140-3p, miR-628-5p, miR-2467-5p | Regulation of cell proliferation and differentiation | Saliva |
| Sledziowska M et al. | [46] | 2023 | UK | Human | Prader Willi | spa-lncRNAs, sno-lncRNAs | SNHG14, RPL4, RPS17, SNURF-SNRPN, PWAR6, IPW, SPA1, SPA2, sno lncRNA1, sno lncRNA2, sno lncRNA3, sno lncRNA4, sno lncRNA5 | Neuron polarity, synaptogenesis, synaptic transmission, disturbed proliferation, increased apoptosis, cell-cell signalling | Human induced pluripotent cells |
| Su Z et al. | [63] | 2020 | USA | Animal | Autism | tRFs, miRNAs | 5ʹ halves from tRNAAsp, tRNAGly, tRNAGlu, tRNAVal, tRNAiMet, tRNALeu tRNASeC and tRNACys, 5ʹ halves from tRNAGly and tRNAGlu, tRF-3Tyr, tRF-3Gln, tRF-3Thr, tRF-3Leu, tRF-3Ser, tRF-3Trp and tRF-3Ala, miR-291a/b-5p, Mmu-miR-146b-5p, Mmu-miR-215-5p, | Cytokine pathways, embryo development | Mouse placenta/decidua |
| Sun JJ et al. | [47] | 2022 | China | Human | Autism | lncRNAs, miRNAs | 49 lncRNAs, 30 miRNAs | Mitochondrial-energy metabolism, cellular communication, transcriptional regulation, and glial-cell fate, | Brain tissue, blood |
| Taheri M et al. | [48] | 2021 | Iran | Human | Autism | lncRNAs | CCAT1 and CCAT2 | Immune-related pathways, T cell function | Blood |
| Taheri M et al. | [49] | 2021 | Iran | Human | Autism | lncRNAS | MEG3 | Neuronal synaptic plasticity, neuronal apoptosis | Blood |
| Tamizkar KH et al. | [50] | 2021 | Iran | Human | Autism | lncRNAs | DISC2, PRKAR2A-AS1, LRRC2-AS1, LOC101928237 | Dopaminergic activity, cerebral hypoperfusion | Blood |
| Tan Q et al. | [51] | 2020 | Canada | Human | Prader-Willi | snoRNAs | SNORD116 cluster | - | Blood |
| Wang Z et al. | [52] | 2022 | China | Human | Autism | miRNAs | hsa-miR-191-5p, hsa-miR-151a-3p, hsa-miR139-5p, hsa-miR-181a-5p, hsa-miR-432-5p | Dendritic spine formation, axon growth, neural differentiation, synaptogenesis | Blood |
| Wong A et al. | [53] | 2022 | USA | Human | Autism | miRNAs | Hsa-miR-6780a-3p, hsa-miR-1225-5p, hsa-miR-2277-3p, hsa-miR-548j-5p, hsa-miR-100-5p | Regulation of neuronal death, dendritic spine formation, early forebrain dorsal-ventral pattern formation | Blood |
| Zarantonello G et al. | [54] | 2021 | Italy, USA | In vitro, human cells | Autism | antisense lncRNAs | RAB11B-AS1 | Vesicular trafficking, synaptic activity | Human induced neural progenitor cells |
| Zhang S et al. | [64] | 2020 | China | Animal | ADHD | lncRNAs | NONRATT027852.2, NONRATT005132.2, NONRATT004982.2 NONRATT027914.2, NON-RATT006598.2, NONRATT002035.2, NON-RATT016515.2 | Synaptic plasticity regulation | Hippocampus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaivazoglou, K.; Triantos, C.; Aggeletopoulou, I. Non-Coding RNAs in Neurodevelopmental Disorders—From Diagnostic Biomarkers to Therapeutic Targets: A Systematic Review. Biomedicines 2025, 13, 1808. https://doi.org/10.3390/biomedicines13081808
Karaivazoglou K, Triantos C, Aggeletopoulou I. Non-Coding RNAs in Neurodevelopmental Disorders—From Diagnostic Biomarkers to Therapeutic Targets: A Systematic Review. Biomedicines. 2025; 13(8):1808. https://doi.org/10.3390/biomedicines13081808
Chicago/Turabian StyleKaraivazoglou, Katerina, Christos Triantos, and Ioanna Aggeletopoulou. 2025. "Non-Coding RNAs in Neurodevelopmental Disorders—From Diagnostic Biomarkers to Therapeutic Targets: A Systematic Review" Biomedicines 13, no. 8: 1808. https://doi.org/10.3390/biomedicines13081808
APA StyleKaraivazoglou, K., Triantos, C., & Aggeletopoulou, I. (2025). Non-Coding RNAs in Neurodevelopmental Disorders—From Diagnostic Biomarkers to Therapeutic Targets: A Systematic Review. Biomedicines, 13(8), 1808. https://doi.org/10.3390/biomedicines13081808









