An Integrative Genomics Approach for the Discovery of Potential Clinically Actionable Diagnostic and Prognostic Biomarkers in Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
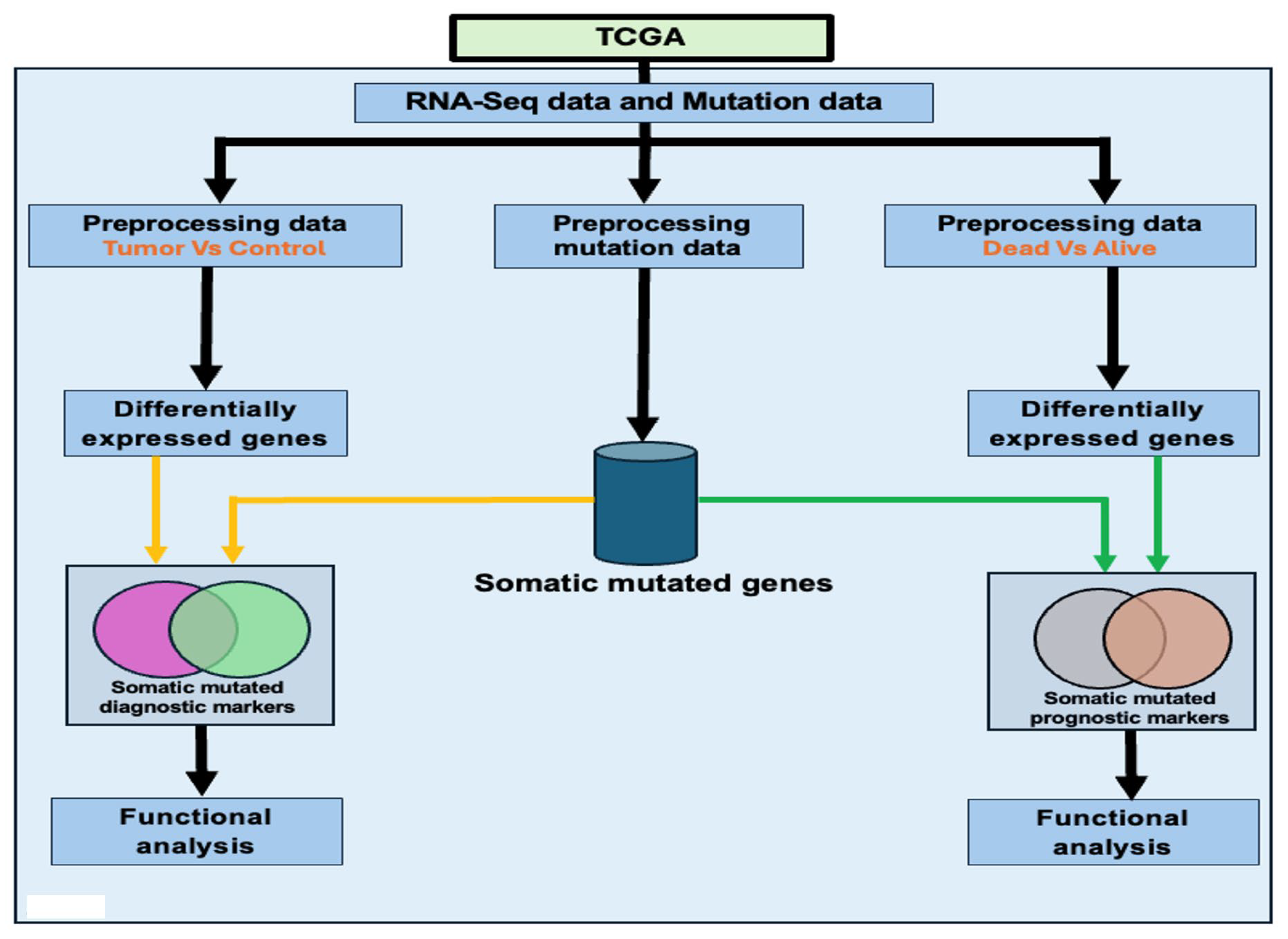
2.1. Overall Study Design and Execution Strategy
2.2. Sources of Gene Expression and Somatic Mutation Data
2.3. Bioinformatics Analysis of RNA-Seq Data
2.4. Integrating Gene Expression with Somatic Mutation Information
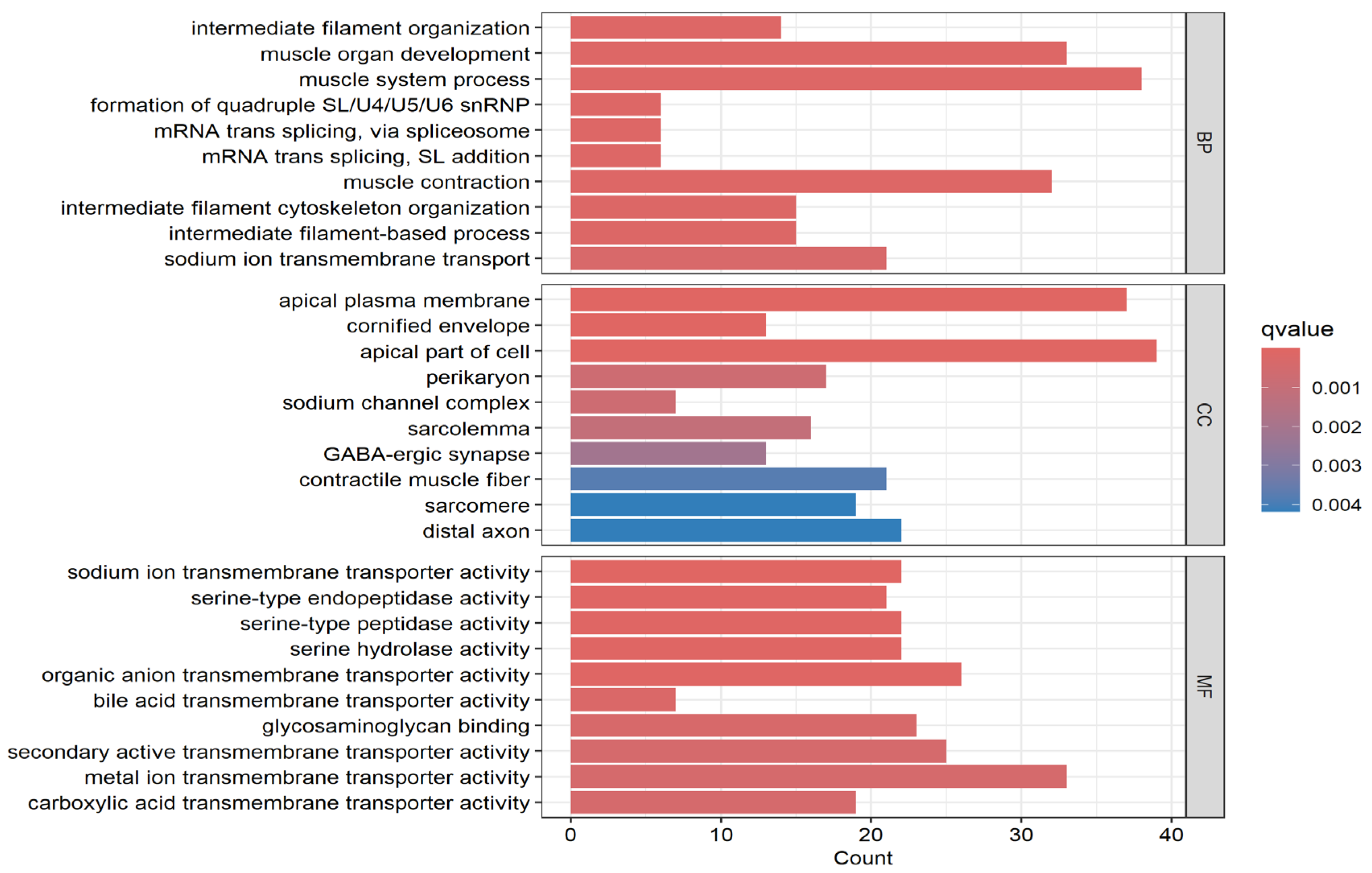
2.5. Functional and Enrichment Analysis
3. Results
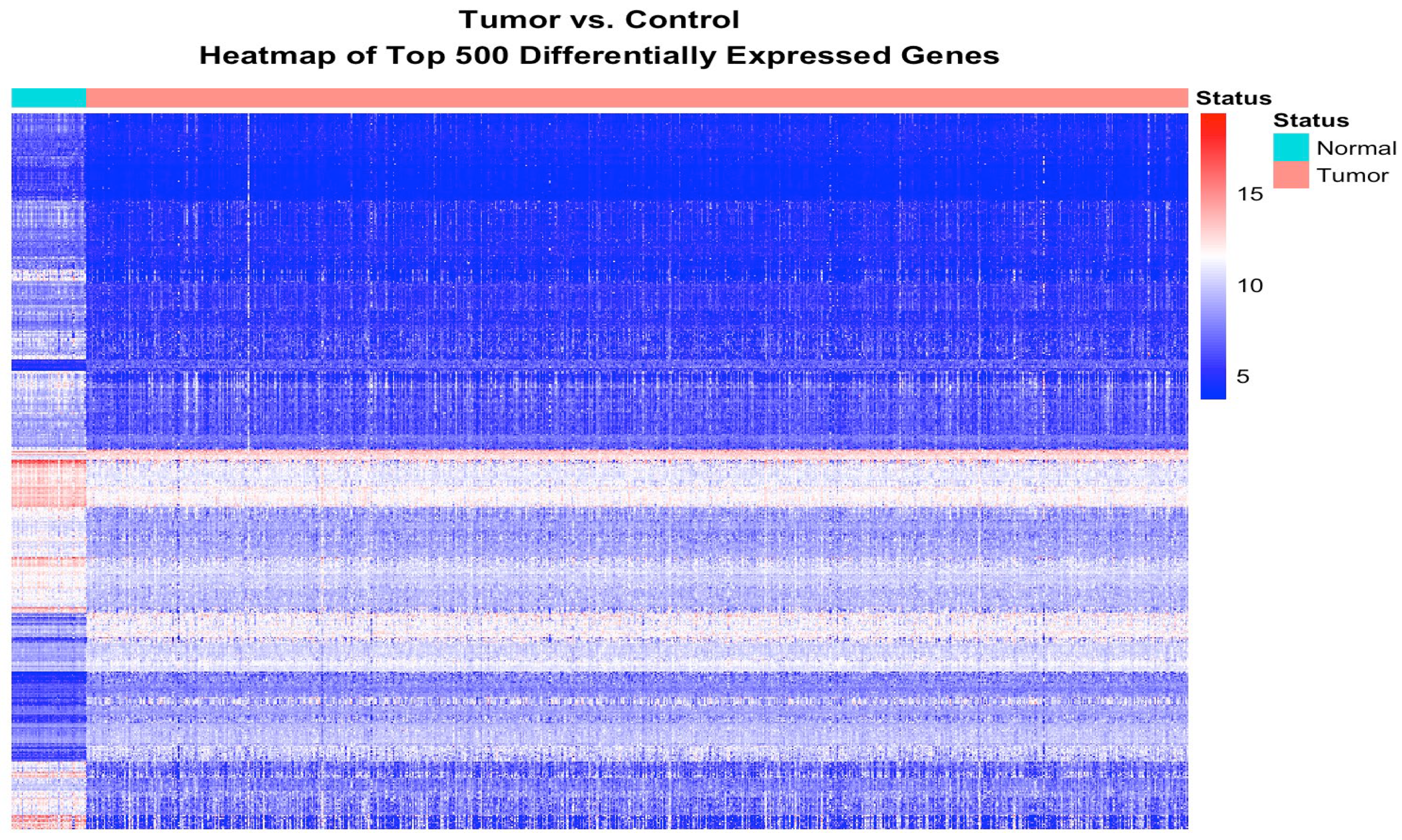
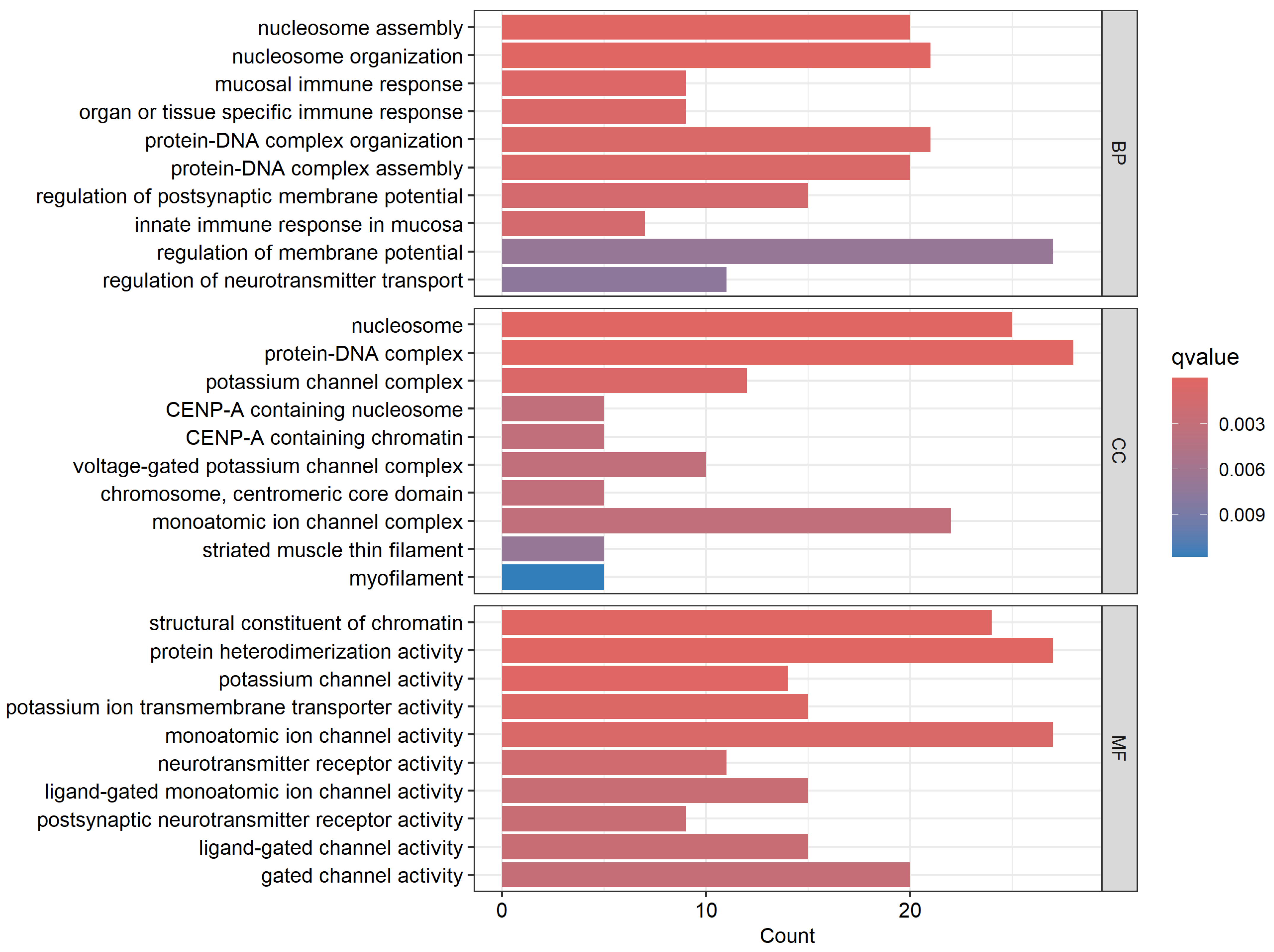
3.1. Discovery of a Signature of Genes Transcriptionally Associated with CRC
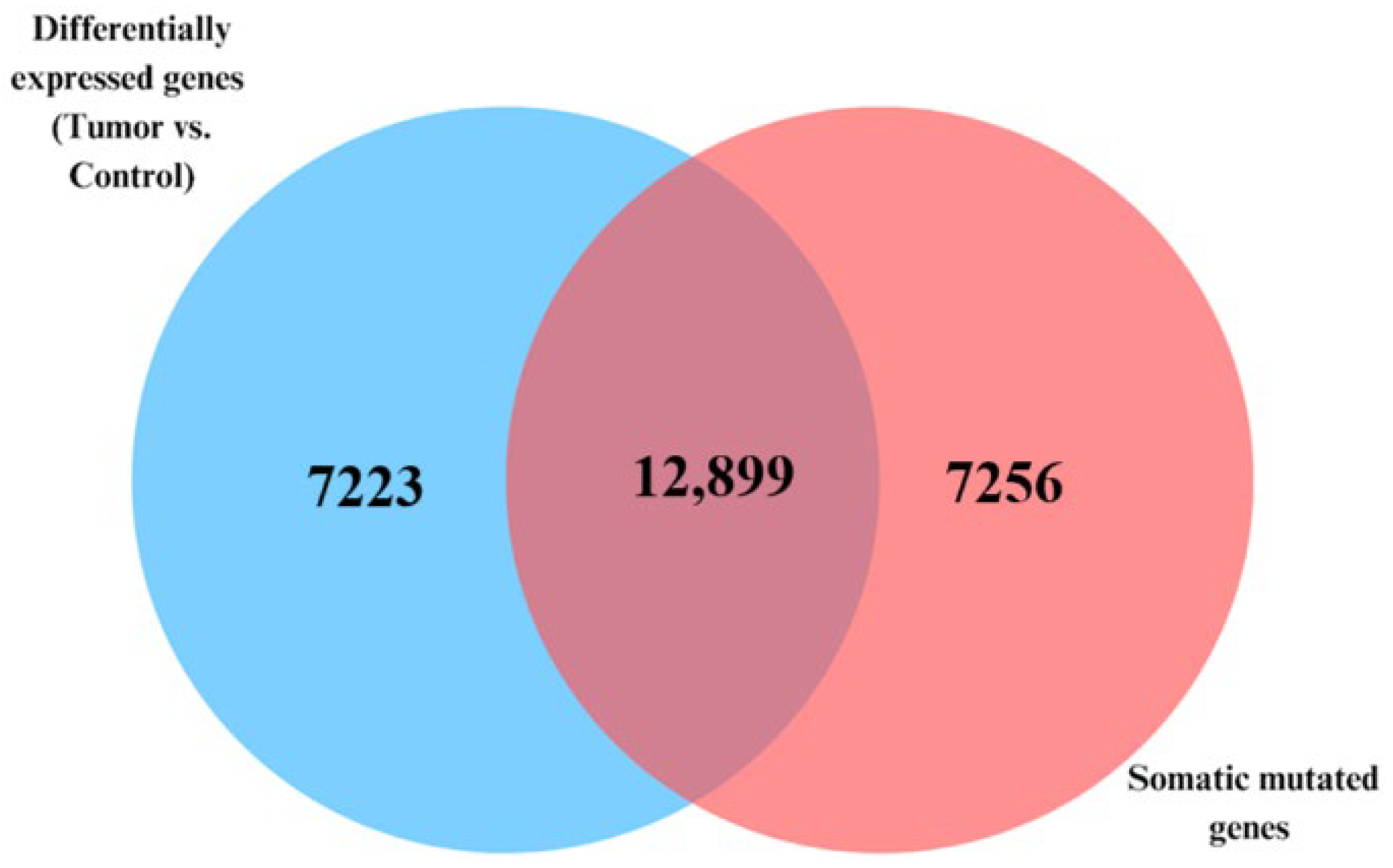
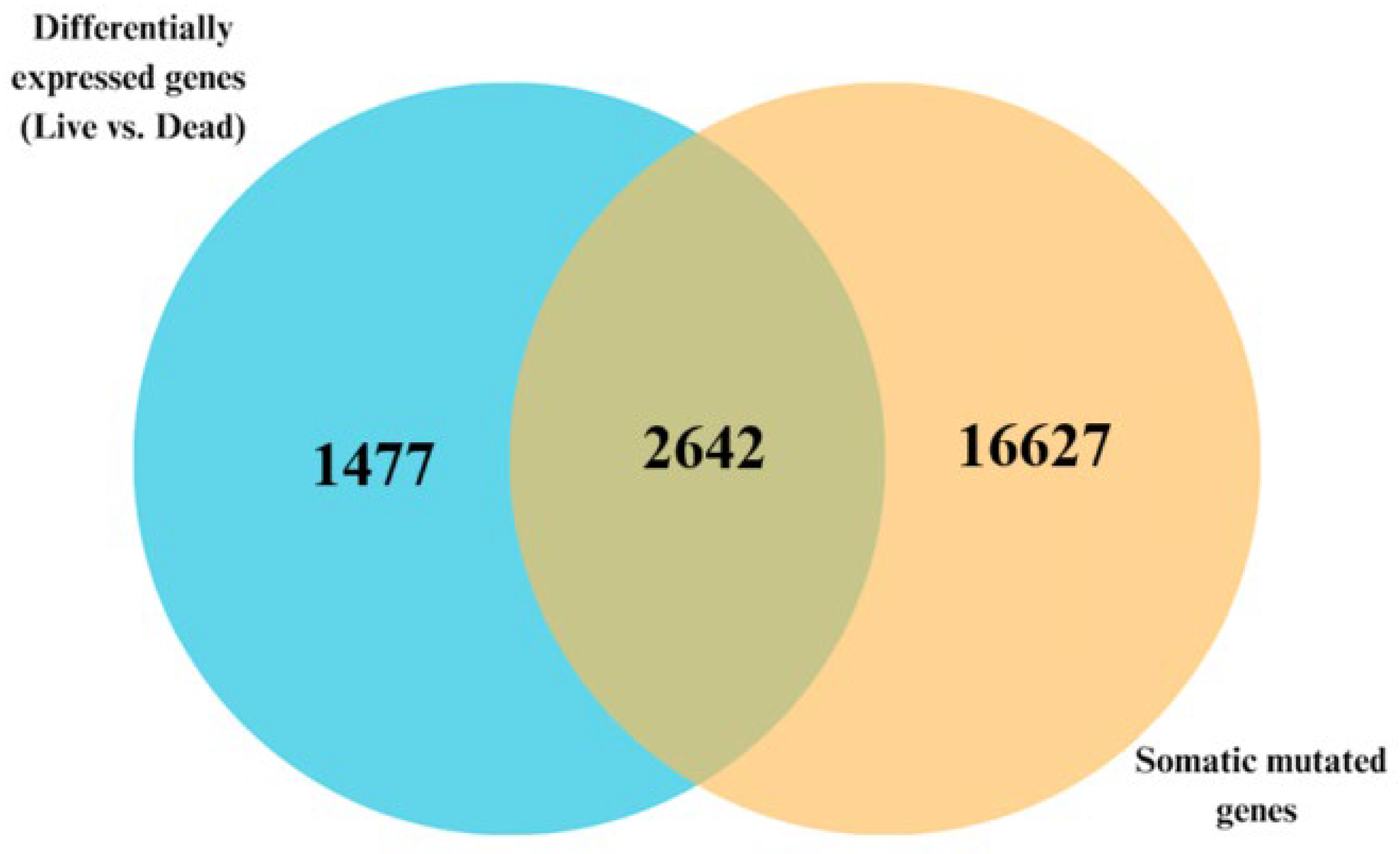
3.2. Signature of Somatic Mutated Genes Transcriptionally Associated with CRC
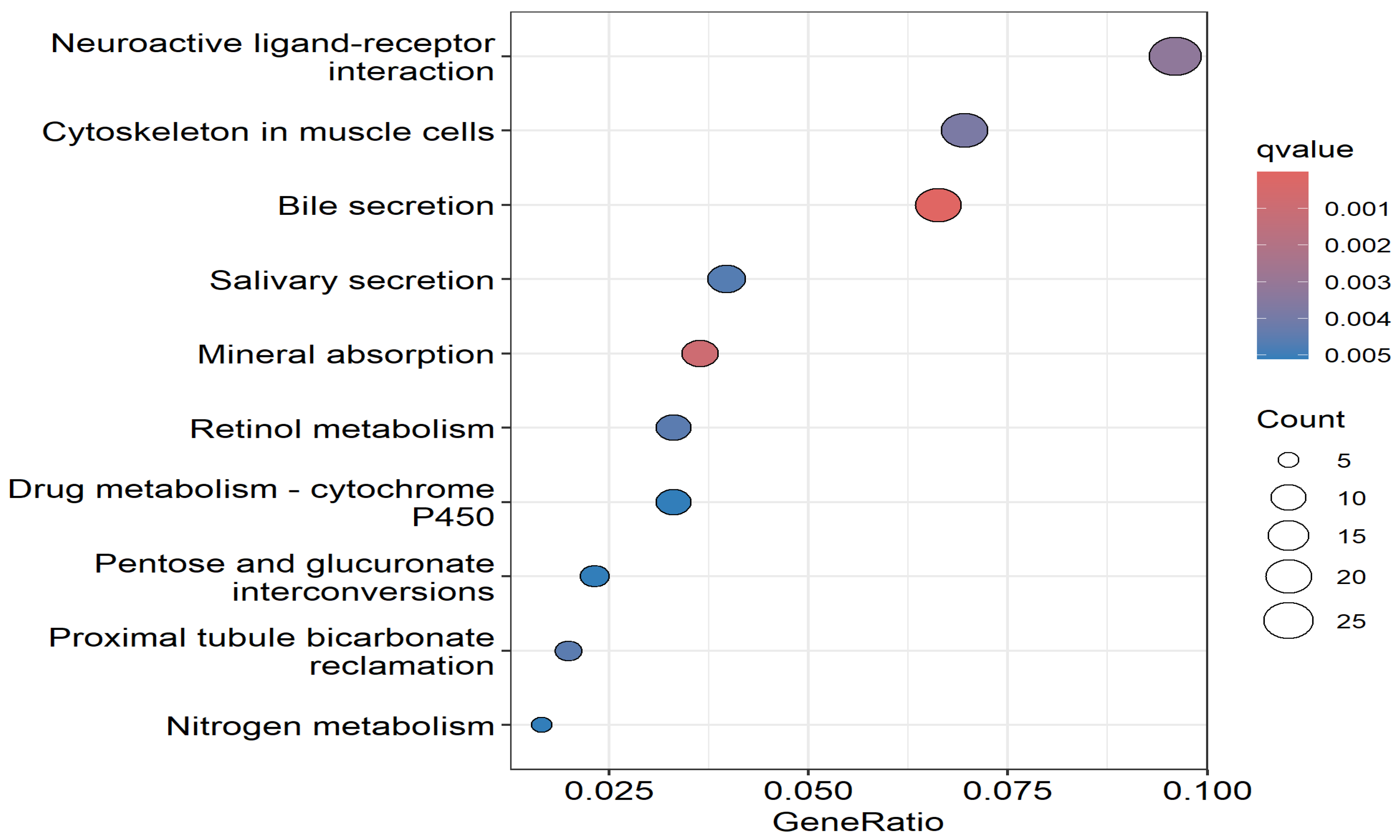
3.3. Discovery of Molecular Drivers of CRC as Potential Therapeutic Targets
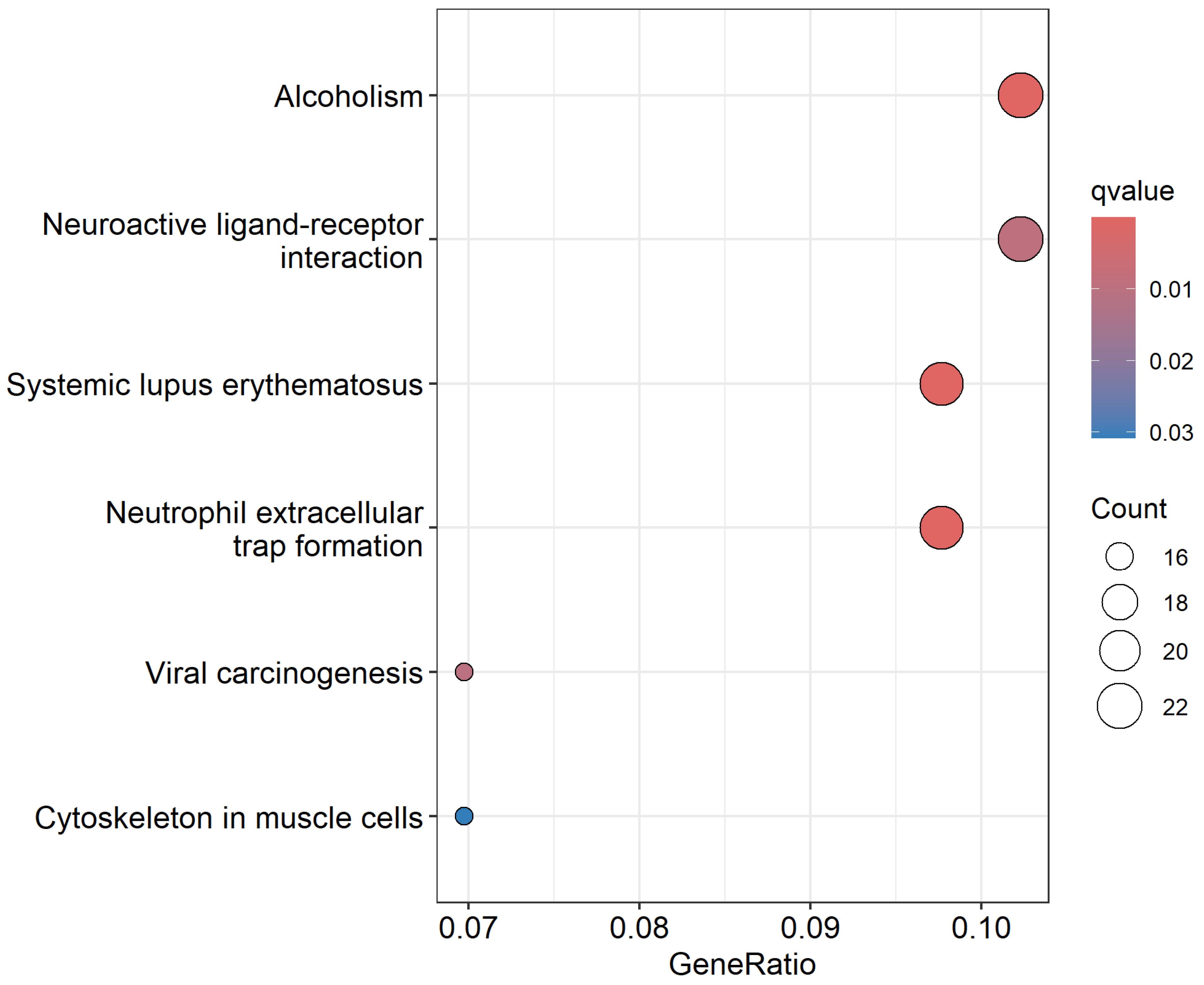
3.4. Discovery of a Signature of Differentially Expressed Somatic Mutated Genes Distinguishing Dead from Alive
3.5. Discovery of Potential Drivers of CRC and Potential Therapeutic Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| AACR | American Association of Cancer Research (AACR) |
| GLOBOCAN | The Global Cancer Observatory |
| TCGA | Cancer Genome Atlas |
| ICGC | International Cancer Genome Consortium |
| RNA-Seq | RNA sequencing |
| GDC | Genomics Data Commons |
| SNP | Single-nucleotide polymorphisms |
| INS | Insert |
| DEL | Deletion |
| Indel | Insert and deletion |
| MAF | Mutation Annotation Format |
| N | Number of samples |
| VST | variance stabilizing transformation |
| GLM | Generalized linear model |
| DEGs | Differentially expressed genes |
| Log2FC | Log base 2 fold changes |
| FDR | False discover rate |
| GO | Gene Ontology |
| IPA | Ingenuity pathway analysis |
| REF | Reference cited |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular function |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F.; et al. Global Burden of Colorectal Cancer in 2020 and 2040: Incidence and Mortality Estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, D.J.; John, A.; Lim, Y.C.; Kibria, K.M.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef]
- Dang, Q.; Chen, Y.; Bai, X.; Liu, H.; Zhang, J.; Fan, Y.; Luo, A.; Sun, K.; Li, X. Molecular Subtypes of Colorectal Cancer in the Era of Precision Oncotherapy: Current Inspirations and Future Challenges. Cancer Med. 2024, 13, e70041. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Lathrop, M.; Gut, I.; Heath, S.; Tost, J.; Gress, T.; Hudson, T. International Network of Cancer Genome Projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef]
- Ullah, I.; Yang, L.; Yin, F.T.; Sun, Y.; Li, X.H.; Li, J.; Wang, X.J. Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives. Cancers 2022, 14, 5545. [Google Scholar] [CrossRef]
- Lee, J.H.; Ahn, B.K.; Baik, S.S.; Lee, K.H. Comprehensive Analysis of Somatic Mutations in Colorectal Cancer with Peritoneal Metastasis. Vivo 2019, 33, 447–452. [Google Scholar] [CrossRef]
- Hassan, S.; Khatoon, A.; Bukhari, U.; Mirza, T. Analysis of Common Somatic Mutations in Colorectal Carcinoma and Associated Dysregulated Pathways. J. Ayub Med. Coll. Abbottabad 2023, 35, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Osakabe, M.; Uesugi, N.; Yanagawa, N.; Matsumoto, T.; Suzuki, H.; Sugai, T. Genome-Wide Analysis of Colorectal Cancer Based on Gene-Based Somatic Copy Number Alterations during Neoplastic Progression within the Same Tumor. Cancer Med. 2023, 12, 4446–4454. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.; Diosdado, B.; Terhaar Sive Droste, J.S.; Bolijn, A.S.; Komor, M.A.; de Wit, M.; Bosch, L.J.W.; van Burink, M.; Dekker, E.; Kuipers, E.J.; et al. Evaluation of Cancer-Associated DNA Copy Number Events in Colorectal (Advanced) Adenomas. Cancer Prev. Res. 2018, 11, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- National Cancer Institute. Genomic Data Commons (GDC) Data Portal. 2024. Available online: https://gdc.cancer.gov/ (accessed on 24 June 2023).
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Mamidi, T.K.K.; Wu, J.; Hicks, C. Elucidation of the Genomic-Epigenomic Interaction Landscape of Aggressive Prostate Cancer. BioMed Res. Int. 2021, 2021, 6641429. [Google Scholar] [CrossRef]
- Smedley, D.; Haider, S.; Durinck, S.; Pandini, L.; Provero, P.; Allen, J.; Arnaiz, O.; Awedh, M.H.; Baldock, R.; Barbiera, G.; et al. The BioMart Community Portal: An Innovative Alternative to Large, Centralized Data Repositories. Nucleic Acids Res. 2015, 43, W589–W598. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wald, A. Tests of Statistical Hypotheses Concerning Several Parameters When the Number of Observations is Large. Trans. Am. Math. Soc. 1943, 54, 426–482. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Raivo Kolde Pheatmap: Pretty Heatmaps 2010, 1.0.12. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 12 September 2024).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- QIAGEN. Ingenuity Pathways Analysis (IPA) System. 2024. Available online: http://www.ingenuity.com/ (accessed on 20 November 2024).
- Kumara, H.M.C.S.; Bellini, G.A.; Caballero, O.L.; Herath, S.A.C.; Su, T.; Ahmed, A.; Njoh, L.; Cekic, V.; Whelan, R.L. P-Cadherin (CDH3) is overexpressed in colorectal tumors and has potential as a serum marker for colorectal cancer monitoring. Oncoscience 2017, 4, 139–147. [Google Scholar] [CrossRef]
- Lin, J.; Fan, X.; Chen, J.; Xie, X.; Yu, H. Small Interfering RNA-Mediated Knockdown of KRT80 Suppresses Colorectal Cancer Proliferation. Exp. Ther. Med. 2020, 20, 176. [Google Scholar] [CrossRef]
- Fonseca, A.S.; Ramão, A.; Bürger, M.C.; De Souza, J.E.S.; Zanette, D.L.; De Molfetta, G.A.; De Araújo, L.F.; De Barros E Lima Bueno, R.; Aguiar, G.M.; Plaça, J.R.; et al. ETV4 plays a role on the primary events during the adenoma-adenocarcinoma progression in colorectal cancer. BMC Cancer 2021, 21, 207. [Google Scholar] [CrossRef]
- Yang, L.; Dong, Z.; Li, S.; Chen, T. ESM1 promotes angiogenesis in colorectal cancer by activating PI3K/Akt/mTOR pathway, thus accelerating tumor progression. Aging 2023, 15, 2920–2936. [Google Scholar] [CrossRef]
- Kaneda, H.; Arao, T.; Tanaka, K.; Tamura, D.; Aomatsu, K.; Kudo, K.; Sakai, K.; De Velasco, M.A.; Matsumoto, K.; Fujita, Y.; et al. FOXQ1 Is Overexpressed in Colorectal Cancer and Enhances Tumorigenicity and Tumor Growth. Cancer Res. 2010, 70, 2053–2063. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Jun, S.; Lee, S.-H.; Sharma, A.; Park, J.-I. Wnt2 Complements Wnt/Beta-Catenin Signaling in Colorectal Cancer. Oncotarget 2015, 6, 37257–37268. [Google Scholar] [CrossRef]
- Nakagawa, S.; Miyoshi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Sekimoto, M.; Doki, Y.; Mori, M. Expression of CLDN1 in Colorectal Cancer: A Novel Marker for Prognosis. Int. J. Oncol. 2011, 39, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zou, X.; Xu, Y.; Zhou, F.; Kuai, R.; Li, J.; Yang, D.; Chu, Y.; Peng, H. AJUBA Transactivates N-Cadherin Expression in Colorectal Cancer Cells through Interaction with Twist. J. Cell. Mol. Med. 2021, 25, 8006–8014. [Google Scholar] [CrossRef] [PubMed]
- Saliba, J.; Coutaud, B.; Makhani, K.; Epstein Roth, N.; Jackson, J.; Park, J.-Y.; Gagnon, N.; Costa, P.; Jeyakumar, T.; Bury, M.; et al. Loss of NFE2L3 Protects against Inflammation-Induced Colorectal Cancer through Modulation of the Tumor Microenvironment. Oncogene 2022, 41, 1563–1575. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Ye, W.; Zhang, Y.; Yang, X.; Liu, F.; Wang, J.; Ding, X.; Yang, Y.; Zhang, R.; Zhao, Y.; et al. Oncogenic Potential of BEST4 in Colorectal Cancer via Activation of PI3K/Akt Signaling. Oncogene 2022, 41, 1166–1177. [Google Scholar] [CrossRef]
- Kong, H.J.; Kang, D.H.; Ahn, T.S.; Kim, K.S.; Kim, T.W.; Lee, S.H.; Lee, D.W.; Ryu, J.S.; Beak, M.J. The Role of CPNE7 (Copine-7) in Colorectal Cancer Prognosis and Metastasis. Int. J. Mol. Sci. 2023, 24, 16704. [Google Scholar] [CrossRef]
- Okano, M.; Yamamoto, H.; Ohkuma, H.; Kano, Y.; Kim, H.; Nishikawa, S.; Konno, M.; Kawamoto, K.; Haraguchi, N.; Takemasa, I.; et al. Significance of INHBA expression in human colorectal cancer. Oncol. Rep. 2013, 30, 2903–2908. [Google Scholar] [CrossRef]
- Agarwal, S.; Behring, M.; Hale, K.; Al Diffalha, S.; Wang, K.; Manne, U.; Varambally, S. MTHFD1L, A Folate Cycle Enzyme, Is Involved in Progression of Colorectal Cancer. Transl. Oncol. 2019, 12, 1461–1467. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, L.; Zhou, F. Clinical Significance of MMP7 Levels in Colorectal Cancer Patients Receiving FOLFOX4 Chemotherapy Treatment. Int. J. Gen. Med. 2023, 16, 2671–2678. [Google Scholar] [CrossRef]
- Yan, B.; Cao, L.; Gao, L.; Wei, S.; Wang, M.; Tian, Y.; Yang, J.; Chen, E. PEX26 Functions as a Metastasis Suppressor in Colorectal Cancer. Dig. Dis. Sci. 2024, 69, 112–122. [Google Scholar] [CrossRef]
- Christodoulou, S.; Alexopoulou, D.K.; Kontos, C.K.; Scorilas, A.; Papadopoulos, I.N. Kallikrein-Related Peptidase-6 (KLK6) mRNA Expression Is an Independent Prognostic Tissue Biomarker of Poor Disease-Free and Overall Survival in Colorectal Adenocarcinoma. Tumor Biol. 2014, 35, 4673–4685. [Google Scholar] [CrossRef]
- Shang, S.; Yang, Y.-W.; Chen, F.; Yu, L.; Shen, S.-H.; Li, K.; Cui, B.; Lv, X.-X.; Zhang, C.; Yang, C.; et al. TRIB3 Reduces CD8+ T Cell Infiltration and Induces Immune Evasion by Repressing the STAT1-CXCL10 Axis in Colorectal Cancer. Sci. Transl. Med. 2022, 14, eabf0992. [Google Scholar] [CrossRef] [PubMed]
- Birkenkamp-Demtröder, K.; Hahn, S.A.; Mansilla, F.; Thorsen, K.; Maghnouj, A.; Christensen, R.; Øster, B.; Ørntoft, T.F. Keratin23 (KRT23) Knockdown Decreases Proliferation and Affects the DNA Damage Response of Colon Cancer Cells. PLoS ONE 2013, 8, e73593. [Google Scholar] [CrossRef] [PubMed]
- Domanegg, K.; Sleeman, J.P.; Schmaus, A. CEMIP, a Promising Biomarker That Promotes the Progression and Metastasis of Colorectal and Other Types of Cancer. Cancers 2022, 14, 5093. [Google Scholar] [CrossRef]
- Agarwal, S.; Behring, M.; Kim, H.G.; Chandrashekar, D.S.; Chakravarthi, B.V.S.K.; Gupta, N.; Bajpai, P.; Elkholy, A.; Al Diffalha, S.; Datta, P.K.; et al. TRIP13 Promotes Metastasis of Colorectal Cancer Regardless of P53 and Microsatellite Instability Status. Mol. Oncol. 2020, 14, 3007–3029. [Google Scholar] [CrossRef]
- Wu, S.; Xu, X.; Sun, C.; Wen, F.; He, S.; Gao, X.; Liu, Y.; Liu, L. Expression of PHLPP2 Correlates with Clinicopathologic Characteristics and Prognosis in Colorectal Cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 2909–2919. [Google Scholar]
- Chen, X.; Yi, G.; Zhou, Y.; Hu, W.; Xi, L.; Han, W.; Wang, F. Prognostic Biomarker SLCO4A1 Is Correlated with Tumor Immune Infiltration in Colon Adenocarcinoma. Mediat. Inflamm. 2023, 2023, 4926474. [Google Scholar] [CrossRef]
- Ma, D.; Liu, S.; Liu, K.; Kong, L.; Xiao, L.; Xin, Q.; Jiang, C.; Wu, J. MDFI Promotes the Proliferation and Tolerance to Chemotherapy of Colorectal Cancer Cells by Binding ITGB4/LAMB3 to Activate the AKT Signaling Pathway. Cancer Biol. Ther. 2024, 25, 2314324. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Kim, D.; Kim, J.; Lee, H.; Ghim, J.; Kang, B.-J.; Song, P.; Suh, P.-G.; Ryu, S.H.; Lee, T.G. NOTUM Is Involved in the Progression of Colorectal Cancer. Cancer Genom. Proteom. 2018, 15, 485–497. [Google Scholar] [CrossRef]
- Cui, Y.; Yang, J.; Bai, Y.; Li, Q.; Yao, Y.; Liu, C.; Wu, F.; Zhang, J.; Zhang, Y. ENC1 Facilitates Colorectal Carcinoma Tumorigenesis and Metastasis via JAK2/STAT5/AKT Axis-Mediated Epithelial Mesenchymal Transition and Stemness. Front. Cell Dev. Biol. 2021, 9, 616887. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Q.; Wu, H.; Guo, J.; Wu, H. VWA2 Protein Molecular Mechanism Predicts Colorectal Cancer: Promoting Cell Invasion and Migration by Inhibiting NK Cell Activation. Int. J. Biol. Macromol. 2024, 279 Pt 3, 135394. [Google Scholar] [CrossRef]
- Dietinger, V.; García de Durango, C.R.; Wiechmann, S.; Boos, S.L.; Michl, M.; Neumann, J.; Hermeking, H.; Kuster, B.; Jung, P. Wnt-Driven LARGE2 Mediates Laminin-Adhesive O-Glycosylation in Human Colonic Epithelial Cells and Colorectal Cancer. Cell Commun. Signal. 2020, 18, 102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Zhu, X.; Wu, H.; Wang, J.; Zhang, M.; Xiang, J.; Xia, S.; Shi, T.; Xi, Q. B7-H3 Promotes the Migration and Invasion of Colorectal Cancer Cells via Regulating the Actin Cytoskeleton and RhoA/ROCK1/LIMK1 Signaling Pathway. Tissue Cell 2024, 90, 102518. [Google Scholar] [CrossRef] [PubMed]
- Caliceti, C.; Punzo, A.; Silla, A.; Simoni, P.; Roda, G.; Hrelia, S. New Insights into Bile Acids Related Signaling Pathways in the Onset of Colorectal Cancer. Nutrients 2022, 14, 2964. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, F.; Jiang, J.; Zhang, H.; Zhou, M. Screening Key Genes and Signaling Pathways in Colorectal Cancer by Integrated Bioinformatics Analysis. Mol. Med. Rep. 2019, 20, 1259–1269. [Google Scholar] [CrossRef]
- Zuo, L.; Yang, X.; Lu, M.; Hu, R.; Zhu, H.; Zhang, S.; Zhou, Q.; Chen, F.; Gui, S.; Wang, Y. All-Trans Retinoic Acid Inhibits Human Colorectal Cancer Cells RKO Migration via Downregulating Myosin Light Chain Kinase Expression through MAPK Signaling Pathway. Nutr. Cancer 2016, 68, 1225–1233. [Google Scholar] [CrossRef]
- Cao, L.; Ba, Y.; Chen, F.; Li, D.; Zhang, S.; Zhang, H. The prognostic significance of epoxide hydrolases in colorectal cancer. Biochem. Biophys. Rep. 2025, 41, 101912. [Google Scholar] [CrossRef]
- Drews, R.M.; Hernando, B.; Tarabichi, M.; Haase, K.; Lesluyes, T.; Smith, P.S.; Morrill Gavarró, L.; Couturier, D.L.; Liu, L.; Schneider, M.; et al. A pan-cancer compendium of chromosomal instability. Nature 2022, 606, 976–983. [Google Scholar] [CrossRef]
- Zhao, L.; Fan, W.; Luo, K.; Xie, S.; Wang, R.; Guan, J.; Chen, Z.; Jin, S. Construction of a TTN Mutation-Based Prognostic Model for Evaluating Immune Microenvironment, Cancer Stemness, and Outcomes of Colorectal Cancer Patients. Stem Cells Int. 2023, 2023, 6079957. [Google Scholar] [CrossRef]
- Kim, S.T.; Sohn, I.; DO, I.-G.; Jang, J.; Kim, S.H.; Jung, I.H.; Park, J.O.; Park, Y.S.; Talasaz, A.; Lee, J.; et al. Transcriptome Analysis of CD133-Positive Stem Cells and Prognostic Value of CCDC144B Survivin in Colorectal Cancer. Cancer Genom. Proteom. 2014, 11, 259–266. [Google Scholar]
- Ha, Y.M.; Choi, E.J.; Yoo, N.J.; Lee, S.H. Mutational Alterations of TDRD1, 4 and 9 Genes in Colorectal Cancers. Pathol. Oncol. Res. 2020, 26, 2007–2008. [Google Scholar] [CrossRef]
- Huang, L.; Zou, T.; Liang, W.; Mo, C.; Wei, J.; Deng, Y.; Ou, M. High-Throughput Sequencing Reveals That Rotundine Inhibits Colorectal Cancer by Regulating Prognosis-Related Genes. J. Pers. Med. 2023, 13, 550. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Hu, W.; Rao, Z.; Fang, T. The Molecular Basis of the Associations Between Non-Alcoholic Fatty Liver Disease and Colorectal Cancer. Front. Genet. 2022, 13, 1007337. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, T.; Liu, J.; Wang, Y.; Zhang, C.; Guo, L.; Shi, D.; Zhang, T.; Wang, X.; Li, J. FGF19-Induced Inflammatory CAF Promoted Neutrophil Extracellular Trap Formation in the Liver Metastasis of Colorectal Cancer. Adv. Sci. 2023, 10, e2302613. [Google Scholar] [CrossRef] [PubMed]
- Ivancevic, A.; Simpson, D.M.; Joyner, O.M.; Bagby, S.M.; Nguyen, L.L.; Bitler, B.G.; Pitts, T.M.; Chuong, E.B. Endogenous Retroviruses Mediate Transcriptional Rewiring in Response to Oncogenic Signaling in Colorectal Cancer. Sci. Adv. 2024, 10, eado1218. [Google Scholar] [CrossRef]
- Anupriya, S.; Parida, N.; Patnaik, S. SOX4 Induces Cytoskeleton Remodeling and Promotes Cell Motility via N-Wasp/ARP2/3 Pathway in Colorectal Cancer Cells. Exp. Cell Res. 2024, 439, 114059. [Google Scholar] [CrossRef]
- Wei, H.; Ren, K.; Zhang, Q.; Jin, Y.; Cao, B.; Tian, Z.; Mao, T.; Ren, L. Titin as a potential novel therapeutic target in colorectal cancer. J. Cell. Mol. Med. 2023, 27, 2937–2944. [Google Scholar] [CrossRef]
- Leng, Y.; Chen, Z.; Ding, H.; Zhao, X.; Qin, L.; Pan, Y. Overexpression of microRNA-29b inhibits epithelial-mesenchymal transition and angiogenesis of colorectal cancer through the ETV4/ERK/EGFR axis. Cancer Cell Int. 2021, 21, 17. [Google Scholar] [CrossRef]
- Kim, H.; Kim, Y.-M. Pan-cancer analysis of somatic mutations and transcriptomes reveals common functional gene clusters shared by multiple cancer types. Sci. Rep. 2018, 8, 6041. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Zardavas, D.; Phillips, W.A.; Loi, S. PIK3CA mutations in breast cancer: Reconciling findings from preclinical and clinical data. Breast Cancer Res. 2014, 16, 201. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Pellatt, A.J.; Mullany, L.E.; Wolff, R.K.; Herrick, J.S. Gene Expression in Colon Cancer: A Focus on Tumor Site and Molecular Phenotype. Genes Chromosomes Cancer 2015, 54, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Zhang, X.; Ye, X.; Zhou, Z.; Wu, X.; Ni, S.; Song, H. Bioinformatic Analysis of RNA-Seq Data Unveiled Critical Genes in Rectal Adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3017–3025. [Google Scholar]
- Elsayed, A.M.A.; Oweda, M.; Abushady, A.M.; Alhelf, M.; Khalil, S.R.M.; Tawfik, M.S.; Al-Atabany, W.; El-Hadidi, M. Identification of Differentially Expressed Genes in Human Colorectal Cancer Using RNASeq Data Validated on the Molecular Level with Real-Time PCR. Biochem. Genet. 2024, 62, 3260–3284. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, W.; Liu, Z.-Q.; Zhou, H.-H. Associating Transcriptional Modules with Colon Cancer Survival through Weighted Gene Co-Expression Network Analysis. BMC Genom. 2017, 18, 361. [Google Scholar] [CrossRef]
- Li, H.; Peng, J.; Wang, Y.; Jiang, H.; Wang, J. A Novel Nomogram Associated with Regulatory T Cells Infiltration by Weighted Gene Co-Expression Network Analysis for Predicting Survival in Patients with Colon Cancer. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 8073–8086. [Google Scholar]
- Almuzzaini, B.; Alghamdi, J.; Alomani, A.; AlGhamdi, S.; Alsharm, A.A.; Alshieban, S.; Sayed, A.; Alhejaily, A.G.; Aljaser, F.S.; Abudawood, M.; et al. Identification of Novel Mutations in Colorectal Cancer Patients Using AmpliSeq Comprehensive Cancer Panel. J. Pers. Med. 2021, 11, 535. [Google Scholar] [CrossRef]
- Chen, J.; Apizi, A.; Wang, L.; Wu, G.; Zhu, Z.; Yao, H.; Chen, G.; Shi, X.; Shi, B.; Tai, Q.; et al. TCGA Database Analysis of the Tumor Mutation Burden and Its Clinical Significance in Colon Cancer. J. Gastrointest. Oncol. 2021, 12, 2244–2259. [Google Scholar] [CrossRef]
- Kamran, S.C.; Lennerz, J.K.; Margolis, C.A.; Liu, D.; Reardon, B.; Wankowicz, S.A.; Van Seventer, E.E.; Tracy, A.; Wo, J.Y.; Carter, S.L.; et al. Integrative Molecular Characterization of Resistance to Neoadjuvant Chemoradiation in Rectal Cancer. Clin. Cancer Res. 2019, 25, 5561–5571. [Google Scholar] [CrossRef]
- Hankey, W.; Frankel, W.L.; Groden, J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: Implications for therapeutic targeting. Cancer Metastasis Rev. 2018, 37, 159–172. [Google Scholar] [CrossRef]
- Saadh, M.J.; Allela, O.Q.B.; Kareem, R.A.; Baldaniya, L.; Ballal, S.; Vashishth, R.; Parmar, M.; Sameer, H.N.; Hamad, A.K.; Athab, Z.H.; et al. Prognostic Gene Expression Profile of Colorectal Cancer. Gene 2025, 955, 149433. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz, N.A.; Mokhtar, N.M.; Harun, R.; Mollah, M.M.; Rose, I.M.; Sagap, I.; Tamil, A.M.; Ngah, W.Z.; Jamal, R. A 19-Gene Expression Signature as a Predictor of Survival in Colorectal Cancer. BMC Med. Genom. 2016, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Wei, I.H.; Shi, Y.; Jiang, H.; Kumar-Sinha, C.; Chinnaiyan, A.M. RNA-Seq accurately identifies cancer biomarker signatures to distinguish tissue of origin. Neoplasia 2014, 16, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.; Theisen, C.S.; Johnson, K.R.; Wheelock, M.J. R-cadherin Influences Cell Motility via Rho Family GTPases. J. Biol. Chem. 2004, 279, 31041–31049. [Google Scholar] [CrossRef]
- Gonzalez-Pons, M.; Cruz-Correa, M. Colorectal Cancer Biomarkers: Where Are We Now? BioMed Res. Int. 2015, 2015, 149014. [Google Scholar] [CrossRef]
- Kalmár, A.; Wichmann, B.; Galamb, O.; Spisák, S.; Tóth, K.; Leiszter, K.; Nielsen, B.S.; Barták, B.K.; Tulassay, Z.; Molnár, B. Gene-Expression Analysis of a Colorectal Cancer-Specific Discriminatory Transcript Set on Formalin-Fixed, Paraffin-Embedded (FFPE) Tissue Samples. Diagn. Pathol. 2015, 10, 126. [Google Scholar] [CrossRef]
- Khaneh, A.; Piri, H.; Alaei, M.; Parvani, N.; Vakilzadeh, I.; Javadi, S.; Moradian Haft Cheshmeh, Z.; Razzaghi, Z.; Robati, R.M.; Zamanian Azodi, M.; et al. Classification and Diagnostic Prediction of Colorectal Cancer Mortality Based on Machine Learning Algorithms: A Multicenter National Study. Asian Pac. J. Cancer Prev. 2024, 25, 333–342. [Google Scholar][Green Version]
- Maurya, N.S.; Kushwaha, S.; Vetukuri, R.R.; Mani, A. Unlocking the Potential of the CA2, CA7, and ITM2C Gene Signatures for the Early Detection of Colorectal Cancer: A Comprehensive Analysis of RNA-Seq Data by Utilizing Machine Learning Algorithms. Genes 2023, 14, 1836. [Google Scholar] [CrossRef]
- Li, J.; Wang, P.; Zhou, Y.; Liang, H.; Luan, K. Different Machine Learning and Deep Learning Methods for the Classification of Colorectal Cancer Lymph Node Metastasis Images. Front. Bioeng. Biotechnol. 2021, 8, 620257. [Google Scholar] [CrossRef]
- Kassab, R.; Khalil, M.A.; Kassab, J.; Kourie, H.R. Immune Checkpoint Inhibitors in BRAF-Mutated Advanced Colorectal Cancer. Future Oncol. 2023, 19, 2417–2424. [Google Scholar] [CrossRef]
- Ghidini, M.; Fusco, N.; Salati, M.; Khakoo, S.; Tomasello, G.; Petrelli, F.; Trapani, D.; Petrillo, A. The Emergence of Immune-Checkpoint Inhibitors in Colorectal Cancer Therapy. Curr. Drug Targets 2021, 22, 1021–1033. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Jhaveri, D.J.; Marshall, V.M.; Bauer, D.C.; Edson, J.; Narayanan, R.K.; Robinson, G.J.; Lundberg, A.E.; Bartlett, P.F.; Wray, N.R.; et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS ONE 2014, 9, e103207. [Google Scholar] [CrossRef]
| Gene Symbol | Chromosome Position | Log2FC | p-Value | Correlation | Total Mutations | Involvement in CRC [REF] |
|---|---|---|---|---|---|---|
| CDH3 | 16q22.1 | 5.66 | 1.00 × 10−300 | 0.95 | 16 | [28] |
| KRT80 | 12q13.13 | 6.56 | 1.00 × 10−300 | 0.93 | 9 | [29] |
| ETV4 | 17q21.31 | 5.14 | 3.57 × 10−257 | 0.93 | 18 | [30] |
| ESM1 | 5q11.2 | 5.71 | 3.57 × 10−257 | 0.92 | 10 | [31] |
| FOXQ1 | 6p25.3 | 6.32 | 6.37 × 10−238 | 0.90 | 11 | [32] |
| SIM2 | 21q22.13 | 7.40 | 4.62 × 10−225 | 0.88 | 15 | |
| WNT2 | 7q31.2 | 5.63 | 5.59 × 10−225 | 0.90 | 16 | [33] |
| CLDN1 | 3q28 | 4.72 | 1.54 × 10−222 | 0.92 | 2 | [34] |
| AJUBA | 14q11.2 | 2.96 | 5.42 × 10−199 | 0.92 | 9 | [35] |
| NFE2L3 | 7p15.2 | 2.73 | 2.50 × 10−175 | 0.91 | 26 | [36] |
| BEST4 | 1p34.1 | −5.84 | 1.72 × 10−168 | −0.94 | 12 | [37] |
| CPNE7 | 16q24.3 | 5.73 | 5.78 × 10−166 | 0.87 | 20 | [38] |
| INHBA | 7p14.1 | 5.43 | 6.09 × 10−160 | 0.89 | 46 | [39] |
| CST1 | 20p11.21 | 8.2 | 9.70 × 10−157 | 0.82 | 19 | |
| EPHX4 | 1p22.1 | 4.42 | 1.30 × 10−156 | 0.87 | 18 | |
| APPL2 | 12q23.3 | −1.86 | 8.28 × 10−155 | −0.86 | 18 | |
| MTHFD1L | 6q25.1 | 2.15 | 1.23 × 10−154 | 0.90 | 37 | [40] |
| MMP7 | 11q22.2 | 7.09 | 1.95 × 10−147 | 0.85 | 15 | [41] |
| PEX26 | 22q11.21 | −1.74 | 2.86 × 10−147 | −0.80 | 5 | [42] |
| C6orf223 | 6p21. | 14.50 | 7.15 × 10−147 | 0.89 | 5 | |
| SLC51A | 3q29 | −3.90 | 1.16 × 10−141 | −0.74 | 12 | |
| GRIN2D | 19q13.33 | 4.81 | 1.04 × 10−138 | 0.84 | 38 | |
| KLK6 | 19q13.41 | 9.70 | 5.94 × 10−136 | 0.77 | 16 | [43] |
| LRP8 | 1p32.3 | 2.89 | 7.07 × 10−134 | 0.89 | 23 | |
| TRIB3 | 20p13 | 3.75 | 5.24 × 10−132 | 0.87 | 13 | [44] |
| GLTP | 12q24.11 | −1.63 | 1.06 × 10−130 | −0.87 | 4 | |
| KRT23 | 17q21.2 | 7.32 | 2.84 × 10−128 | 0.76 | 15 | [45] |
| COL11A1 | 1p21.1 | 5.99 | 1.52 × 10−127 | 0.84 | 106 | |
| CEMIP | 15q25.1 | 4.12 | 2.22 × 10−127 | 0.88 | 41 | [46] |
| TRIP13 | 5p15.33 | 2.15 | 1.35 × 10−121 | 0.88 | 7 | [47] |
| PHLPP2 | 16q22.2 | −2.58 | 2.89 × 10−121 | −0.85 | 30 | [48] |
| SLC25A34 | 1p36.21 | −3.79 | 5.15 × 10−120 | −0.88 | 11 | |
| SLCO4A1 | 20q13.33 | 3.41 | 6.26 × 10−120 | 0.87 | 23 | [49] |
| MDFI | 6p21.1 | 3.58 | 6.26 × 10−120 | 0.86 | 4 | [50] |
| NOTUM | 17q25.3 | 8.67 | 1.72 × 10−119 | 0.73 | 18 | [51] |
| ENC1 | 5q13.3 | 2.01 | 3.55 × 10−119 | 0.88 | 28 | [52] |
| VWA2 | 10q25.3 | 3.94 | 1.25 × 10−118 | 0.84 | 26 | [53] |
| SLC22A5 | 5q31.1 | −1.93 | 2.19 × 10−117 | −0.82 | 8 | |
| LARGE2 | 11p11.2 | 4.37 | 3.04 × 10−117 | 0.84 | 17 | [54] |
| ETFDH | 4q32.1 | −1.84 | 7.55 × 10−116 | −0.90 | 13 |
| Gene | Chromosome Position | Adjusted p-Value | Log2FC | Correlation | Mutations in Alive (n = 495) | Mutations in Deceased (n = 127) [RF] |
|---|---|---|---|---|---|---|
| H3C2 | 6p22.2 | 2.19 × 10−49 | −4.68 | −0.20 | 6 | 3 |
| H2BC13 | 6p22.1 | 1.86 × 10−47 | −3.88 | −0.24 | 6 | 1 |
| H1-3 | 6p22.2 | 6.03 × 10−46 | −4.44 | −0.18 | 7 | 3 |
| H2BC17 | 6p22.1 | 7.36 × 10−41 | −3.99 | −0.21 | 1 | 0 |
| H2AC13 | 6p22.1 | 2.03 × 10−38 | −3.42 | −0.23 | 3 | 0 |
| H1-4 | 6p22.2 | 2.31 × 10−38 | −3.97 | −0.19 | 7 | 1 |
| ZBTB20 | 3q13.31 | 4.66 × 10−36 | −2.82 | −0.16 | 39 | 15 |
| ZNF460 | 19q13.43 | 6.90 × 10−34 | −2.58 | −0.20 | 11 | 6 |
| H4C5 | 6p22.2 | 2.43 × 10−30 | −2.98 | −0.16 | 2 | 2 |
| LRRTM2 | 5q31.2 | 2.92 × 10−30 | −2.82 | −0.17 | 14 | 1 |
| H1-5 | 6p22.1 | 4.73 × 10−30 | −3.57 | −0.19 | 15 | 0 |
| OMG | 17q11.2 | 3.82 × 10−28 | −2.80 | −0.16 | 4 | 0 |
| ANKRD36C | 2q11.1 | 2.70 × 10−24 | −2.17 | −0.14 | 3 | 0 |
| H2AC20 | 1q21.2 | 5.25 × 10−24 | −2.30 | −0.20 | 7 | 2 |
| H2BC7 | 6p22.2 | 4.08 × 10−22 | −2.53 | −0.15 | 2 | 4 |
| H2BC18 | 1q21.2 | 7.01 × 10−21 | −2.32 | −0.11 | 5 | 1 |
| GSN-AS1 | 9q33.2 | 3.24 × 10−20 | −2.09 | −0.16 | 5 | 2 |
| NBEAL1 | 2q33.2 | 3.76 × 10−20 | −1.59 | −0.15 | 29 | 9 |
| GPR82 | Xp11.4 | −1.19 × 10−19 | −1.73 | −0.23 | 5 | 0 |
| ANKRD36B | 2q11.2 | 1.18 × 10−18 | −1.65 | −0.16 | 10 | 1 |
| TTN | 2q31.2 | 1.90 × 10−18 | −1.86 | −0.12 | 360 | 95 [62] |
| GRIK1 | 21q21.3 | 4.26 × 10−18 | −1.99 | −0.16 | 35 | 4 |
| GPR18 | 13q32.3 | 6.94 × 10−18 | −1.82 | −0.16 | 6 | 2 |
| SYCP3 | 12q23.2 | 7.08 × 10−18 | −1.83 | −0.13 | 5 | 0 |
| CLDN20 | 6q25.3 | 1.10 × 10−17 | −1.97 | −0.15 | 3 | 1 |
| INSYN2A | 10q26.2 | 1.12 × 10−17 | −2.03 | −0.14 | 8 | 6 |
| CCDC144B | 17p11.2 | 3.37 × 10−17 | −2.57 | −0.13 | 4 | 1 [63] |
| GNAT2 | 1p13.3 | 4.38 × 10−17 | −1.42 | −0.25 | 10 | 3 |
| H2BC4 | 6p22.2 | 6.24 × 10−17 | −1.88 | −0.16 | 8 | 2 |
| GCNT7 | 20q13.31 | 1.53 × 10−16 | −1.88 | −0.14 | 3 | 1 |
| H2BC6 | 6p22.2 | 2.50 × 10−16 | −1.86 | −0.15 | 3 | 3 |
| KCNC1 | 10q25.3 | 3.75 × 10−16 | −1.91 | −0.11 | 19 | 8 |
| TDRD1 | 10q25.3 | 4.57 × 10−16 | −2.15 | −0.10 | 16 | 5 [64] |
| SPDYE1 | 7p13 | 4.61 × 10−16 | −1.74 | −0.12 | 7 | 2 |
| ANKRD36 | 2q11.2 | 2.29 × 10−15 | −1.49 | −0.13 | 4 | 2 [65] |
| LRRN3 | 7q31.1 | 2.66 × 10−15 | −1.81 | −0.15 | 32 | 7 |
| MAK | 6p24.2 | 2.94 × 10−15 | −1.57 | −0.13 | 15 | 6 |
| H2AC11 | 6p22.1 | 4.82 × 10−15 | −1.53 | −0.17 | 8 | 2 |
| SHOC1 | 9q31.3 | 9.09 × 10−15 | −2.30 | −0.14 | 20 | 9 |
| COL6A6 | 3q22.1 | 1.76 × 10−14 | −1.90 | −0.13 | 67 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fertel, M.; Alawad, D.M.; Hicks, C. An Integrative Genomics Approach for the Discovery of Potential Clinically Actionable Diagnostic and Prognostic Biomarkers in Colorectal Cancer. Biomedicines 2025, 13, 1651. https://doi.org/10.3390/biomedicines13071651
Fertel M, Alawad DM, Hicks C. An Integrative Genomics Approach for the Discovery of Potential Clinically Actionable Diagnostic and Prognostic Biomarkers in Colorectal Cancer. Biomedicines. 2025; 13(7):1651. https://doi.org/10.3390/biomedicines13071651
Chicago/Turabian StyleFertel, Mark, Duaa Mohammad Alawad, and Chindo Hicks. 2025. "An Integrative Genomics Approach for the Discovery of Potential Clinically Actionable Diagnostic and Prognostic Biomarkers in Colorectal Cancer" Biomedicines 13, no. 7: 1651. https://doi.org/10.3390/biomedicines13071651
APA StyleFertel, M., Alawad, D. M., & Hicks, C. (2025). An Integrative Genomics Approach for the Discovery of Potential Clinically Actionable Diagnostic and Prognostic Biomarkers in Colorectal Cancer. Biomedicines, 13(7), 1651. https://doi.org/10.3390/biomedicines13071651





