Targeting CDK4/6 in Cancer: Molecular Docking and Cytotoxic Evaluation of Thottea siliquosa Root Extract
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Authentication of Plant Material
2.2. Phytochemical Extraction
2.3. GC-MS Analysis
2.4. Cell Culture and Cytotoxicity (MTT Assay)
2.4.1. HCT116 Cell Lines
2.4.2. L929 Cell Lines
2.5. Cell Migration Assay
2.5.1. HCT116 Cell Lines
2.5.2. L929 Cell Lines
2.6. Cell Cycle Analysis
2.7. Selection and Preparation of Ligand Molecule
2.8. Target Retrieval
2.9. Molecular Docking
2.10. ADME Profiling
2.11. Statistical Analysis
3. Results
3.1. GC-MS Analysis
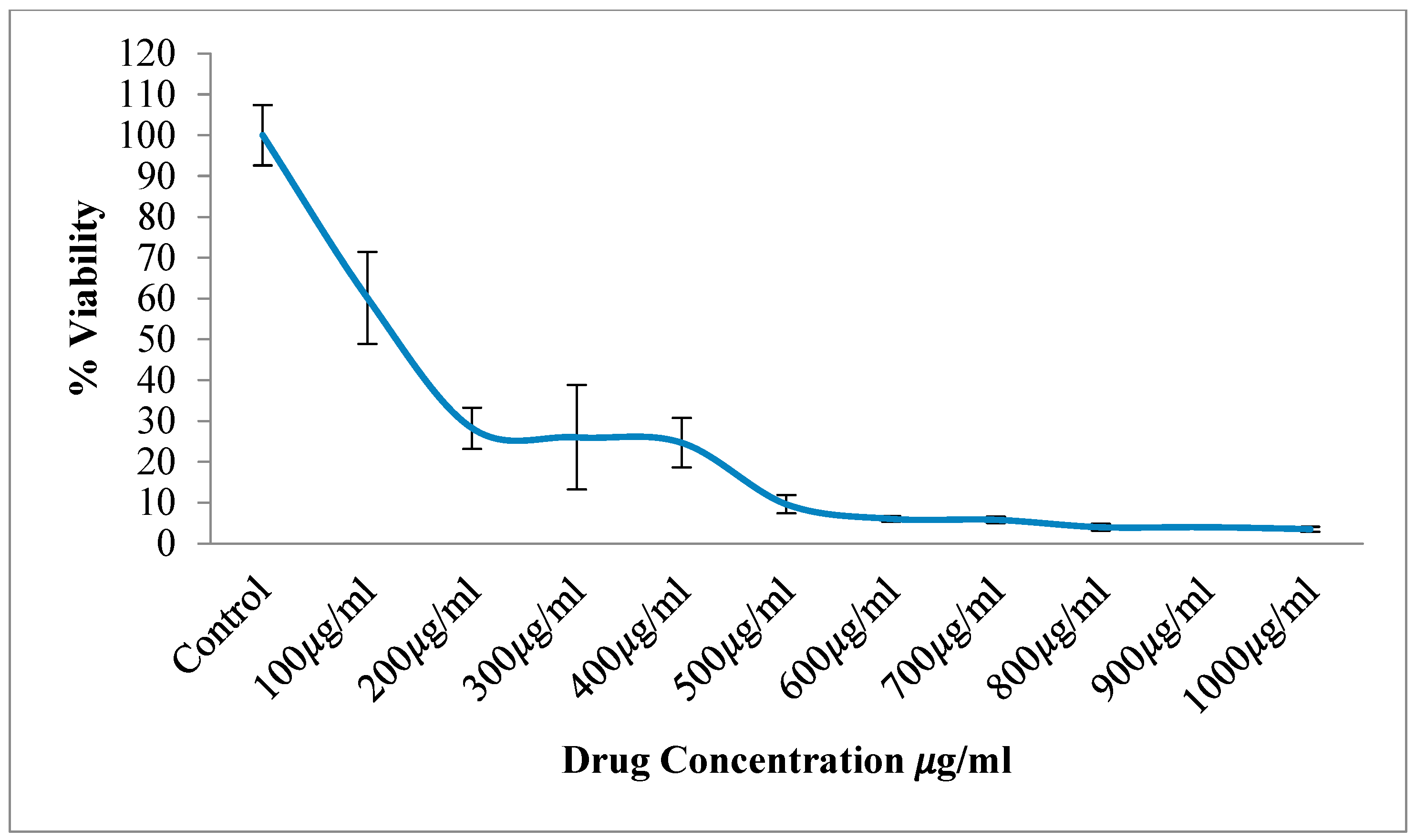
3.2. Cell Viability and Cytotoxicity (MTT Assay)
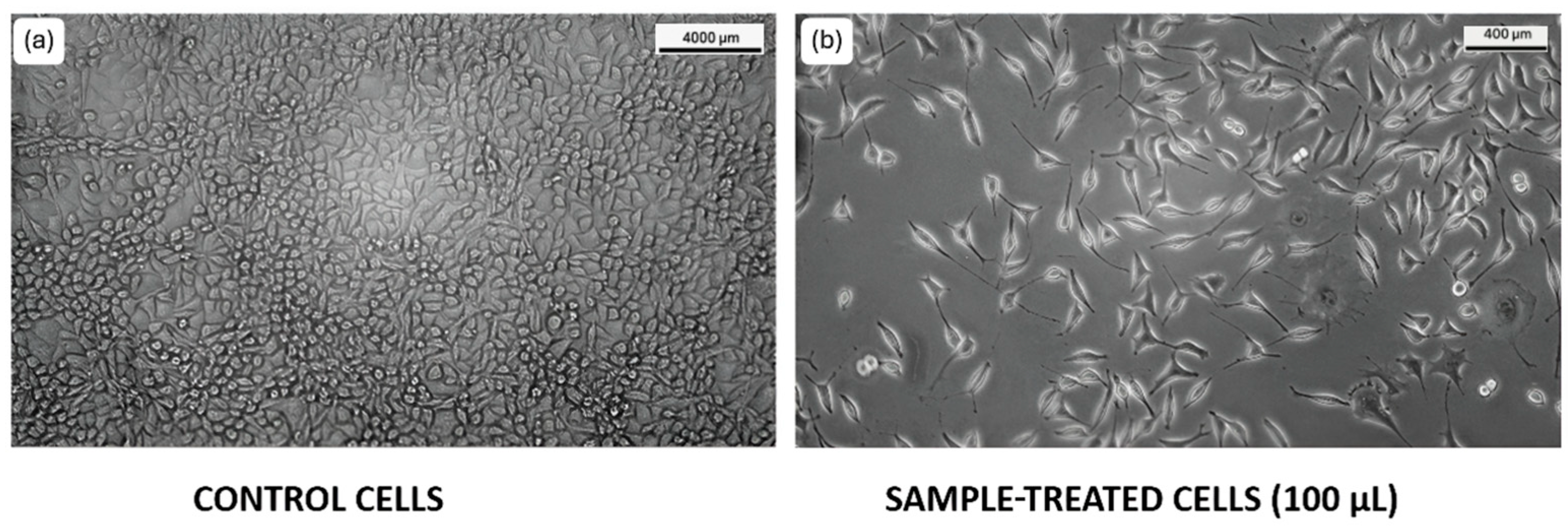
3.2.1. HCT116 Cell Lines
3.2.2. L929 Cell Lines
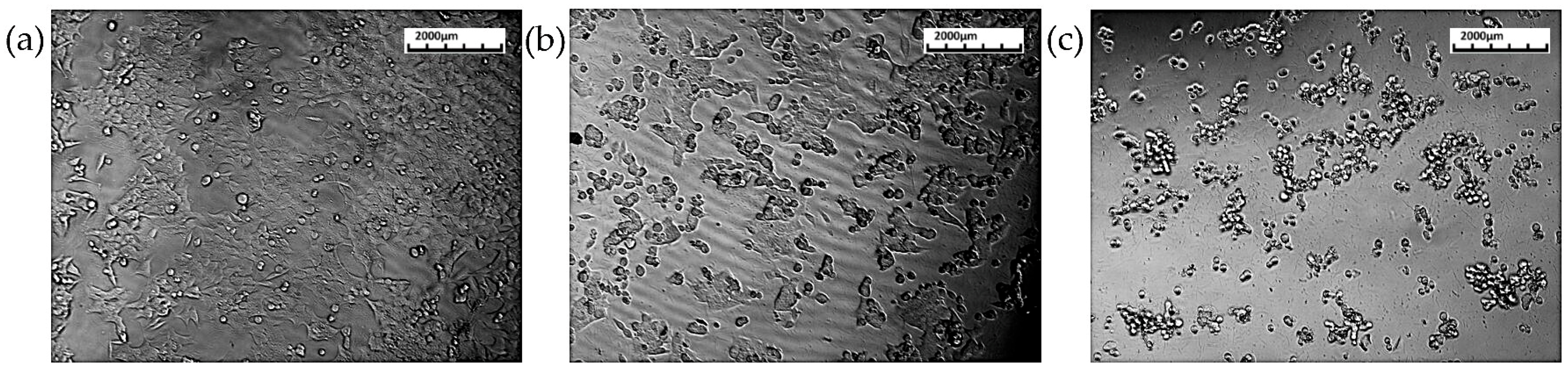
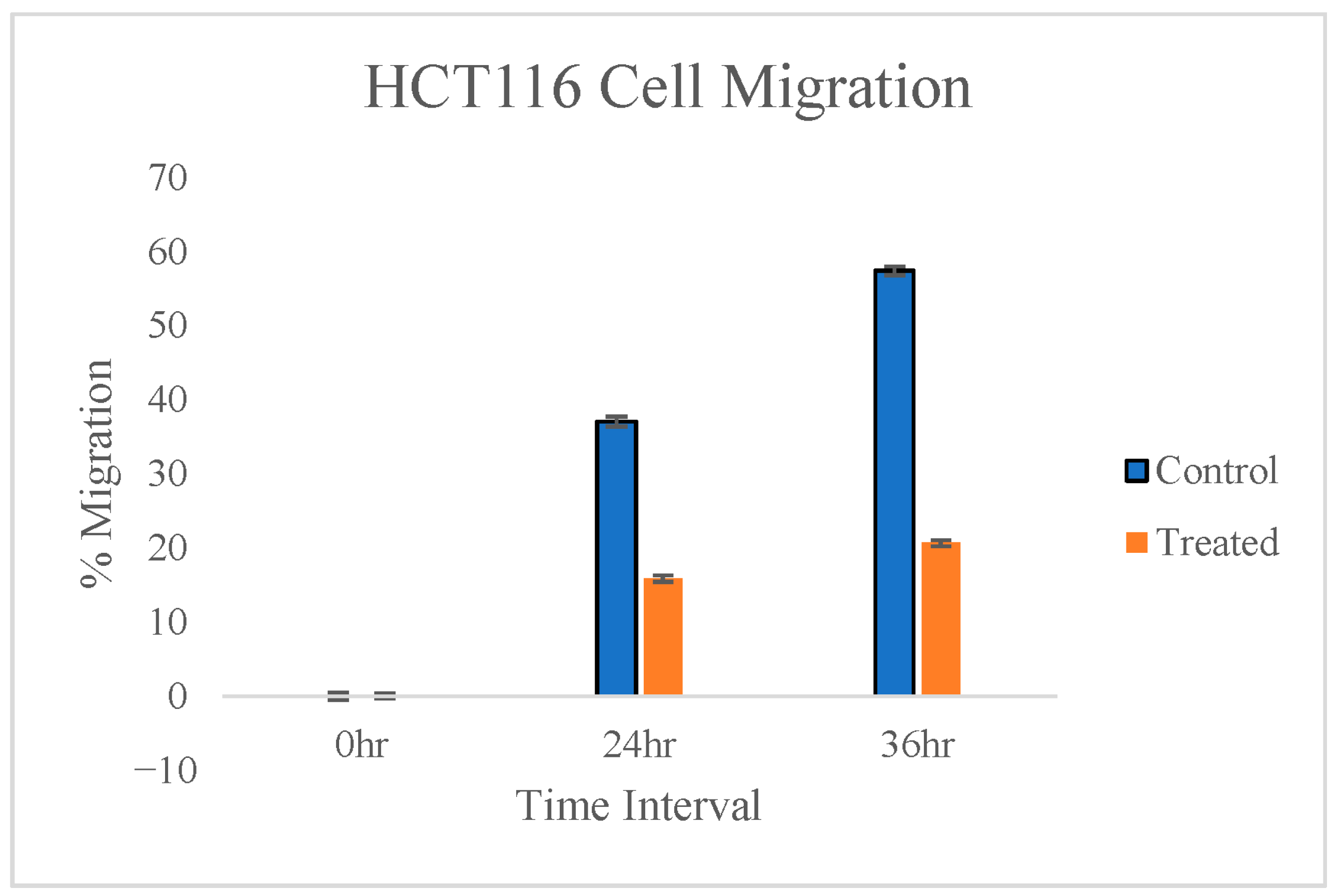
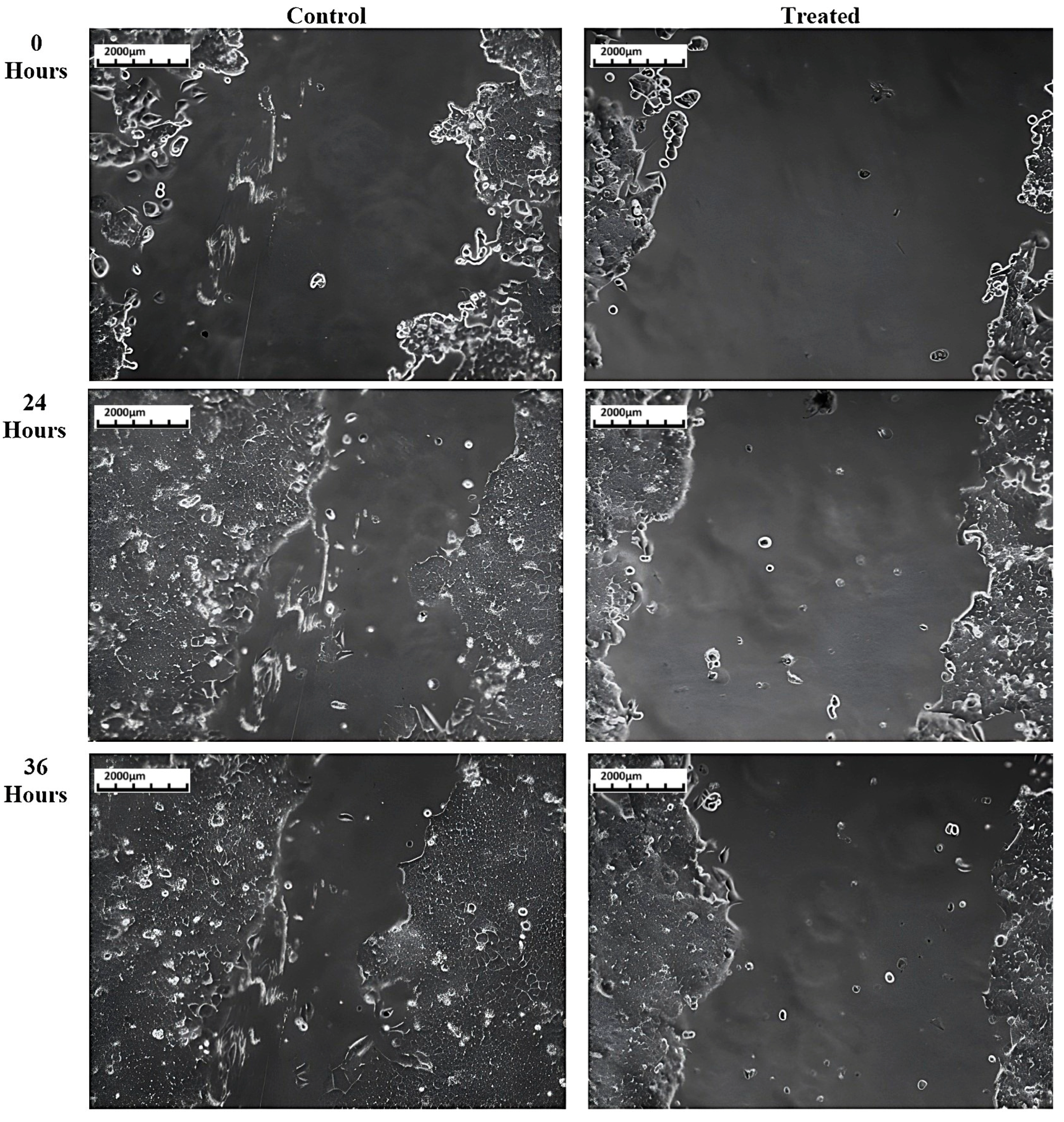
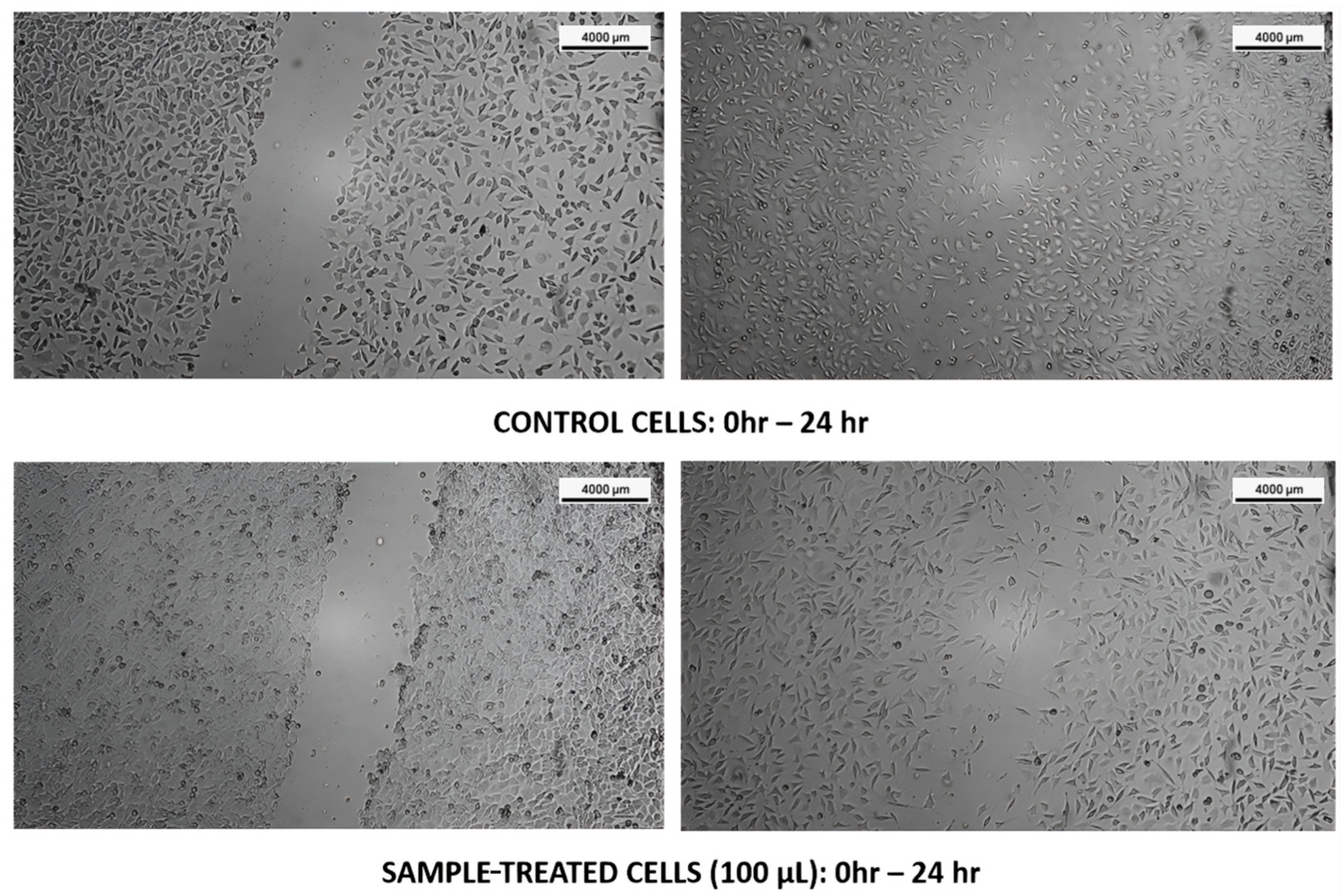
3.3. Cell Migration Assay
3.3.1. HCT116 Cell Lines
3.3.2. L929 Cell Lines
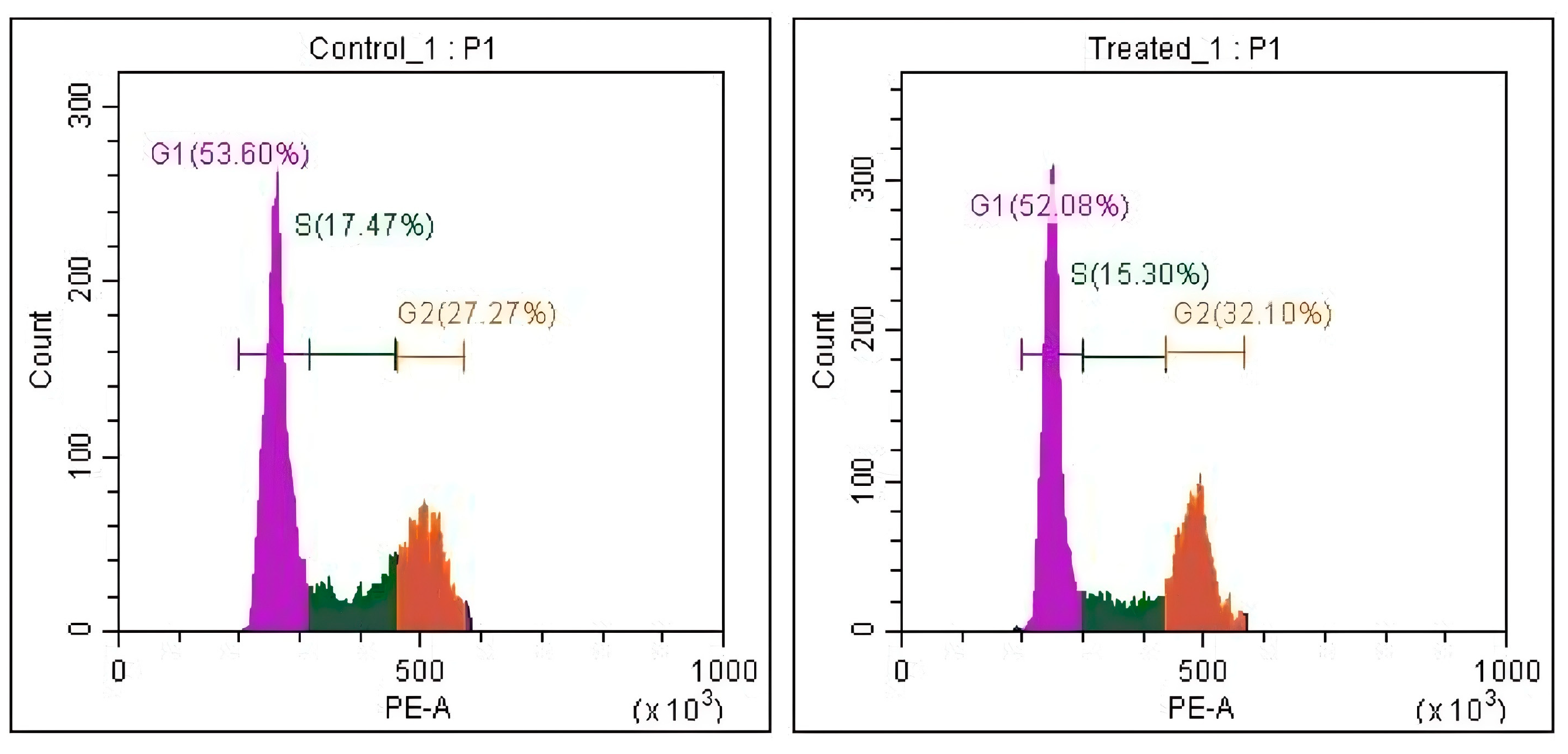
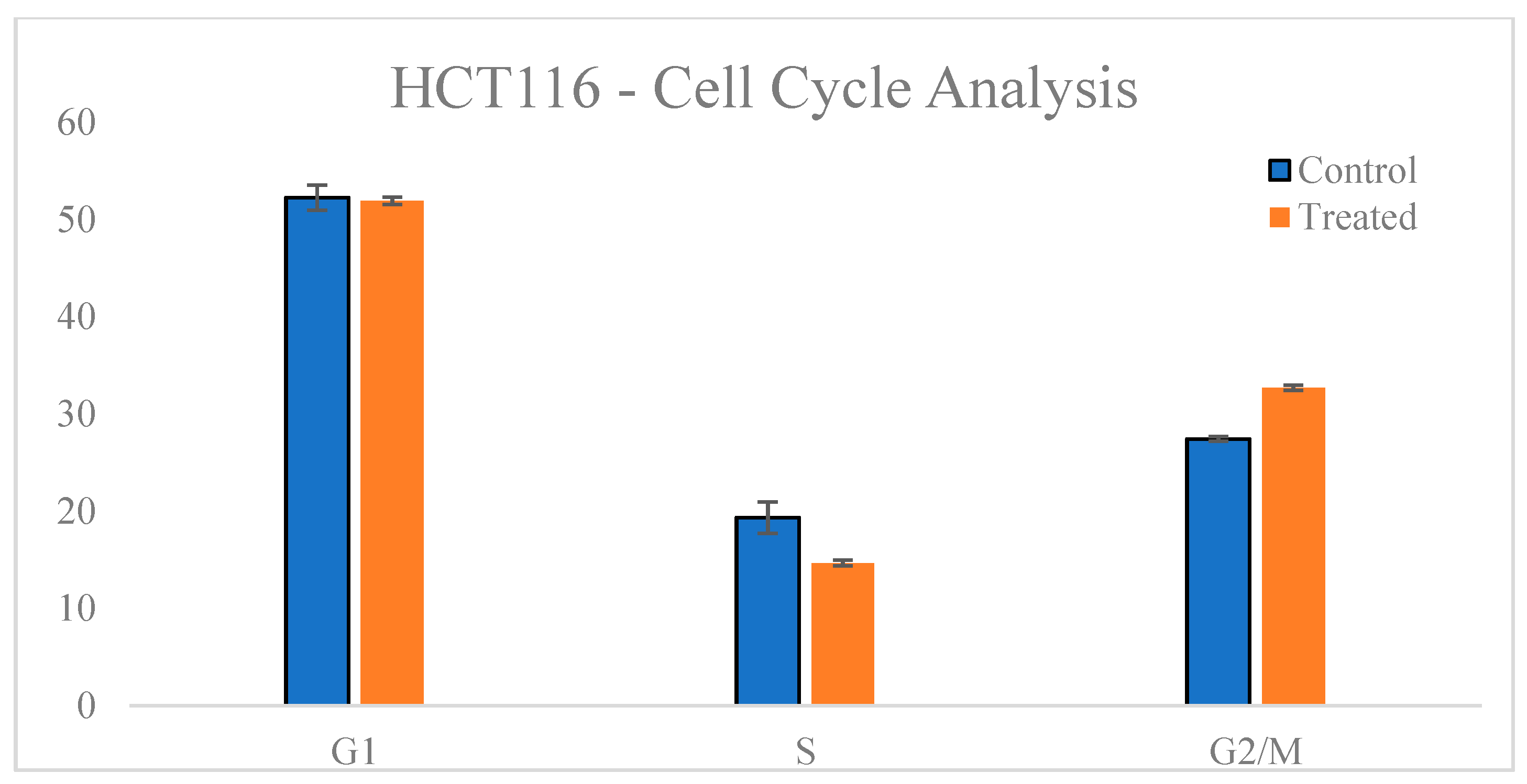
3.4. Cell Cycle Analysis
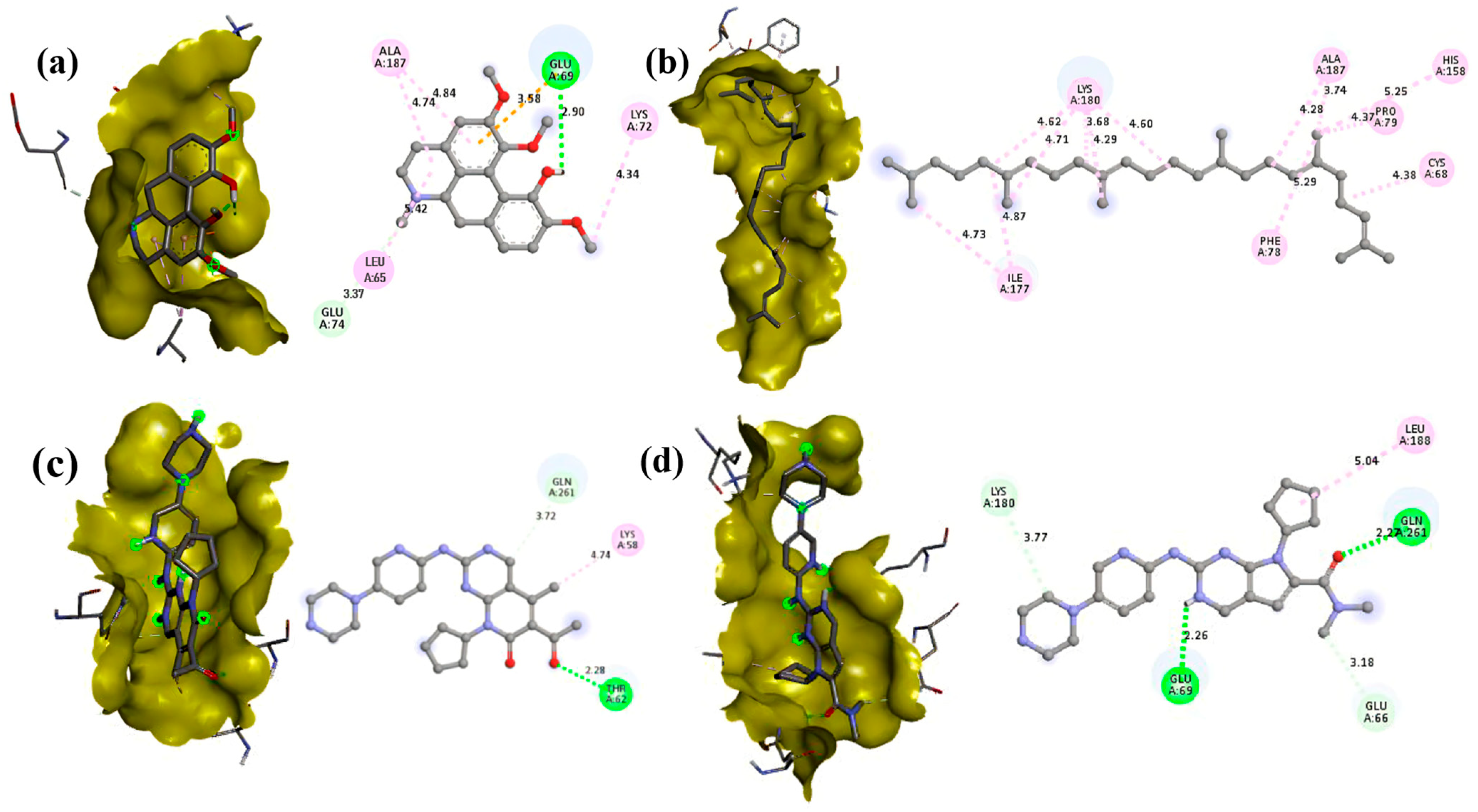
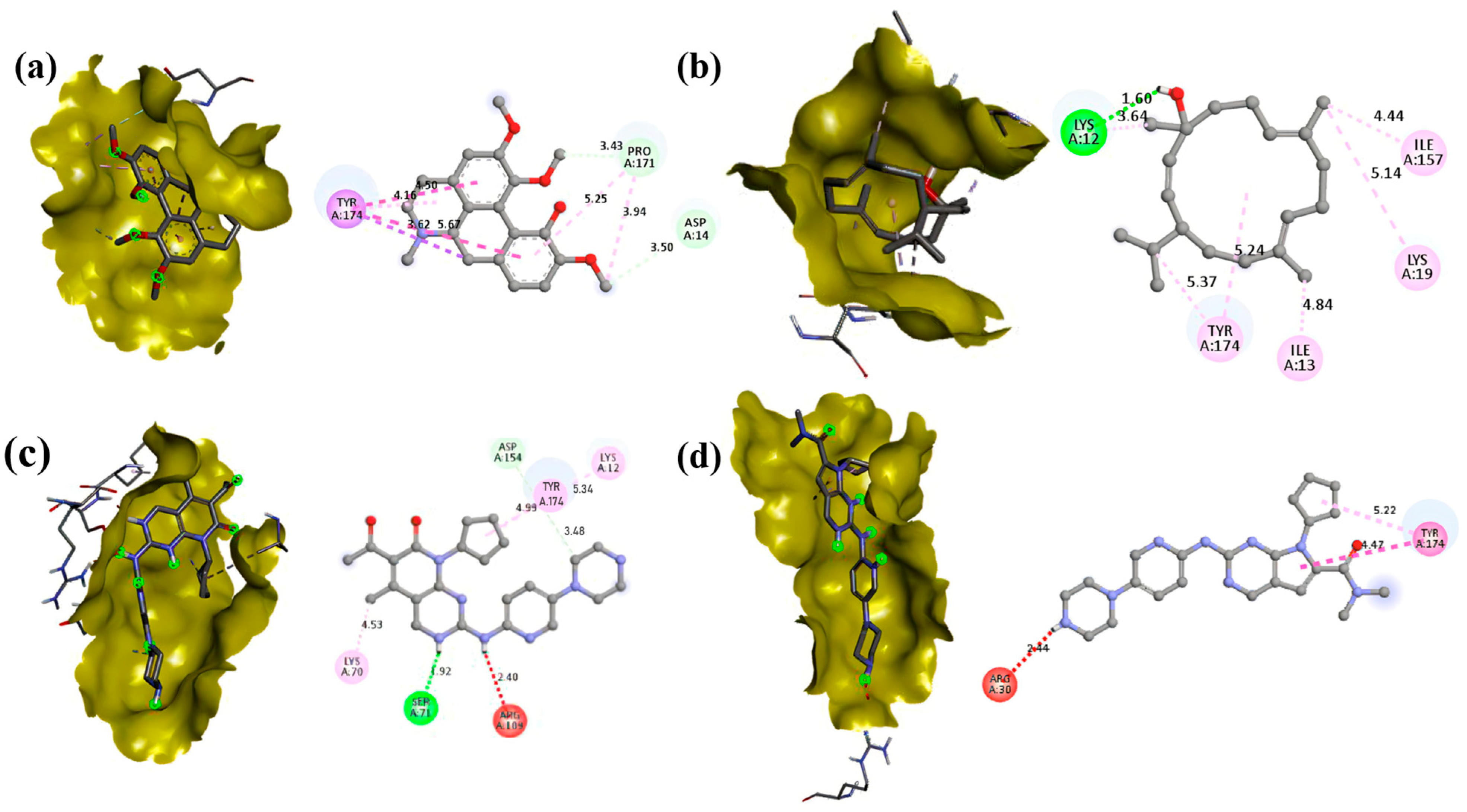
3.5. Molecular Docking
3.6. ADME Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDK | Cyclin-Dependent Kinase |
| ADME | Absorption, Distribution, Metabolism, and Excretion |
| GC-MS | Gas Chromatography-Mass Spectrometry |
| HCT116 | Human Colonic Tumor—116 Cancer Cell Line |
| MTT | 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide |
| IC50 | Half-Maximal Inhibitory Concentration |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DMSO | Dimethyl Sulfoxide |
| PBS | Phosphate-Buffered Saline |
| PI | Propidium Iodide |
| RCSB | Research Collaboratory for Structural Bioinformatics |
| AD4 | AutoDock4 |
| FDA | Food and Drug Administration |
| G1 phase | Gap1 |
| S phase | Synthesis Phase |
| G2 phase | Gap2 |
| M phase | Mitosis |
References
- Pellarin, I.; Dall’Acqua, A.; Favero, A.; Segatto, I.; Rossi, V.; Crestan, N.; Karimbayli, J.; Belletti, B.; Baldassarre, G. Cyclin-Dependent Protein Kinases and Cell Cycle Regulation in Biology and Disease. Signal Transduct. Target. Ther. 2025, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Longworth, S.; Damania, B. Modulation of Cell Cycle Kinases by Kaposi’s Sarcoma-Associated Herpesvirus. J. Med. Virol. 2025, 97, e70157. [Google Scholar] [CrossRef] [PubMed]
- Oner, M.; Cheng, Y.-C.; Soong, S.-W.; Cheng, P.-T.; Wang, Y.-H.; Yang, S.-F.; Tsai, S.C.-S.; Lin, H. Dinaciclib Interrupts Cell Cycle and Induces Apoptosis in Oral Squamous Cell Carcinoma: Mechanistic Insights and Therapeutic Potential. Int. J. Mol. Sci. 2025, 26, 2197. [Google Scholar] [CrossRef]
- Priyadharshini, G.; Sukumaran, G.; Dilipan, E.; Ramani, P. Targeting Oral Cancer: In Silico Docking Studies of Phytochemicals on Oncogenic Molecular Markers. Asian Pac. J. Cancer Prev. 2024, 25, 2069–2075. [Google Scholar] [CrossRef]
- Zhang, W.; Hong, W. Upregulation of miR-519d-3p Inhibits Viability, Proliferation, and G1/S Cell Cycle Transition of Oral Squamous Cell Carcinoma Cells Through Targeting CCND1. Cancer Biother. Radiopharm. 2024, 39, 153–163. [Google Scholar] [CrossRef]
- Dehelean, C.A.; Marcovici, I.; Soica, C.; Mioc, M.; Coricovac, D.; Iurciuc, S.; Cretu, O.M.; Pinzaru, I. Plant-Derived Anticancer Compounds as New Perspectives in Drug Discovery and Alternative Therapy. Molecules 2021, 26, 1109. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, L.; Shivaji, S.; Anandakumar, S. Phytochemical screening, cytotoxic activity and molecular docking studies of Eclipta alba leaves extract against oral cancer. Rasayan J. Chem. 2022, 15, 676–685. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A View on Drug Resistance in Cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Nusaiba, S.A.W.; Murugan, K. In Vitro Analysis on Bactericidal Screening and Antioxidant Potentiality of Leaf and Root Extracts of Thottea siliquosa (Lam.) Ding Hou. An Ethnobotanical Plant. Asian Pac. J. Trop. Biomed. 2013, 3, 859–865. [Google Scholar] [CrossRef]
- Vrundha, C.P.K.; Thomas, T.D. Control of Media Browning during Micropropagation and Assessment of Biochemical and Clonal Fidelity of in Vitro-Derived and Mother Plants in Thottea siliquosa (Lamk.) Ding Hou., an Important Ethnomedicinal Shrub. J. Genet. Eng. Biotechnol. 2023, 21, 70. [Google Scholar] [CrossRef]
- de Angelo, R.M.; de Sousa, D.S.; da Silva, A.P.; Chiari, L.P.A.; da Silva, A.B.F.; Honorio, K.M. Molecular Docking: State-of-the-Art Scoring Functions and Search Algorithms. In Computer-Aided and Machine Learning-Driven Drug Design: From Theory to Applications; Maltarollo, V.G., Ed.; Springer Nature: Cham, Switzerland, 2024; pp. 163–198. ISBN 978-3-031-76718-0. [Google Scholar]
- Abubakar, A.R.; Haque, M. Preparation of Medicinal Plants: Basic Extraction and Fractionation Procedures for Experimental Purposes. J. Pharm. Bioallied Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.S.; Khaled, N.B.; Ye, L.; Wimmer, R.; Hammann, L.; Weich, A.; Suppan, C.; Mahajan, U.M.; Jung, A.; Kumbrink, J.; et al. Efficacy of CDK4/6 Inhibition in Colorectal Cancer and the Role of P16 Expression in Predicting Drug Resistance. Cell Oncol. 2025, 1–13. [Google Scholar] [CrossRef]
- Ravi, S.; Gaddamedi, S.; Gopal Agrawal, H.; Soni, A.; Kumar Mishra, A.; Narayan Rath, S. Design, Synthesis, Biological Evaluation, and Molecular Docking Studies of N3-Functionalized TZD-Indole Conjugates as Potential Anticancer Agents. ChemistrySelect 2024, 9, e202400123. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Chikowe, I.; Bwaila, K.D.; Ugbaja, S.C.; Abouzied, A.S. GC–MS Analysis, Molecular Docking, and Pharmacokinetic Studies of Multidentia Crassa Extracts’ Compounds for Analgesic and Anti-Inflammatory Activities in Dentistry. Sci. Rep. 2024, 14, 1876. [Google Scholar] [CrossRef]
- Goel, S.; Bergholz, J.S.; Zhao, J.J. Targeting CDK4 and CDK6 in Cancer. Nat. Rev. Cancer. 2022, 22, 356–372. [Google Scholar] [CrossRef]
- Wu, F.; Tian, M.; Sun, Y.; Wu, C.; Liu, X. Efficacy, Chemical Composition, and Pharmacological Effects of Herbal Drugs Derived from Fritillaria Cirrhosa D. Don and Fritillaria Thunbergii Miq. Front. Pharmacol. 2022, 13, 985935. [Google Scholar] [CrossRef]
- Kocanci, F.; Aslim, B. Chemical Composition and Neurotherapeutic Potential of Glaucium Corniculatum Extracts. Pharmacogn. Mag. 2021, 17, 67. [Google Scholar] [CrossRef]
- Madriwala, B.; Suma, B.V.; Jays, J. Molecular Docking Study Of Hentriacontane For Anticancer And Antitubercular Activity. Int. J. Chem. Res. 2022, 6, 1–4. [Google Scholar] [CrossRef]
- Dong, J.-W.; Li, X.-J.; Xu, X.-X.; Gu, S.-F.; Zhao, H.; Wang, X.-X. Solid-State Fermentation of Aspergillus Sydowii G12, an Approach to Produce Isocorydine. Process Biochem. 2024, 147, 147–151. [Google Scholar] [CrossRef]
- Luo, J.; Wang, N.; Hua, L.; Deng, F.; Liu, D.; Zhou, J.; Yuan, Y.; Ouyang, F.; Chen, X.; Long, S.; et al. The Anti-Sepsis Effect of Isocorydine Screened from Guizhou Ethnic Medicine Is Closely Related to Upregulation of Vitamin D Receptor Expression and Inhibition of NFκB P65 Translocation into the Nucleus. J. Inflamm. Res. 2022, 15, 5649–5664. [Google Scholar] [CrossRef]
- Song, L.; Zhao, F.; Liu, Y.; Guo, X.; Wu, C.; Liu, J. Effects of 8-Amino-Isocorydine, a Derivative of Isocorydine, on Gastric Carcinoma Cell Proliferation and Apoptosis. Curr. Ther. Res. 2021, 94, 100624. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-R.; Lin, Y.-K.; Fang, J.-Y. Biological and Pharmacological Activities of Squalene and Related Compounds: Potential Uses in Cosmetic Dermatology. Molecules 2009, 14, 540–554. [Google Scholar] [CrossRef]
- Micera, M.; Botto, A.; Geddo, F.; Antoniotti, S.; Bertea, C.M.; Levi, R.; Gallo, M.P.; Querio, G. Squalene: More than a Step toward Sterols. Antioxidants 2020, 9, 688. [Google Scholar] [CrossRef] [PubMed]
- Muzalevskaya, E.N.; Miroshnichenko, L.A.; Nikolaevskii, V.A.; Ushakov, I.B.; Chernov, Y.N.; Alabovskii, V.V.; Batishcheva, G.A.; Buzlama, A.V. Squalene: Physiological and pharmacological properties. Eksperimental Naia I Klin. Farmakol. 2015, 78, 30–36. [Google Scholar]
- Yuefan, L.I.; Yuanyuan, W.; Gaiqin, M.A.; Qi, L.I.; Xiuzhu, Y.U. Research progress on sources, extraction and functional properties of squalene. J. Henan Univ. Technol. 2022, 43, 19–29. [Google Scholar] [CrossRef]
- Jokar, E.; Sateei, A.; Ebadi, M.; Ahmadi Golsefidi, M. Changes in the medicinal and antimicrobial compounds, methyl palmitate, methyl stearate, and bis (2-ethylhexyl) phthalate in American agave (Agave Americana L. cv Marginata) under urea treatment. Iran. J. Plant Physiol. 2023, 4, 4637. [Google Scholar] [CrossRef]
- Olivia, N.U.; Goodness, U.C.; Obinna, O.M. Phytochemical Profiling and GC-MS Analysis of Aqueous Methanol Fraction of Hibiscus Asper Leaves. Future J. Pharm. Sci. 2021, 7, 59. [Google Scholar] [CrossRef]
- Chellappan, B.M.; Venkatachalam, G.; Reddy, C.U.; Kumar, M.; Venketeswarawarlu, B.S. GC-MS Profiling and in Vitro Assessment of Antioxidant and Neuroprotective Properties of Ethanolic and Acetone Extracts of Oldenlandia corymbosa L. Asian J. Life Sci. 2022, 11, 188–192. [Google Scholar] [CrossRef]
- Xu, C.; Zhao, S.; Li, M.; Dai, Y.; Tan, L.; Liu, Y. Chemical Composition, Antimicrobial and Antioxidant Activities of Essential Oil from Flue-Cured Tobacco Flower Bud. Biotechnol. Biotechnol. Equip. 2016, 30, 1026–1030. [Google Scholar] [CrossRef]
- Boța, M.; Vlaia, L.; Jîjie, A.-R.; Marcovici, I.; Crişan, F.; Oancea, C.; Dehelean, C.A.; Mateescu, T.; Moacă, E.-A. Exploring Synergistic Interactions between Natural Compounds and Conventional Chemotherapeutic Drugs in Preclinical Models of Lung Cancer. Pharmaceuticals 2024, 17, 598. [Google Scholar] [CrossRef]
- Chaachouay, N. Synergy, Additive Effects, and Antagonism of Drugs with Plant Bioactive Compounds. Drugs Drug Candidates 2025, 4, 4. [Google Scholar] [CrossRef]
- Childs, R.W.; Carlsten, M. Therapeutic Approaches to Enhance Natural Killer Cell Cytotoxicity against Cancer: The Force Awakens. Nat. Rev. Drug. Discov. 2015, 14, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Ghadimi, P.; Ghorbian, S. Effects of Melatonin on the Expression of Invasion-Related Markers (MMP2 and MMP9) in Breast Cancer Cells. J. Cell. Biochem. 2025, 126, e70010. [Google Scholar] [CrossRef]
- Nabi, N.; Nisa, M.U.; Khan, M.S.; Gillani, S.Q. Role of Signaling Pathways Regulating Cell Cycle Progression in Cancer. In Cell Signaling Pathways and Their Therapeutic Implication in Cancers; Rehman, M.U., Khan, M.S., Eds.; Springer Nature: Singapore, 2025; pp. 357–393. ISBN 978-981-9627-63-9. [Google Scholar]
- Yang, L.; Zhu, Y.; Ding, X.; Sun, J.; Cao, X.; Zhang, Z.; Li, L.; Lin, J. M7: A Novel in Vitro CDK1 Inhibitor Suppressing Colorectal Cancer Cell Proliferation, Migration and Promoting Apoptosis. Eur. J. Med. Chem. Rep. 2025, 14, 100271. [Google Scholar] [CrossRef]
- Guo, L.; Qi, J.; Wang, H.; Jiang, X.; Liu, Y. Getting under the Skin: The Role of CDK4/6 in Melanomas. Eur. J. Med. Chem. 2020, 204, 112531. [Google Scholar] [CrossRef]
- Nardone, V.; Barbarino, M.; Angrisani, A.; Correale, P.; Pastina, P.; Cappabianca, S.; Reginelli, A.; Mutti, L.; Miracco, C.; Giannicola, R.; et al. CDK4, CDK6/Cyclin-D1 Complex Inhibition and Radiotherapy for Cancer Control: A Role for Autophagy. Int. J. Mol. Sci. 2021, 22, 8391. [Google Scholar] [CrossRef]
- Tadesse, S.; Yu, M.; Kumarasiri, M.; Le, B.T.; Wang, S. Targeting CDK6 in Cancer: State of the Art and New Insights. Cell Cycle 2015, 14, 3220–3230. [Google Scholar] [CrossRef]
- Azad, I. Molecular Docking in the Study of Ligand-Protein Recognition: An Overview. In Molecular Docking—Recent Advances; IntechOpen: London, UK, 2023; ISBN 978-1-80356-468-5. [Google Scholar]
- Spassov, D.S. Binding Affinity Determination in Drug Design: Insights from Lock and Key, Induced Fit, Conformational Selection, and Inhibitor Trapping Models. Int. J. Mol. Sci. 2024, 25, 7124. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, R.; Paul, S.; Alam, M.S.; Rahman, A.F.M.M. Synthesis, Biological Properties, In Silico ADME, Molecular Docking Studies, and FMO Analysis of Chalcone Derivatives as Promising Antioxidant and Antimicrobial Agents. ACS Omega 2025, 10, 4367–4387. [Google Scholar] [CrossRef]
- Lin, W.; Zeng, Y.; Weng, L.; Yang, J.; Zhuang, W. Comparative Analysis of Adverse Events Associated with CDK4/6 Inhibitors Based on FDA’s Adverse Event Reporting System: A Case Control Pharmacovigilance Study. BMC Pharmacol. Toxicol. 2024, 25, 47. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, Y.; Zhou, X.; Jia, X.; Zhong, Y. A Disproportionality Analysis of CDK4/6 Inhibitors in the FDA Adverse Event Reporting System (FAERS). Expert Opin. Drug Saf. 2025, 24, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Luo, P.; Xu, J. Adverse Event Profiles of CDK4/6 Inhibitors: Data Mining and Disproportionality Analysis of the FDA Adverse Event Reporting System. Ther. Adv. Drug Saf. 2024, 15, 20420986241278498. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, I.-M.; Parosanu, A.I.; Nitipir, C. An Overview of the Safety Profile and Clinical Impact of CDK4/6 Inhibitors in Breast Cancer—A Systematic Review of Randomized Phase II and III Clinical Trials. Biomolecules 2023, 13, 1422. [Google Scholar] [CrossRef]
- Yang, L.; Xue, J.; Yang, Z.; Wang, M.; Yang, P.; Dong, Y.; He, X.; Bao, G.; Peng, S. Side Effects of CDK4/6 Inhibitors in the Treatment of HR+/HER2− Advanced Breast Cancer: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Ann. Palliat. Med. 2021, 10, 5590599–5595599. [Google Scholar] [CrossRef]
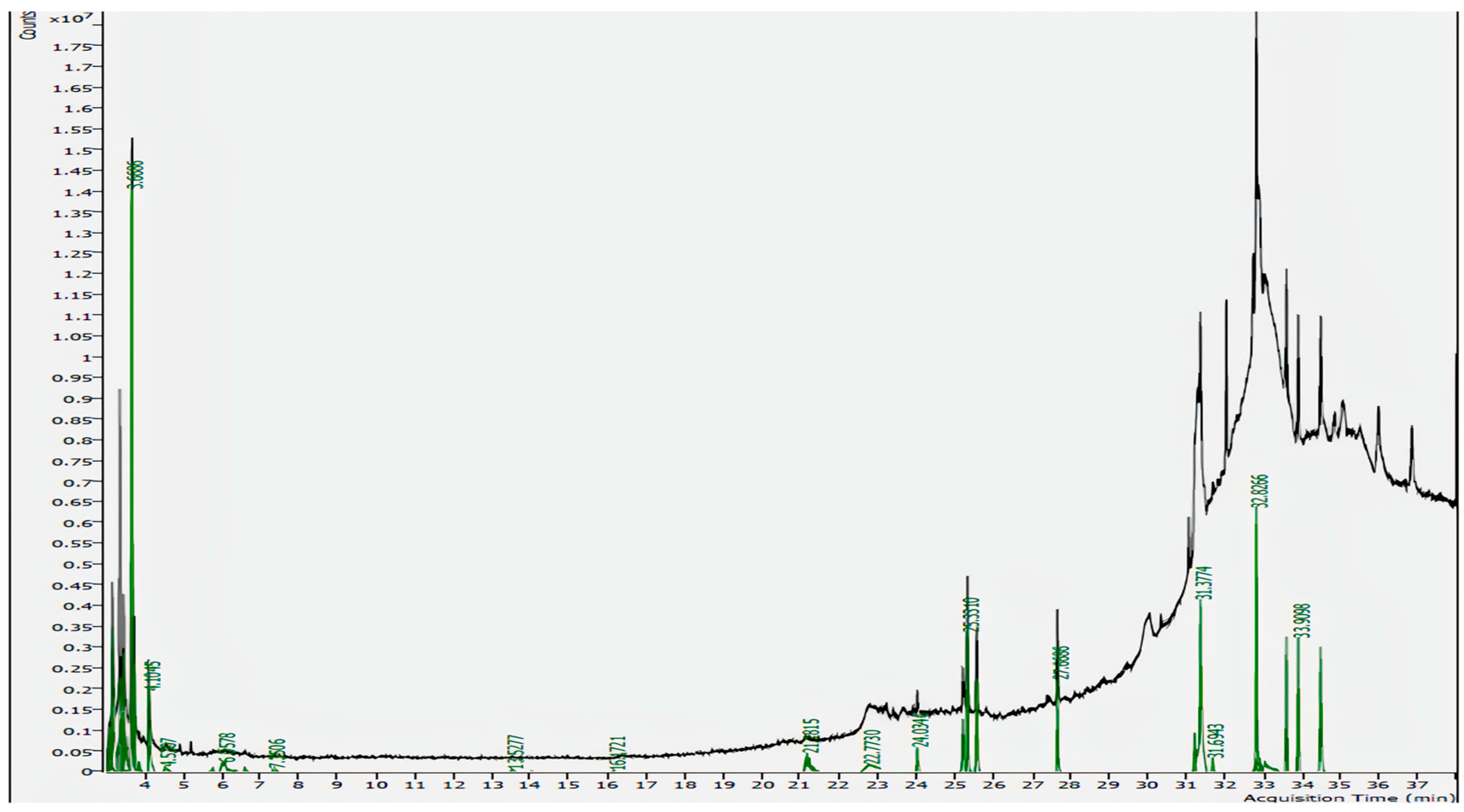
| RT (Min) | Molecular Formula | Molecular Mass (g/mol) | Compound Name | Peak Area% | Match Factor |
|---|---|---|---|---|---|
| 31.3774 | C19H38O4 | 330.50 | 2-palmitoylglycerol | 19.23% | 81.4 |
| 32.8266 | C31H64 | 436.85 | Hentriacontane | 15.97% | 90.1 |
| 34.4893 | C20H23NO4 | 341.4 | Isocorydine | 8.73% | 79.6 |
| 33.9098 | C30H50 | 410.73 | Squalene | 7.69% | 85.5 |
| 25.3310 | C17H34O2 | 270.45 | Methyl Palmitate | 7.63% | 94.8 |
| 25.5736 | C20H34O | 290.5 | Thunbergol | 5.77% | 77.9 |
| 27.6686 | C19H38O2 | 298.50 | Methyl Stearate | 5.04% | 93.0 |
| Concentrations (µg/mL) | % of Viability ± SD | |
|---|---|---|
| Control cells | 100 ± 6.04 | |
| Sample-treated cells | 100 | 60.18 ± 9.21 |
| 200 | 28.24 ± 4.12 | |
| 300 | 26.02 ± 10.43 | |
| 400 | 24.70 ± 4.93 | |
| 500 | 9.64 ± 1.82 | |
| 600 | 6.06 ± 0.58 | |
| 700 | 5.82 ± 0.63 | |
| 800 | 3.99 ± 0.67 | |
| 900 | 4.02 ± 0.43 | |
| 1000 | 3.53 ± 0.50 |
| % Migration | ||
|---|---|---|
| Control Cells | Treated Cells | |
| 0 h | 0 | 0 |
| 24 h | 34.93 | 27.01 |
| 36 h | 57.26 | 30.78 |
| Group | G1 Phase (%) | S Phase (%) | G2 Phase (%) |
|---|---|---|---|
| Control | 52.25 ± 0.66 | 19.34 ± 1.22 | 27.44 ± 0.24 |
| Treated | 51.95 ± 0.36 | 14.67 ± 0.60 | 32.70 ± 0.45 |
| Compound | PubChem ID | Binding Energy (kcal/mol) | |
|---|---|---|---|
| CDK4 (2W9F) | CDK6 (1JOW) | ||
| Isocorydine | 10143 | −7.4 | −7.2 |
| Thunbergol | 5363523 | −6.5 | −7 |
| Squalene | 638072 | −7.1 | −4.9 |
| 2-Palmitoylglycerol | 123409 | −5.2 | −4.9 |
| Methyl palmitate | 8181 | −4.5 | −5.5 |
| Methyl stearate | 8201 | −4.4 | −4.7 |
| Hentriacontane | 12410 | −4.8 | −3.9 |
| Palbociclib | 5330286 | −7.2 | −8.3 |
| Ribociclib | 44631912 | −7.1 | −8.1 |
| 10143 | 5363523 | 638072 | 123409 | 8181 | 8201 | 12410 | 5330286 (Standard) | 4431912 (Standard) |
|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | Lys58 | - |
| - | - | - | - | Gln183 | Gln183 (Pi-Sigma) | - | - | - |
| Glu69 (Pi-Alkyl) | - | - | - | - | - | - | - | Glu69 (Pi-Sigma) |
| - | - | Phe78 | - | Phe78 (Pi-Alkyl) | - | - | - | - |
| - | - | - | - | - | - | - | Thr62 (Pi-Alkyl) | - |
| Ala187 | Ala187 | Ala187 (Pi-Alkyl) | Ala187 | Ala187 | Ala187 | Ala187 | - | - |
| - | - | - | Ser258 | - | - | - | - | - |
| - | - | - | - | - | Cys73 (Pi-Alkyl) | - | - | - |
| - | - | - | - | - | - | - | - | Glu66 (Alkyl) |
| - | - | Ile177 | - | - | - | - | - | - |
| - | - | Pro79 | - | Pro79 (Pi-Sigma) | - | - | - | - |
| - | Leu188 | - | - | - | - | Leu188 (Pi-Sigma) | - | Leu188 |
| Glu74 (Pi-Alkyl) | - | - | - | - | - | - | - | - |
| - | - | Cys68 (Pi-Sigma) | Cys68 (Pi-Alkyl) | Cys68 | - | Cys68 | - | - |
| - | Gln261 (Pi-Alkyl) | - | Gln261 | - | - | - | Gln261 | Gln261 |
| Leu65 | Leu65 | - | Leu65 | - | - | Leu65 | - | - |
| - | - | Lys180 | - | Lys180 (Pi-Alkyl) | Lys180 | Lys180 | - | Lys180 (Pi-Sigma) |
| Lys72 | - | - | - | - | - | - | - | |
| - | - | His158 (Pi-Sigma) | - | - | - | - | - | - |
| 10143 | 5363523 | 638072 | 123409 | 8181 | 8201 | 12410 | 5330286 (Standard) | 4431912 (Standard) |
|---|---|---|---|---|---|---|---|---|
| - | - | - | - | Leu185 (Pi-Alkyl) | - | - | - | - |
| Tyr174 | Tyr174 | - | Tyr174 | - | Tyr174 | - | Tyr174 (Pi-Sigma) | Tyr174 |
| - | - | - | - | Leu32 | - | - | - | - |
| - | - | Trp58 | - | - | - | Trp58 (Pi-Sigma) | - | - |
| - | Ile13 | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | Arg30 |
| Asp14 | - | - | - | - | - | - | - | - |
| - | - | - | Pro158 (Pi-Alkyl) | - | Pro158 (Pi-Sigma) | - | - | - |
| - | - | - | Asp154 | - | Asp154 | - | Asp154 | - |
| - | - | Ile54 | - | - | - | Ile54 | - | - |
| - | - | - | - | Leu34 (Pi-Sigma) | - | - | - | - |
| - | Lys12 | - | - | - | - | - | Lys12 | - |
| - | - | Leu254 | - | - | - | Leu254 | - | - |
| - | - | - | - | - | - | - | Lys70 (Pi-Alkyl) | - |
| - | Lys19 | - | Lys19 | - | - | - | - | - |
| - | - | Leu55 (Pi-Alkyl) | - | - | - | Leu55 | - | - |
| - | Ile157 | - | Ile157 | - | Ile157 | - | - | - |
| - | - | - | - | - | - | - | Arg109 | - |
| - | - | - | - | Phe37 | - | - | - | - |
| - | - | Lys91 | - | - | - | Lys91 | - | - |
| - | - | - | - | Pro195 | - | - | - | - |
| Pro171 | - | - | Pro171 | - | Pro171 | - | - | - |
| - | - | - | - | - | - | - | Ser71 (Pi-Alkyl) | - |
| - | - | - | - | Ile198 | - | - | - | - |
| - | - | Leu94 (Pi-Alkyl) | - | - | - | Leu94 | - | - |
| Ligands | TPSA (Å2) | Water Solubility | GI Absorption | BBB Permeation | Drug Likeliness | Lipinski’s Rule of Five | PAINS Alert |
|---|---|---|---|---|---|---|---|
| Isocorydine | 51.16 | Soluble | High | Yes | Yes | 0 violation | 0 |
| Thunbergol | 20.23 | Moderate | High | No | Yes | 1 violation (LogP = 5.64) | 0 |
| Squalene | 0.00 | Poor | Low | No | Yes | 1 violation (LogP = 10.74) | 0 |
| 2-palmitoylglycerol | 66.76 | Moderate | High | Yes | Yes | 0 violation | 0 |
| Methyl palmitate | 26.30 | Moderate | High | Yes | Yes | 1 violation (LogP = 6.34) | 0 |
| Methyl stearate | 26.30 | Moderate | High | No | Yes | 1 violation (LogP = 7.15) | 0 |
| Hentriacontane | 0.00 | Insoluble | Low | No | Yes | 1 violation (LogP = 12.29) | 0 |
| Palbociclib | 105.04 | Soluble | High | No | Yes | 0 violation | 0 |
| Ribociclib | 91.21 | Soluble | High | No | Yes | 0 violation | 0 |
| Compound Name | Pharmacological Activities | References |
|---|---|---|
| 2-palmitoylglycerol | Anti-inflammatory, Prevents apoptosis | [20,21] |
| Hentriacontane | Anticancer, Antioxidant, Anti-inflammatory, Anti-tubercular, Immunomodulator, Hepatoprotective, Antimicrobial | [22] |
| Isocorydine | Anticancer, Antioxidant, Anti-inflammatory, Antisepsis, Anti-arrhythmia, Vasodilation, Antitumor, | [23,24,25] |
| Squalene | Anticancer, Antioxidant, Anti-inflammatory, Cardioprotective, Hepatoprotective, Promotes skin health | [26,27,28,29] |
| Methyl Palmitate | Anticancer, Antioxidant, Anti-inflammatory, Antimicrobial, Hypocholesterolemic, Hemolytic | [30,31] |
| Thunbergol | Antioxidant, Antimicrobial, Neuro-protective | [32,33] |
| Methyl Stearate | Anticancer, Anti-inflammatory, Antimicrobial | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elakkiya, M.R.; Krishnasreya, M.; Tharani, S.; Arun, M.; Vijayalakshmi, L.; Lim, J.; Ghfar, A.A.; Chithradevi, B. Targeting CDK4/6 in Cancer: Molecular Docking and Cytotoxic Evaluation of Thottea siliquosa Root Extract. Biomedicines 2025, 13, 1658. https://doi.org/10.3390/biomedicines13071658
Elakkiya MR, Krishnasreya M, Tharani S, Arun M, Vijayalakshmi L, Lim J, Ghfar AA, Chithradevi B. Targeting CDK4/6 in Cancer: Molecular Docking and Cytotoxic Evaluation of Thottea siliquosa Root Extract. Biomedicines. 2025; 13(7):1658. https://doi.org/10.3390/biomedicines13071658
Chicago/Turabian StyleElakkiya, Maruthamuthu Rathinam, Mohandas Krishnasreya, Sureshkumar Tharani, Muthukrishnan Arun, L. Vijayalakshmi, Jiseok Lim, Ayman A. Ghfar, and Balasundaramsaraswathy Chithradevi. 2025. "Targeting CDK4/6 in Cancer: Molecular Docking and Cytotoxic Evaluation of Thottea siliquosa Root Extract" Biomedicines 13, no. 7: 1658. https://doi.org/10.3390/biomedicines13071658
APA StyleElakkiya, M. R., Krishnasreya, M., Tharani, S., Arun, M., Vijayalakshmi, L., Lim, J., Ghfar, A. A., & Chithradevi, B. (2025). Targeting CDK4/6 in Cancer: Molecular Docking and Cytotoxic Evaluation of Thottea siliquosa Root Extract. Biomedicines, 13(7), 1658. https://doi.org/10.3390/biomedicines13071658








