The Anti-Inflammatory, Immunomodulatory, and Pro-Autophagy Activities of Probiotics for Colorectal Cancer Prevention and Treatment: A Narrative Review
Abstract
1. Introduction
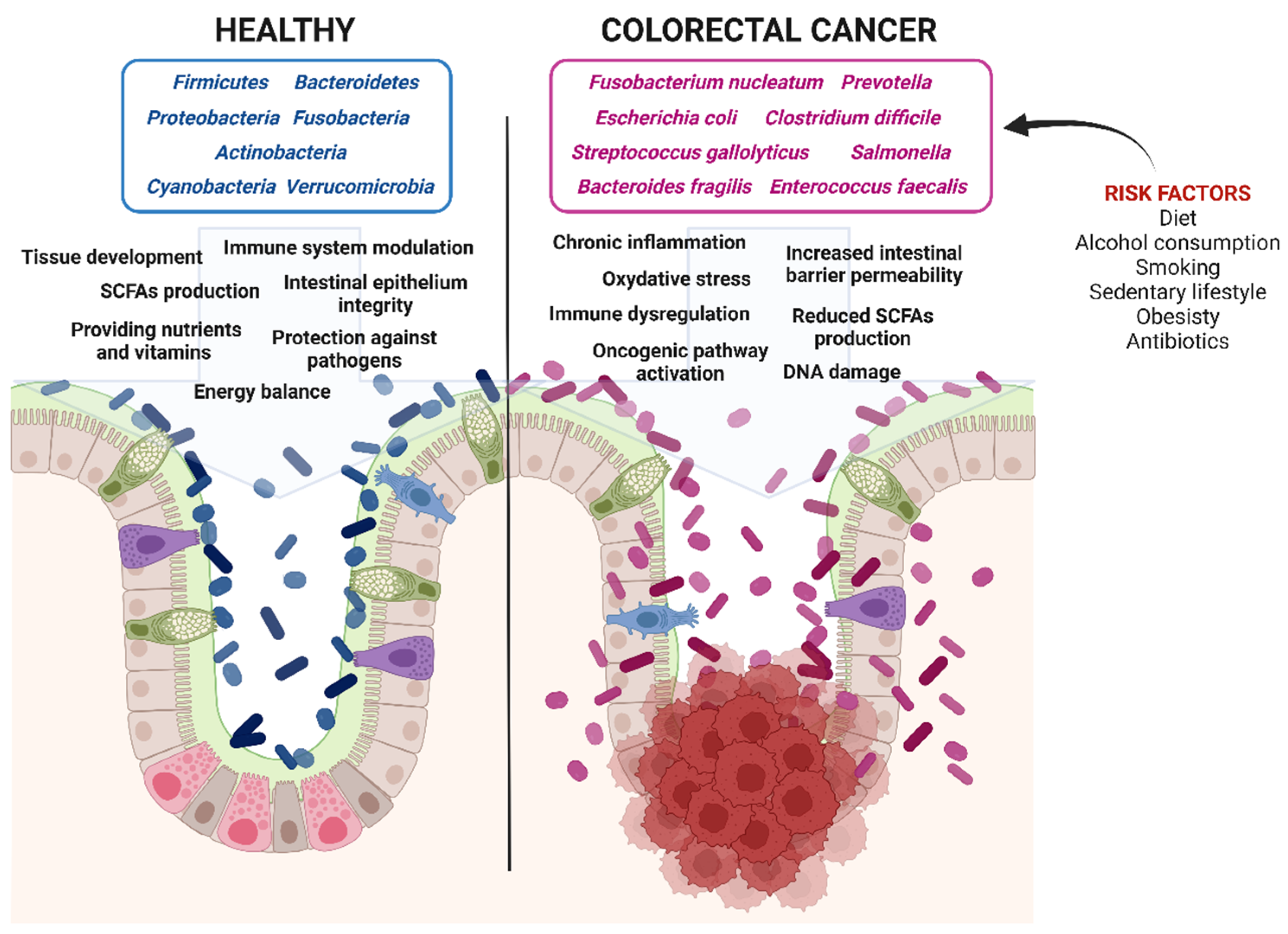
1.1. Colorectal Cancer: Epidemiology, Risk Factors, Prevention, and Therapy at a Glance
1.2. Microbiota and Colorectal Cancerogenesis: The Role of Genetics and Epigenetics
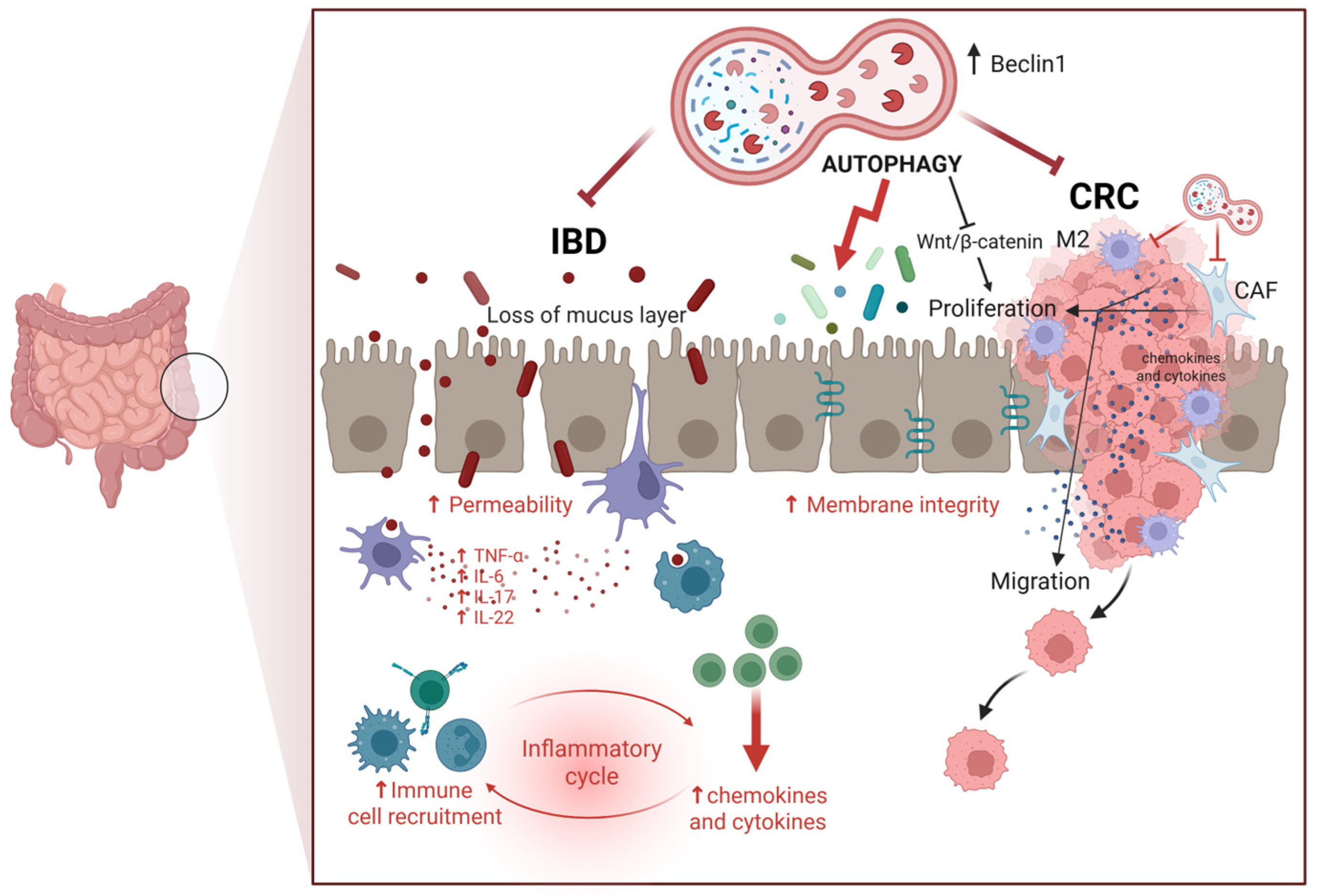
1.3. Colorectal Cancerogenesis: The Role of Autophagy
1.4. Colorectal Cancerogenesis: The Role of Inflammation and Its Interplay with Immune Cells
1.5. Dysbiosis and Colorectal Cancerogenesis: The Role of Diet and Microbiota
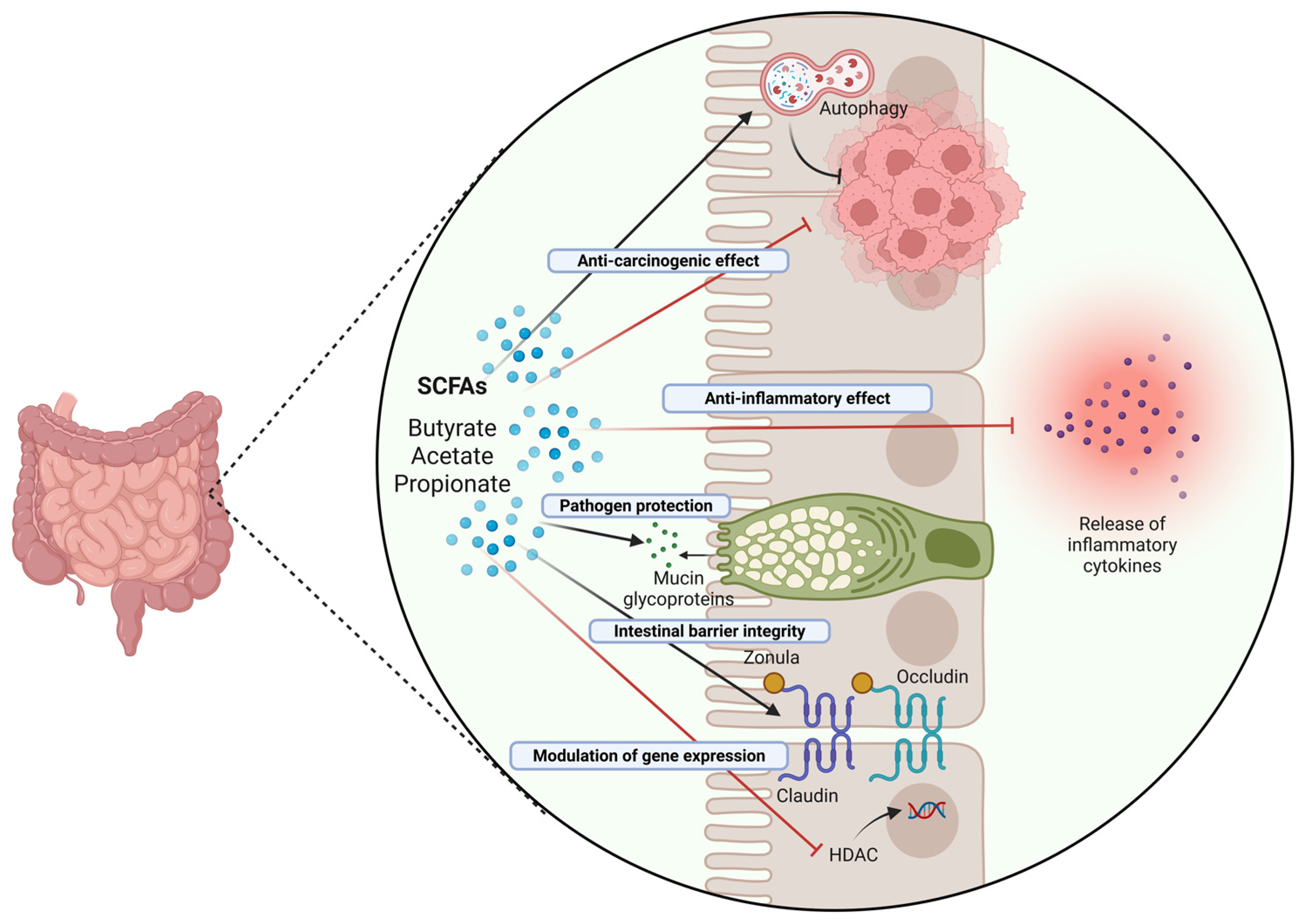
1.6. Beneficial Effects of Probiotics and Postbiotics in Colorectal Cancer: Impact on Inflammation and Autophagy
1.7. Challenges in Probiotic-Based Therapy and Future Directions
2. Highlights and Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Valle, L.; Vilar, E.; Tavtigian, S.V.; Stoffel, E.M. Genetic predisposition to colorectal cancer: Syndromes, genes, classification of genetic variants and implications for precision medicine. J. Pathol. 2019, 247, 574–588. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fantini, M.C.; Guadagni, I. From inflammation to colitis-associated colorectal cancer in inflammatory bowel disease: Pathogenesis and impact of current therapies. Dig. Liver Dis. 2021, 53, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, G.; Ghasemi-Kebria, F.; Malekzadeh, R. Colorectal Cancer: Epidemiology, Risk Factors, and Prevention. Cancers 2024, 16, 1530. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fadlallah, H.; El Masri, J.; Fakhereddine, H.; Youssef, J.; Chemaly, C.; Doughan, S.; Abou-Kheir, W. Colorectal cancer: Recent advances in management and treatment. World J. Clin. Oncol. 2024, 15, 1136–1156. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, A.; Gautam, V.; Sandhu, A.; Rawat, K.; Sharma, A.; Saha, L. Current and emerging therapeutic approaches for colorectal cancer: A comprehensive review. World J. Gastrointest. Surg. 2023, 15, 495–519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target Ther. 2020, 5, 22. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Tsuruya, A.; Kuwahara, A.; Saito, Y.; Yamaguchi, H.; Tsubo, T.; Suga, S.; Inai, M.; Aoki, Y.; Takahashi, S.; Tsutsumi, E.; et al. Ecophysiological consequences of alcoholism on human gut microbiota: Implications for ethanol-related pathogenesis of colon cancer. Sci. Rep. 2016, 6, 27923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Munteanu, C.; Schwartz, B. Interactions between Dietary Antioxidants, Dietary Fiber and the Gut Microbiome: Their Putative Role in Inflammation and Cancer. Int. J. Mol. Sci. 2024, 25, 8250. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vernia, F.; Longo, S.; Stefanelli, G.; Viscido, A.; Latella, G. Dietary Factors Modulating Colorectal Carcinogenesis. Nutrients 2021, 13, 143. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parizadeh, M.; Arrieta, M.C. The global human gut microbiome: Genes, lifestyles, and diet. Trends Mol. Med. 2023, 29, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Quaglio, A.E.V.; Grillo, T.G.; De Oliveira, E.C.S.; Di Stasi, L.C.; Sassaki, L.Y. Gut microbiota, inflammatory bowel disease and colorectal cancer. World J. Gastroenterol. 2022, 28, 4053–4060. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ionescu, V.A.; Gheorghe, G.; Georgescu, T.F.; Buica, V.; Catanescu, M.-S.; Cercel, I.-A.; Budeanu, B.; Budan, M.; Bacalbasa, N.; Diaconu, C. Exploring the Role of the Gut Microbiota in Colorectal Cancer Development. Gastrointest. Disord. 2024, 6, 526–537. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moreira, M.M.; Carriço, M.; Capelas, M.L.; Pimenta, N.; Santos, T.; Ganhão-Arranhado, S.; Mäkitie, A.; Ravasco, P. The impact of pre-, pro- and synbiotics supplementation in colorectal cancer treatment: A systematic review. Front. Oncol. 2024, 14, 1395966. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak-Siedlecka, K.; Marano, L.; Merola, E.; Roviello, F.; Połom, K. Sodium butyrate in both prevention and supportive treatment of colorectal cancer. Front. Cell Infect. Microbiol. 2022, 12, 1023806. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singh, V.; Lee, G.; Son, H.; Koh, H.; Kim, E.S.; Unno, T.; Shin, J.H. Butyrate producers, “The Sentinel of Gut”: Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 2023, 13, 1103836. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cao, Q.; Tian, Y.; Deng, Z.; Yang, F.; Chen, E. Epigenetic Alteration in Colorectal Cancer: Potential Diagnostic and Prognostic Implications. Int. J. Mol. Sci. 2024, 25, 3358. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jansen, M.; Menko, F.H.; Brosens, L.A.; Giardiello, F.M.; Offerhaus, G.J. Establishing a clinical and molecular diagnosis for hereditary colorectal cancer syndromes: Present tense, future perfect? Gastrointest. Endosc. 2014, 80, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jeong, S. Mutation Hotspots in the β-Catenin Gene: Lessons from the Human Cancer Genome Databases. Mol. Cells 2019, 42, 8–16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gan, W.J. Wnt/β-Catenin Signaling Pathway in the Development and Progression of Colorectal Cancer. Cancer Manag. Res. 2023, 15, 435–448. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt signaling in colorectal cancer: Pathogenic role and therapeutic target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ionescu, V.A.; Gheorghe, G.; Bacalbasa, N.; Chiotoroiu, A.L.; Diaconu, C. Colorectal Cancer: From Risk Factors to Oncogenesis. Medicina 2023, 59, 1646. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dziubańska-Kusibab, P.J.; Berger, H.; Battistini, F.; Bouwman, B.A.M.; Iftekhar, A.; Katainen, R.; Cajuso, T.; Crosetto, N.; Orozco, M.; Aaltonen, L.A.; et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 2020, 26, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; de’Angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, J.; Chen, S.; Zang, D.; Sun, H.; Sun, Y.; Chen, J. Butyrate as a promising therapeutic target in cancer: From pathogenesis to clinic (Review). Int. J. Oncol. 2024, 64, 44. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allen, J.; Hao, S.; Sears, C.L.; Timp, W. Epigenetic Changes Induced by Bacteroides fragilis Toxin. Infect. Immun. 2019, 87, e00447-18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xing, J.; Liao, Y.; Zhang, H.; Zhang, W.; Zhang, Z.; Zhang, J.; Wang, D.; Tang, D. Impacts of MicroRNAs Induced by the Gut Microbiome on Regulating the Development of Colorectal Cancer. Front. Cell Infect. Microbiol. 2022, 12, 804689. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ambalam, P.; Dave, J.M.; Nair, B.M.; Vyas, B.R. In vitro mutagen binding and antimutagenic activity of human Lactobacillus rhamnosus 231. Anaerobe 2011, 17, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Pithva, S.P.; Ambalam, P.S.; Ramoliya, J.M.; Dave, J.M.; Vyas, B.R. Antigenotoxic and Antimutagenic Activities of Probiotic Lactobacillus rhamnosus Vc against N-Methyl-N’-Nitro-N-Nitrosoguanidine. Nutr. Cancer 2015, 67, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Heydari, Z.; Rahaie, M.; Alizadeh, A.M.; Agah, S.; Khalighfard, S.; Bahmani, S. Effects of Lactobacillus acidophilus and Bifidobacterium bifidum Probiotics on the Expression of MicroRNAs 135b, 26b, 18a and 155, and Their Involving Genes in Mice Colon Cancer. Probiotics Antimicrob. Proteins 2019, 11, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, C.A.; Gamal-Eldeen, A.M.; El-Hussieny, E.A.; Raafat, B.M.; Mehanna, N.S.; Talaat, R.M.; Shaaban, M.T. Bifidobacterium longum Suppresses Murine Colorectal Cancer through the Modulation of oncomiRs and Tumor Suppressor miRNAs. Nutr. Cancer 2019, 71, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Mederle, A.L.; Semenescu, A.; Drăghici, G.A.; Dehelean, C.A.; Vlăduț, N.-V.; Nica, D.V. Sodium Butyrate: A Multifaceted Modulator in Colorectal Cancer Therapy. Medicina 2025, 61, 136. [Google Scholar] [CrossRef]
- Yu, G.; Klionsky, D.J. Life and Death Decisions-The Many Faces of Autophagy in Cell Survival and Cell Death. Biomolecules 2022, 12, 866. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vidoni, C.; Ferraresi, A.; Secomandi, E.; Vallino, L.; Dhanasekaran, D.N.; Isidoro, C. Epigenetic targeting of autophagy for cancer prevention and treatment by natural compounds. Semin. Cancer Biol. 2020, 66, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Vidoni, C.; Vallino, L.; Ferraresi, A.; Secomandi, E.; Salwa, A.; Chinthakindi, M.; Galetto, A.; Dhanasekaran, D.N.; Isidoro, C. Epigenetic control of autophagy in women’s tumors: Role of non-coding RNAs. J. Cancer Metastasis Treat. 2021, 7, 4. [Google Scholar] [CrossRef]
- Thuwajit, C.; Ferraresi, A.; Titone, R.; Thuwajit, P.; Isidoro, C. The metabolic cross-talk between epithelial cancer cells and stromal fibroblasts in ovarian cancer progression: Autophagy plays a role. Med. Res. Rev. 2018, 38, 1235–1254. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ferraresi, A.; Girone, C.; Esposito, A.; Vidoni, C.; Vallino, L.; Secomandi, E.; Dhanasekaran, D.N.; Isidoro, C. How Autophagy Shapes the Tumor Microenvironment in Ovarian Cancer. Front. Oncol. 2020, 10, 599915. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ferraresi, A.; Girone, C.; Maheshwari, C.; Vallino, L.; Dhanasekaran, D.N.; Isidoro, C. Ovarian Cancer Cell-Conditioning Medium Induces Cancer-Associated Fibroblast Phenoconversion through Glucose-Dependent Inhibition of Autophagy. Int. J. Mol. Sci. 2024, 25, 5691. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shao, L.N.; Zhu, B.S.; Xing, C.G.; Yang, X.D.; Young, W.; Cao, J.P. Effects of autophagy regulation of tumor-associated macrophages on radiosensitivity of colorectal cancer cells. Mol. Med. Rep. 2016, 13, 2661–2670. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.R.; Kim, M.S.; Oh, J.E.; Kim, Y.R.; Song, S.Y.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J. Pathol. 2009, 217, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, H.; Lv, L.; Lu, K.; Li, H.; Zhang, W.; Cui, T. Autophagy: Dual roles and perspective for clinical treatment of colorectal cancer. Biochimie 2023, 206, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, S.; Muhammad, J.S.; Maghazachi, A.A.; Hamid, Q. Autophagy: A Versatile Player in the Progression of Colorectal Cancer and Drug Resistance. Front. Oncol. 2022, 12, 924290. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jo, Y.K.; Kim, S.C.; Park, I.J.; Park, S.J.; Jin, D.H.; Hong, S.W.; Cho, D.H.; Kim, J.C. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS ONE 2012, 7, e52705. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jo, Y.K.; Roh, S.A.; Lee, H.; Park, N.Y.; Choi, E.S.; Oh, J.H.; Park, S.J.; Shin, J.H.; Suh, Y.A.; Lee, E.K.; et al. Polypyrimidine tract-binding protein 1-mediated down-regulation of ATG10 facilitates metastasis of colorectal cancer cells. Cancer Lett. 2017, 385, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, N.; Faried, A.; Tsutsumi, S.; Kuwano, H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur. J. Cancer 2010, 46, 1900–1909. [Google Scholar] [CrossRef] [PubMed]
- Qureshi-Baig, K.; Kuhn, D.; Viry, E.; Pozdeev, V.I.; Schmitz, M.; Rodriguez, F.; Ullmann, P.; Koncina, E.; Nurmik, M.; Frasquilho, S.; et al. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy 2020, 16, 1436–1452. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lévy, J.; Cacheux, W.; Bara, M.A.; L’Hermitte, A.; Lepage, P.; Fraudeau, M.; Trentesaux, C.; Lemarchand, J.; Durand, A.; Crain, A.M.; et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat. Cell Biol. 2015, 17, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Lu, Y.; Zhang, Q.; Qu, X. Heterozygous deletion of ATG5 in Apc(Min/+) mice promotes intestinal adenoma growth and enhances the antitumor efficacy of interferon-gamma. Cancer Biol. Ther. 2015, 16, 383–391. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lucas, C.; Salesse, L.; Hoang, M.H.T.; Bonnet, M.; Sauvanet, P.; Larabi, A.; Godfraind, C.; Gagnière, J.; Pezet, D.; Rosenstiel, P.; et al. Autophagy of Intestinal Epithelial Cells Inhibits Colorectal Carcinogenesis Induced by Colibactin-Producing Escherichia coli in ApcMin/+ Mice. Gastroenterology 2020, 158, 1373–1388. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, J.; Zhou, L.; Han, J.; Liu, R.; Zhang, H.; Ning, T.; Gao, Z.; Liu, B.; Chen, X.; et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics 2020, 10, 1981–1996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhai, H.; Song, B.; Xu, X.; Zhu, W.; Ju, J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene 2013, 32, 1570–1579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tan, S.; Shi, H.; Ba, M.; Lin, S.; Tang, H.; Zeng, X.; Zhang, X. miR-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int. J. Mol. Med. 2016, 37, 1030–1038. [Google Scholar] [CrossRef] [PubMed]
- Trincheri, N.F.; Follo, C.; Nicotra, G.; Peracchio, C.; Castino, R.; Isidoro, C. Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis 2008, 29, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Cai, L.D.; Liu, Y.H.; Li, S.; Gan, W.J.; Li, X.M.; Wang, J.R.; Guo, P.D.; Zhou, Q.; Lu, X.X.; et al. Ube2v1-mediated ubiquitination and degradation of Sirt1 promotes metastasis of colorectal cancer by epigenetically suppressing autophagy. J. Hematol. Oncol. 2018, 11, 95. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huo, H.Z.; Zhou, Z.Y.; Wang, B.; Qin, J.; Liu, W.Y.; Gu, Y. Dramatic suppression of colorectal cancer cell growth by the dual mTORC1 and mTORC2 inhibitor AZD-2014. Biochem. Biophys. Res. Commun. 2014, 443, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Li, C.Y.; Peng, R.Q.; Wu, X.J.; Wang, H.Y.; Wan, D.S.; Zhu, X.F.; Zhang, X.S. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy 2009, 5, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.A.; Lee, C.T.; Lee, J.C.; Wang, Y.W.; Huang, C.T.; Lan, S.H.; Lin, P.C.; Lin, B.W.; Tian, Y.F.; Liu, H.S.; et al. MiR-338-5p promotes metastasis of colorectal cancer by inhibition of phosphatidylinositol 3-kinase, catalytic subunit type 3-mediated autophagy pathway. EBioMedicine 2019, 43, 270–281. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zheng, S.; Zhong, Y.F.; Tan, D.M.; Xu, Y.; Chen, H.X.; Wang, D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J. Biosci. 2019, 44, 92. [Google Scholar] [CrossRef] [PubMed]
- Garavaglia, B.; Vallino, L.; Amoruso, A.; Pane, M.; Ferraresi, A.; Isidoro, C. The role of gut microbiota, immune system, and autophagy in the pathogenesis of inflammatory bowel disease: Molecular mechanisms and therapeutic approaches. Asp. Mol. Med. 2024, 4, 100056. [Google Scholar] [CrossRef]
- Hooper, K.M.; Barlow, P.G.; Henderson, P.; Stevens, C. Interactions Between Autophagy and the Unfolded Protein Response: Implications for Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Iida, T.; Onodera, K.; Nakase, H. Role of autophagy in the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2017, 23, 1944–1953. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, M.; Xu, W.; Wang, J.; Yan, J.; Shi, Y.; Zhang, C.; Ge, W.; Wu, J.; Du, P.; Chen, Y. Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-κB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine 2018, 35, 345–360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nighot, P.K.; Hu, C.A.; Ma, T.Y. Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J. Biol. Chem. 2015, 290, 7234–7246. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wong, M.; Ganapathy, A.S.; Suchanec, E.; Laidler, L.; Ma, T.; Nighot, P. Intestinal epithelial tight junction barrier regulation by autophagy-related protein ATG6/beclin 1. Am. J. Physiol. Cell Physiol. 2019, 316, C753–C765. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, C.; Yan, J.; Xiao, Y.; Shen, Y.; Wang, J.; Ge, W.; Chen, Y. Inhibition of Autophagic Degradation Process Contributes to Claudin-2 Expression Increase and Epithelial Tight Junction Dysfunction in TNF-α Treated Cell Monolayers. Int. J. Mol. Sci. 2017, 18, 157. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, L.; Liu, C.; Zhao, W.; He, C.; Ding, J.; Dai, R.; Xu, K.; Xiao, L.; Luo, L.; Liu, S.; et al. Impaired Autophagy in Intestinal Epithelial Cells Alters Gut Microbiota and Host Immune Responses. Appl. Environ. Microbiol. 2018, 84, e00880-18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, X.; Yin, L.; Shen, S.; Hou, Y. Inflammation and cancer: Paradoxical roles in tumorigenesis and implications in immunotherapies. Genes Dis. 2021, 10, 151–164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Borowczak, J.; Szczerbowski, K.; Maniewski, M.; Kowalewski, A.; Janiczek-Polewska, M.; Szylberg, A.; Marszałek, A.; Szylberg, Ł. The Role of Inflammatory Cytokines in the Pathogenesis of Colorectal Carcinoma-Recent Findings and Review. Biomedicines 2022, 10, 1670. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muthusami, S.; Ramachandran, I.K.; Babu, K.N.; Krishnamoorthy, S.; Guruswamy, A.; Queimado, L.; Chaudhuri, G.; Ramachandran, I. Role of Inflammation in the Development of Colorectal Cancer. Endocr. Metab. Immune Disord. Drug. Targets 2021, 21, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhu, L.; Liu, H.; Feng, X.; Zhang, C.; Liu, B.; Li, T.; Liu, L.; Chang, H.; Sun, J.; et al. Altered gut metabolites and metabolic reprogramming involved in the pathogenesis of colitis-associated colorectal cancer and the transition of colon “inflammation to cancer”. J. Pharm. Biomed. Anal. 2025, 253, 116553. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.C.; Itzkowitz, S.H. Colorectal Cancer in Inflammatory Bowel Disease: Mechanisms and Management. Gastroenterology 2022, 162, 715–730.e3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sato, Y.; Tsujinaka, S.; Miura, T.; Kitamura, Y.; Suzuki, H.; Shibata, C. Inflammatory Bowel Disease and Colorectal Cancer: Epidemiology, Etiology, Surveillance, and Management. Cancers 2023, 15, 4154. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fanizza, J.; Bencardino, S.; Allocca, M.; Furfaro, F.; Zilli, A.; Parigi, T.L.; Fiorino, G.; Peyrin-Biroulet, L.; Danese, S.; D’Amico, F. Inflammatory Bowel Disease and Colorectal Cancer. Cancers 2024, 16, 2943. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heo, G.; Lee, Y.; Im, E. Interplay between the Gut Microbiota and Inflammatory Mediators in the Development of Colorectal Cancer. Cancers 2021, 13, 734. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Turano, M.; Cammarota, F.; Duraturo, F.; Izzo, P.; De Rosa, M. Role of IL-6/IL-6R in the Development and Management of Colon Cancer. Membranes 2021, 11, 312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mager, L.F.; Wasmer, M.H.; Rau, T.T.; Krebs, P. Cytokine-Induced Modulation of Colorectal Cancer. Front. Oncol. 2016, 6, 96. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nagasaki, T.; Hara, M.; Nakanishi, H.; Takahashi, H.; Sato, M.; Takeyama, H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: Anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br. J. Cancer 2014, 110, 469–478. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, Q.; Shang, Y.; Sun, F.; Dong, X.; Niu, J.; Li, F. Interleukin-6 Promotes Epithelial-Mesenchymal Transition and Cell Invasion through Integrin β6 Upregulation in Colorectal Cancer. Oxid. Med. Cell Longev. 2020, 2020, 8032187. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, T.; Liu, L.; Lai, W.; Zeng, Y.; Xu, H.; Lan, Q.; Su, P.; Chu, Z. Interaction with tumor-associated macrophages promotes PRL-3-induced invasion of colorectal cancer cells via MAPK pathway-induced EMT and NF-κB signaling-induced angiogenesis. Oncol. Rep. 2019, 41, 2790–2802. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Knüpfer, H.; Preiss, R. Serum interleukin-6 levels in colorectal cancer patients--a summary of published results. Int. J. Colorectal Dis. 2010, 25, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, S.; Liu, G.; Li, Y.; Pan, Y. Metabolic Reprogramming Induces Macrophage Polarization in the Tumor Microenvironment. Front. Immunol. 2022, 13, 840029. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wan, G.; Xie, M.; Yu, H.; Chen, H. Intestinal dysbacteriosis activates tumor-associated macrophages to promote epithelial-mesenchymal transition of colorectal cancer. Innate. Immun. 2018, 24, 480–489. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, D.; Li, Y. Gut microbiota and tumor-associated macrophages: Potential in tumor diagnosis and treatment. Gut Microbes 2023, 15, 2276314. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kikuchi, T.; Mimura, K.; Ashizawa, M.; Okayama, H.; Endo, E.; Saito, K.; Sakamoto, W.; Fujita, S.; Endo, H.; Saito, M.; et al. Characterization of tumor-infiltrating immune cells in relation to microbiota in colorectal cancers. Cancer Immunol. Immunother. 2020, 69, 23–32. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, H.; Wang, Y.; Zhao, J.; Jiang, J.; Zhou, Y.; Shi, P.; Liu, Q.; Sun, Y. Dietary Nondigestible Polysaccharides Ameliorate Colitis by Improving Gut Microbiota and CD4+ Differentiation, as Well as Facilitating M2 Macrophage Polarization. JPEN J. Parenter. Enteral. Nutr. 2019, 43, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R. A revisited concept. Tumors: Wounds that do not heal. Crit. Rev. Oncol. Hematol. 2018, 128, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Veettil, S.K.; Wong, T.Y.; Loo, Y.S.; Playdon, M.C.; Lai, N.M.; Giovannucci, E.L.; Chaiyakunapruk, N. Role of Diet in Colorectal Cancer Incidence: Umbrella Review of Meta-analyses of Prospective Observational Studies. JAMA Netw. Open 2021, 4, e2037341. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lewandowska, A.; Rudzki, G.; Lewandowski, T.; Stryjkowska-Góra, A.; Rudzki, S. Risk Factors for the Diagnosis of Colorectal Cancer. Cancer Control 2022, 29, 10732748211056692. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rubio, K.; Hernández-Cruz, E.Y.; Rogel-Ayala, D.G.; Sarvari, P.; Isidoro, C.; Barreto, G.; Pedraza-Chaverri, J. Nutriepigenomics in Environmental-Associated Oxidative Stress. Antioxidants 2023, 12, 771. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Conti, L.; Corn, M.D.; Scazzocchio, B.; Varì, R.; D’Archivio, M.; Varano, B.; Masella, R.; Gessani, S. Dietary fatty acids and adipose tissue inflammation at the crossroad between obesity and colorectal cancer. J. Cancer Metastasis Treat. 2019, 5, 64. [Google Scholar] [CrossRef]
- Torres-Maravilla, E.; Boucard, A.S.; Mohseni, A.H.; Taghinezhad, S.S.; Cortes-Perez, N.G.; Bermúdez-Humarán, L.G. Role of Gut Microbiota and Probiotics in Colorectal Cancer: Onset and Progression. Microorganisms 2021, 9, 1021. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rosenberg, E. Diversity of bacteria within the human gut and its contribution to the functional unity of holobionts. NPJ Biofilms Microbiomes 2024, 10, 134. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ahmad, A.; Mahmood, N.; Raza, M.A.; Mushtaq, Z.; Saeed, F.; Afzaal, M.; Hussain, M.; Amjad, H.W.; Al-Awadi, H.M. Gut microbiota and their derivatives in the progression of colorectal cancer: Mechanisms of action, genome and epigenome contributions. Heliyon 2024, 10, e29495. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7618. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, P.; Wang, Y.; Yang, G.; Zhang, Q.; Meng, L.; Xin, Y.; Jiang, X. The role of short-chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol. Res. 2021, 165, 105420. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Bhutia, Y.D.; Yang, S.; Ganapathy, V. Short-Chain Fatty Acid Transporters: Role in Colonic Homeostasis. Compr. Physiol. 2017, 8, 299–314. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal. Transduct. Target Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Tran Van Nhieu, J.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ye, X.; Wang, R.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Adoni, H.; Ajami, N.J.; Wong, M.C.; Smith, D.P.; et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev. Res. 2017, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lim, K.C.; Huang, J.; Saidi, R.F.; Sears, C.L. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 1998, 95, 14979–14984. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.; Thiele Orberg, E.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lu, R.; Wu, S.; Zhang, Y.G.; Xia, Y.; Liu, X.; Zheng, Y.; Chen, H.; Schaefer, K.L.; Zhou, Z.; Bissonnette, M.; et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis 2014, 3, e105. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Drewes, J.L.; Chen, J.; Markham, N.O.; Knippel, R.J.; Domingue, J.C.; Tam, A.J.; Chan, J.L.; Kim, L.; McMann, M.; Stevens, C.; et al. Human Colon Cancer-Derived Clostridioides difficile Strains Drive Colonic Tumorigenesis in Mice. Cancer Discov. 2022, 12, 1873–1885. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, R.; Herold, J.L.; Schady, D.; Davis, J.; Kopetz, S.; Martinez-Moczygemba, M.; Murray, B.E.; Han, F.; Li, Y.; Callaway, E.; et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 2017, 13, e1006440. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Balamurugan, R.; Rajendiran, E.; George, S.; Samuel, G.V.; Ramakrishna, B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008, 23, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Nemati, M.; Omrani, G.R.; Ebrahimi, B.; Montazeri-Najafabady, N. The Beneficial Effects of Probiotics via Autophagy: A Systematic Review. Biomed Res. Int. 2021, 2021, 2931580. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of Action of Probiotics. Adv. Nutr. 2019, 10, S49–S66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kahouli, I.; Tomaro-Duchesneau, C.; Prakash, S. Probiotics in colorectal cancer (CRC) with emphasis on mechanisms of action and current perspectives. J. Med. Microbiol. 2013, 62, 1107–1123. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Hua, H.; Liu, L.; Mao, Y.; Wang, R. Interactions between toll-like receptors signaling pathway and gut microbiota in host homeostasis. Immun. Inflamm. Dis. 2024, 12, e1356. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yesudhas, D.; Gosu, V.; Anwar, M.A.; Choi, S. Multiple roles of toll-like receptor 4 in colorectal cancer. Front. Immunol. 2014, 5, 334. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lowe, E.L.; Crother, T.R.; Rabizadeh, S.; Hu, B.; Wang, H.; Chen, S.; Shimada, K.; Wong, M.H.; Michelsen, K.S.; Arditi, M. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS ONE 2010, 5, e13027. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jang, Y.J.; Kim, W.K.; Han, D.H.; Lee, K.; Ko, G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes 2019, 10, 696–711. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shi, J.; Li, H.; Liang, S.; Evivie, S.; Huo, G.; Li, B.; Liu, F. Selected lactobacilli strains inhibit inflammation in LPS-induced RAW264.7 macrophages by suppressing the TLR4-mediated NF-κB and MAPKs activation. Food Sci. Technol. 2022, 42, e107621. [Google Scholar] [CrossRef]
- Molska, M.; Reguła, J. Potential Mechanisms of Probiotics Action in the Prevention and Treatment of Colorectal Cancer. Nutrients 2019, 11, 2453. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Torkamaneh, M.; Torfeh, M.; Jouriani, F.H.; Sepehr, A.; Ashrafian, F.; Aghamohammad, S.; Rohani, M. Investigating the crucial role of selected Bifidobacterium probiotic strains in preventing or reducing inflammation by affecting the autophagy pathway. Lett. Appl. Microbiol. 2023, 76, ovad135. [Google Scholar] [CrossRef] [PubMed]
- Vallino, L.; Garavaglia, B.; Visciglia, A.; Amoruso, A.; Pane, M.; Ferraresi, A.; Isidoro, C. Cell-free Lactiplantibacillus plantarum OC01 supernatant suppresses IL-6-induced proliferation and invasion of human colorectal cancer cells: Effect on β-Catenin degradation and induction of autophagy. J. Tradit. Complement. Med. 2023, 13, 193–206. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garavaglia, B.; Vallino, L.; Ferraresi, A.; Amoruso, A.; Pane, M.; Isidoro, C. Probiotic-Derived Metabolites from Lactiplantibacillus plantarum OC01 Reprogram Tumor-Associated Macrophages to an Inflammatory Anti-Tumoral Phenotype: Impact on Colorectal Cancer Cell Proliferation and Migration. Biomedicines 2025, 13, 339. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mazziotta, C.; Tognon, M.; Martini, F.; Torreggiani, E.; Rotondo, J.C. Probiotics Mechanism of Action on Immune Cells and Beneficial Effects on Human Health. Cells 2023, 12, 184. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hou, H.; Chen, D.; Zhang, K.; Zhang, W.; Liu, T.; Wang, S.; Dai, X.; Wang, B.; Zhong, W.; Cao, H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022, 526, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Wang, P.Y.; Wang, X.; Wan, Y.L.; Liu, Y.C. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig. Dis. Sci. 2012, 57, 3126–3135. [Google Scholar] [CrossRef] [PubMed]
- Burger-van Paassen, N.; Vincent, A.; Puiman, P.J.; van der Sluis, M.; Bouma, J.; Boehm, G.; van Goudoever, J.B.; van Seuningen, I.; Renes, I.B. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: Implications for epithelial protection. Biochem. J. 2009, 420, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Nakatani, A.; Hasegawa, S.; Irie, J.; Ozawa, K.; Tsujimoto, G.; Suganami, T.; Itoh, H.; Kimura, I. The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS ONE 2017, 12, e0179696. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kibbie, J.J.; Dillon, S.M.; Thompson, T.A.; Purba, C.M.; McCarter, M.D.; Wilson, C.C. Butyrate directly decreases human gut lamina propria CD4 T cell function through histone deacetylase (HDAC) inhibition and GPR43 signaling. Immunobiology 2021, 226, 152126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luo, S.; Li, Z.; Mao, L.; Chen, S.; Sun, S. Sodium butyrate induces autophagy in colorectal cancer cells through LKB1/AMPK signaling. J. Physiol. Biochem. 2019, 75, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, J.; Wang, M.; Yuan, H.; Xing, Y.; Zhou, X.; Ding, M.; Chen, W.; Qu, B.; Zhu, L. CISD2 Promotes Proliferation of Colorectal Cancer Cells by Inhibiting Autophagy in a Wnt/β-Catenin-Signaling-Dependent Pathway. Biochem. Genet. 2023, 61, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, W.; Evans, P.M.; Chen, X.; He, X.; Liu, C. Adenomatous polyposis coli (APC) differentially regulates beta-catenin phosphorylation and ubiquitination in colon cancer cells. J. Biol. Chem. 2006, 281, 17751–17757. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Fu, J.; Huang, Y.; Fu, C. Mechanism of APC truncation involved in colorectal cancer tumorigenesis (Review). Oncol. Lett. 2024, 29, 2. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garavaglia, B.; Vallino, L.; Ferraresi, A.; Esposito, A.; Salwa, A.; Vidoni, C.; Gentilli, S.; Isidoro, C. Butyrate Inhibits Colorectal Cancer Cell Proliferation through Autophagy Degradation of β-Catenin Regardless of APC and β-Catenin Mutational Status. Biomedicines 2022, 10, 1131. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brasiel, P.G.A.; Dutra Luquetti, S.C.P.; Peluzio, M.D.C.G.; Novaes, R.D.; Gonçalves, R.V. Preclinical Evidence of Probiotics in Colorectal Carcinogenesis: A Systematic Review. Dig. Dis. Sci. 2020, 65, 3197–3210. [Google Scholar] [CrossRef] [PubMed]
- Cruz, B.C.S.; Sarandy, M.M.; Messias, A.C.; Gonçalves, R.V.; Ferreira, C.L.L.F.; Peluzio, M.C.G. Preclinical and clinical relevance of probiotics and synbiotics in colorectal carcinogenesis: A systematic review. Nutr. Rev. 2020, 78, 667–687. [Google Scholar] [CrossRef] [PubMed]
- Bertkova, I.; Hijova, E.; Chmelarova, A.; Mojzisova, G.; Petrasova, D.; Strojny, L.; Bomba, A.; Zitnan, R. The effect of probiotic microorganisms and bioactive compounds on chemically induced carcinogenesis in rats. Neoplasma 2010, 57, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, N.K.; Sinha, P.R. Inhibition of 1, 2-dimethylhydrazine induced colon genotoxicity in rats by the administration of probiotic curd. Mol. Biol. Rep. 2010, 37, 1373–1376. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.H.; Shim, Y.Y.; Cha, S.K.; Reaney, M.J.T.; Chee, K.M. Effect of Lactobacillus acidophilus KFRI342 on the development of chemically induced precancerous growths in the rat colon. J. Med. Microbiol. 2012, 61, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Ghanavati, R.; Akbari, A.; Mohammadi, F.; Asadollahi, P.; Javadi, A.; Talebi, M.; Rohani, M. Lactobacillus species inhibitory effect on colorectal cancer progression through modulating the Wnt/β-catenin signaling pathway. Mol. Cell Biochem. 2020, 470, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Appleyard, C.B.; Cruz, M.L.; Isidro, A.A.; Arthur, J.C.; Jobin, C.; De Simone, C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G1004-13. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Do, E.J.; Hwang, S.W.; Kim, S.Y.; Ryu, Y.M.; Cho, E.A.; Chung, E.J.; Park, S.; Lee, H.J.; Byeon, J.S.; Ye, B.D.; et al. Suppression of colitis-associated carcinogenesis through modulation of IL-6/STAT3 pathway by balsalazide and VSL#3. J. Gastroenterol. Hepatol. 2016, 31, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Yu, Y.N.; Wang, J.L.; Lin, Y.W.; Kong, X.; Yang, C.Q.; Yang, L.; Liu, Z.J.; Yuan, Y.Z.; Liu, F.; et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013, 97, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Golkhalkhali, B.; Rajandram, R.; Paliany, A.S.; Ho, G.F.; Wan Ishak, W.Z.; Johari, C.S.; Chin, K.F. Strain-specific probiotic (microbial cell preparation) and omega-3 fatty acid in modulating quality of life and inflammatory markers in colorectal cancer patients: A randomized controlled trial. Asia Pac. J. Clin. Oncol. 2018, 14, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, C.; Millette, M.; Oth, D.; Ruiz, M.T.; Luquet, F.M.; Lacroix, M. Probiotic Lactobacillus acidophilus and L. casei mix sensitize colorectal tumoral cells to 5-fluorouracil-induced apoptosis. Nutr. Cancer 2010, 62, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Osterlund, P.; Ruotsalainen, T.; Korpela, R.; Saxelin, M.; Ollus, A.; Valta, P.; Kouri, M.; Elomaa, I.; Joensuu, H. Lactobacillus supplementation for diarrhea related to chemotherapy of colorectal cancer: A randomised study. Br. J. Cancer 2007, 97, 1028–1034. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- An, J.; Ha, E.M. Combination Therapy of Lactobacillus plantarum Supernatant and 5-Fluouracil Increases Chemosensitivity in Colorectal Cancer Cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Akedo, I.; Otani, T.; Suzuki, T.; Nakamura, T.; Takeyama, I.; Ishiguro, S.; Miyaoka, E.; Sobue, T.; Kakizoe, T. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int. J. Cancer 2005, 116, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qin, H.; Yang, Z.; Xia, Y.; Liu, W.; Yang, J.; Jiang, Y.; Zhang, H.; Yang, Z.; Wang, Y.; et al. Randomised clinical trial: The effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery—A doubleblind study. Aliment. Pharmacol. Ther. 2011, 33, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Nagata, S.; Saito, M.; Shimizu, T.; Yamashiro, Y.; Matsuki, T.; Asahara, T.; Nomoto, K. Effects of the enteral administration of Bifidobacterium breve on patients undergoing chemotherapy for pediatric malignancies. Support. Care Cancer 2010, 18, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Kotzampassi, K.; Stavrou, G.; Damoraki, G.; Georgitsi, M.; Basdanis, G.; Tsaousi, G.; Giamarellos-Bourboulis, E.J. A Four-Probiotics Regimen Reduces Postoperative Complications After Colorectal Surgery: A Randomized, Double-Blind, Placebo-Controlled Study. World J. Surg. 2015, 39, 2776–2783. [Google Scholar] [CrossRef] [PubMed]
- Urbancsek, H.; Kazar, T.; Mezes, I.; Neumann, K. Results of a double-blind, randomized study to evaluate the efficacy and safety of Antibiophilus in patients with radiation-induced diarrhoea. Eur. J. Gastroenterol. Hepatol. 2001, 13, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Chitapanarux, I.; Chitapanarux, T.; Traisathit, P.; Kudumpee, S.; Tharavichitkul, E.; Lorvidhaya, V. Randomized controlled trial of live lactobacillus acidophilus plus bifidobacterium bifidum in prophylaxis of diarrhea during radiotherapy in cervical cancer patients. Radiat. Oncol. 2010, 5, 31. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shakhpazyan, N.K.; Mikhaleva, L.M.; Bedzhanyan, A.L.; Gioeva, Z.V.; Mikhalev, A.I.; Midiber, K.Y.; Pechnikova, V.V.; Biryukov, A.E. Exploring the Role of the Gut Microbiota in Modulating Colorectal Cancer Immunity. Cells 2024, 13, 1437. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Peng, Z.; Cheng, S.; Kou, Y.; Wang, Z.; Jin, R.; Hu, H.; Zhang, X.; Gong, J.F.; Li, J.; Lu, M.; et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020, 8, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Pace, F.; Pace, M.; Quartarone, G. Probiotics in digestive diseases: Focus on Lactobacillus GG. Minerva Gastroenterol. Dietol. 2015, 6, 273–292. [Google Scholar] [PubMed]
- Holzapfel, W.H.; Haberer, P.; Snel, J.; Schillinger, U.; Huis in’t Veld, J.H. Overview of gut flora and probiotics. Int. J. Food Microbiol. 1998, 41, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reid, G.; Jass, J.; Sebulsky, M.T.; McCormick, J.K. Potential uses of probiotics in clinical practice. Clin. Microbiol. Rev. 2003, 16, 658–672. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gianotti, L.; Morelli, L.; Galbiati, F.; Rocchetti, S.; Coppola, S.; Beneduce, A.; Gilardini, C.; Zonenschain, D.; Nespoli, A.; Braga, M. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J. Gastroenterol. 2010, 16, 167–175. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mego, M.; Chovanec, J.; Vochyanova-Andrezalova, I.; Konkolovsky, P.; Mikulova, M.; Reckova, M.; Miskovska, V.; Bystricky, B.; Beniak, J.; Medvecova, L.; et al. Prevention of irinotecan induced diarrhea by probiotics: A randomized double blind, placebo controlled pilot study. Complement. Ther. Med. 2015, 23, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Mego, M.; Kasperova, B.; Chovanec, J.; Danis, R.; Reckova, M.; Bystricky, B.; Konkolovsky, P.; Jurisova, S.; Porsok, S.; Vaclav, V.; et al. The beneficial effect of probiotics in the prevention of irinotecan-induced diarrhea in colorectal cancer patients with colostomy: A pooled analysis of two probiotic trials (Probio-SK-003 and Probio-SK-005) led by Slovak Cooperative Oncology Group. Front Oncol. 2024, 14, 1438657. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, X.; Fruehauf, J.; Goldsmith, J.D.; Xu, H.; Katchar, K.K.; Koon, H.W.; Zhao, D.; Kokkotou, E.G.; Pothoulakis, C.; Kelly, C.P. Saccharomyces boulardii inhibits EGF receptor signaling and intestinal tumor growth in Apc(min) mice. Gastroenterology 2009, 137, 914–923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Azcárate-Peril, M.A.; Sikes, M.; Bruno-Bárcena, J.M. The intestinal microbiota, gastrointestinal environment and colorectal cancer: A putative role for probiotics in prevention of colorectal cancer? Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G401–G424. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Theodoropoulos, G.E.; Memos, N.A.; Peitsidou, K.; Karantanos, T.; Spyropoulos, B.G.; Zografos, G. Synbiotics and gastrointestinal function-related quality of life after elective colorectal cancer resection. Ann. Gastroenterol. 2016, 29, 56–62. [Google Scholar] [PubMed] [PubMed Central]
- Chen, C.C.; Lin, W.C.; Kong, M.S.; Shi, H.N.; Walker, W.A.; Lin, C.Y.; Huang, C.T.; Lin, Y.C.; Jung, S.M.; Lin, T.Y. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br. J. Nutr. 2012, 107, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Hibberd, A.A.; Lyra, A.; Ouwehand, A.C.; Rolny, P.; Lindegren, H.; Cedgård, L.; Wettergren, Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017, 4, e000145. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, H.A.; Kim, H.; Lee, K.W.; Park, K.Y. Dead Nano-Sized Lactobacillus plantarum Inhibits Azoxymethane/Dextran Sulfate Sodium-Induced Colon Cancer in Balb/c Mice. J. Med. Food. 2015, 18, 1400–1405. [Google Scholar] [CrossRef] [PubMed]
- Stene, C.; Xu, J.; Fallone de Andrade, S.; Palmquist, I.; Molin, G.; Ahrné, S.; Thorlacius, H.; Johnson, L.B.; Jeppsson, B. Synbiotics protected radiation-induced tissue damage in rectal cancer patients: A controlled trial. Clin. Nutr. 2025, 49, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Carlini, M.; Grieco, M.; Spoletini, D.; Menditto, R.; Napoleone, V.; Brachini, G.; Mingoli, A.; Marcellinaro, R. Implementation of the gut microbiota prevents anastomotic leaks in laparoscopic colorectal surgery for cancer:the results of the MIRACLe study. Updates Surg. 2022, 74, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Deol, P.K.; Khare, P.; Bishnoi, M.; Kondepudi, K.K.; Kaur, I.P. Coadministration of ginger extract-Lactobacillus acidophilus (cobiotic) reduces gut inflammation and oxidative stress via downregulation of COX-2, i-NOS, and c-Myc. Phytother. Res. 2018, 32, 1950–1956. [Google Scholar] [CrossRef] [PubMed]
- El-Deeb, N.M.; Yassin, A.M.; Al-Madboly, L.A.; El-Hawiet, A. A novel purified Lactobacillus acidophilus 20079 exopolysaccharide, LA-EPS-20079, molecularly regulates both apoptotic and NF-κB inflammatory pathways in human colon cancer. Microb. Cell Fact. 2018, 17, 29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhuo, Q.; Yu, B.; Zhou, J.; Zhang, J.; Zhang, R.; Xie, J.; Wang, Q.; Zhao, S. Lysates of Lactobacillus acidophilus combined with CTLA-4-blocking antibodies enhance antitumor immunity in a mouse colon cancer model. Sci. Rep. 2019, 9, 20128. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lin, P.Y.; Li, S.C.; Lin, H.P.; Shih, C.K. Germinated brown rice combined with Lactobacillus acidophilus and Bifidobacterium animalis subsp. lactis inhibits colorectal carcinogenesis in rats. Food Sci. Nutr. 2018, 7, 216–224. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Desrouillères, K.; Millette, M.; Bagheri, L.; Maherani, B.; Jamshidian, M.; Lacroix, M. The synergistic effect of cell wall extracted from probiotic biomass containing Lactobacillus acidophilus CL1285, L. casei LBC80R, and L. rhamnosus CLR2 on the anticancer activity of cranberry juice-HPLC fractions. J. Food Biochem. 2020, 44, e13195. [Google Scholar] [CrossRef] [PubMed]
- Jacouton, E.; Chain, F.; Sokol, H.; Langella, P.; Bermúdez-Humarán, L.G. Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer. Front Immunol. 2017, 8, 1553. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sharma, M.; Shukla, G. Administration of Metabiotics Extracted From Probiotic Lactobacillus rhamnosus MD 14 Inhibit Experimental Colorectal Carcinogenesis by Targeting Wnt/β-Catenin Pathway. Front Oncol. 2020, 10, 746. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Valiei, A.; Aminian-Dehkordi, J.; Mofrad, M.R.K. Gut-on-a-chip models for dissecting the gut microbiology and physiology. APL Bioeng. 2023, 7, 011502. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Palamà, M.E.F.; Aiello, M.; Borka, G.; Furci, J.; Parodi, I.; Firpo, G.; Scaglione, S. A Dynamic Double-Flow Gut-On-Chip Model for Predictive Absorption Studies In Vitro. Adv. Mater. Technol. 2025, 10, 2401661. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, Y.; Zhao, R.; Shafi, S.; Yang, Y.; Liu, G.; Liu, S.B. Research progress of intestinal microbiota in targeted therapy and immunotherapy of colorectal cancer. J. Cancer Metastasis Treat. 2024, 10, 12. [Google Scholar] [CrossRef]
- Zhang, J.W.; Du, P.; Gao, J.; Yang, B.R.; Fang, W.J.; Ying, C.M. Preoperative probiotics decrease postoperative infectious complications of colorectal cancer. Am. J. Med. Sci. 2012, 343, 199–205. [Google Scholar] [CrossRef] [PubMed]

| Probiotic Strain | Effects and Proposed Mechanism | Preclinical Evidence (Experimental Model) + [Ref] | Clinical Trial (Type, Sample Sizes) + [Ref] | Comments (Dosage, Treatment Durations, Patient Demographics) | Pros/Cons Limitations |
|---|---|---|---|---|---|
| Lactobacillus rhamnosus GG | Enhancement of gut barrier integrity | In vitro CRC model: high adhesion, high resistance against gastric acidity, and high antimicrobial activity against pathogens [175] | NCT00197873 Prospective, multicenter, randomized, double-blind, placebo-controlled study N = 84 | Dose: 20 × 109CFU/daily Duration: 9 weeks/phase (lactobacilli/placebo), twice daily Subjects: advanced CRC patients undergoing chemotherapy (capecitabine, oxaliplatin, and bevacizumab) |
|
| Bifidobacterium longum BB536 + Lactobacillus acidophilus LA1 |
| In vitro CRC model: LA1 boosts the immune system and balances intestinal microflora [176,177,178,179] | NCT00936572 Prospective, randomized, double-blind, parallel-arm trial with three groups (high dose, low dose, placebo) N = 33 [180] | Dose: High = 109 CFU, Low = 107 CFU Duration: 3×/day from pre-operative day −5 to −1 and post-operative day 3 to 8 Subjects: CRC surgery patients (laparoscopy/laparotomy) |
|
| HEXBIO® formulation Lactobacillus acidophilus, L. lactis, L. casei, Bifidobacterium longum, B. bifidum, B. infantis |
| In vivo CRC mouse model: B. longum counteracts systemic inflammation, results in a drop in the aberrant crypt foci number in CRC mice and increased necrosis and fibrosis [47] | NCT04021589 Randomized, double-blind, placebo-controlled study N = 60 | Dose: 30 × 109 CFU/day (twice daily) Duration: 6 months, initiated 4 weeks after surgery Subjects: CRC patients planned for colorectal resection |
|
| Colon Dophilus™ Bifidobacterium breve HA-129 (25%), Bifidobacterium bifidum HA-132 (20%), Bifidobacterium longum HA-135 (14.5%), Lactobacillus acidophilus HA-122 (8%), Lactobacillus casei HA-108 (8%), Lactobacillus plantarum HA-119 (8%), Streptococcus thermophilus HA-110 (6%), Lactobacillus brevis HA-112 (2%), Bifidobacterium infantis HA-116 (0.5%) | Decrease the activity of intestinal beta- D-glucuronidase | N/A | NCT01410955 Randomized, quadruple-blind, placebo-controlled study, parallel assignment N = 46 [181] | Dose: 10 × 109 CFU of bacteria (thrice per day) Duration: 12 weeks during irinotecan-based chemotherapy Subjects: patients with histologically proven CRC starting therapy with irinotecan |
|
| Probio-Tec® BG-VCap-6.5 Bifidobacterium animalis subsp. lactis BB-12® + Lactobacillus rhamnosus GG (LGG®) |
| N/A | NCT02819960 Phase 3 Multicenter, randomized, double-blind, placebo-controlled study N = 233 [182] | Dose: 2.7 × 109 CFU of bacteria in 50%/50% ratio LGG®:BB-12® (thrice per day) Duration: 6 weeks during chemotherapy Subjects: patients starting a new line of irinotecan-based therapy |
|
| Floratil Saccharomyces boulardii |
| In vivo Apc (Min) CRC mouse model: reduced tumor number and volume by 50% [183,184] | NCT01609660 Phase 4, randomized, open-label, parallel-group trial N = 33 | Dose: 100 mg/day orally Duration: 7 days pre-operatively Subjects: patients undergoing elective CRC resection |
|
| Synbiotic Forte™ Pediococcus pentosaceus 5-33:3, Leuconostoc mesenteroides 32-77:1, Lactobacillus paracasei ssp. paracasei 19, and Lactobacillus plantarum 2362 | N/A | N/A | NCT01479907 Double-blinded, prospective, randomized N = 100 [185] |
Dose: 1011 of each of 4 LAB strains/dose (12 g in 250 cc of water) Duration: 15 days Subjects: patients who have undergone colectomy for cancer |
|
| ProBion Clinica Bifidobacterium lactis Bl-04 (ATCC SD5219), Lactobacillus acidophilus NCFM (ATCC 700396) | Epigenetic changes | In vivo CRC rodent model: NCFM attenuates tumor growth by reducing levels of pro-carcinogenic metabolites in the gut [186] | NCT03072641 Randomized, parallel assignment N = 20 [187] | Dose: tablets of 1.4 × 1010 CFU of Bl-04 + 7 × 109 CFU of NCFM Duration: Avg. 31 ± 28 days pre-operatively (range: 8–78 days) Subjects: patients with at least one malignant tumor in the colon |
|
|
HEAL 19 Lactobacillus plantarum |
| In vivo CRC model: nano-sized L. plantarum reduces colonic tumorigenesis, promotes lower colon weight/length ratios, and prevents animal weight loss [188] | NCT03420443 Randomized, parallel assignment, triple-blind N = 30 [189] | Dose: control group: no treatment; oat bran group: 45 g of oat bran and freezing medium, synbiotic group: HEAL 19 (1010 CFU/g), freeze-dried blueberry husks (13 g), and oat bran (22 g) Duration: 2 weeks (1 pre-radiotherapy and 1 during radiotherapy) Subjects: patients with rectal adenocarcinoma |
|
| MIRACle study Streptococcus thermophilus; Bifidobacterium breve; Bifidobacterium longum; Bifidobacterium infantis; Lactobacillus acidophilus; Lactobacillus plantarum; Lactobacillus paracasei; Lactobacillus delbrueckii subsp. Bulgaricus | N/A | N/A | NCT05164887 Case-control, prospective N = 131 [190] | Dose: Pre-, intra- (intraluminal anastomotic), and post-operative administration of probiotics (4.4 g equal to 450 × 109 live bacterial cells/every 12 h) Duration: from day −5 until day +4 Subjects: CRC patients undergoing laparoscopic resections with ileo-colorectal anastomoses |
|
|
Lactobacillus acidophilus, Lactobacillus acidophilus
LA-EPS-20079, and L. acidophilus MTCC 5401 |
| In vivo inflammation-driven CRC rodent model: reversion of the inflammation-induced colonic histological alterations and restoration of colonic permeability [191] In vivo mouse model: cytotoxic effects on cancer cells [192] In vivo mouse model: enhance the anti-tumor activity of CTLA-4 mAb therapy [193] | N/A | N/A | N/A |
| Lactobacillus acidophilus + Bifidobacterium animalis subsp. lactis |
| In vivo CRC rodent model: inhibition of pre-neoplastic lesions [194] | N/A | N/A | N/A |
| Lactobacillus acidophilus CL1285 + Lactobacillus casei LBC80R + Lactobacillus rhamnosus CLR2 | Stimulation of quinone reductase activity | In vitro CRC model: antioxidant activity and inhibition of HT-29 cell proliferation [195] | N/A | N/A | N/A |
| Lactobacillus casei BL23 |
| In vivo CRC mouse model: reduce histological scores and proliferative index values [196] | N/A | N/A | N/A |
| OMNi-BiOTiC® 10AAD Lactobacillus rhamnosus MD14 |
| In vivo CRC rodent model: attenuated early colon carcinogenesis and aberrant crypt foci, restoring to almost normal colon histology [197] | NCT03705442 Prospective, randomized, parallel assignment, placebo-controlled study, double-blind N = 76 | Dose: two capsules (5 × 109 CFU/each) per day, every 12 h Duration: 84 days (six chemotherapy cycles every 14 days) Subjects: histologically confirmed diagnosis of CRC with metastasis, patients starting first line of chemotherapy (FOLFIRI protocol) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garavaglia, B.; Vallino, L.; Ferraresi, A.; Visciglia, A.; Amoruso, A.; Pane, M.; Munteanu, C.; Isidoro, C. The Anti-Inflammatory, Immunomodulatory, and Pro-Autophagy Activities of Probiotics for Colorectal Cancer Prevention and Treatment: A Narrative Review. Biomedicines 2025, 13, 1554. https://doi.org/10.3390/biomedicines13071554
Garavaglia B, Vallino L, Ferraresi A, Visciglia A, Amoruso A, Pane M, Munteanu C, Isidoro C. The Anti-Inflammatory, Immunomodulatory, and Pro-Autophagy Activities of Probiotics for Colorectal Cancer Prevention and Treatment: A Narrative Review. Biomedicines. 2025; 13(7):1554. https://doi.org/10.3390/biomedicines13071554
Chicago/Turabian StyleGaravaglia, Beatrice, Letizia Vallino, Alessandra Ferraresi, Annalisa Visciglia, Angela Amoruso, Marco Pane, Camelia Munteanu, and Ciro Isidoro. 2025. "The Anti-Inflammatory, Immunomodulatory, and Pro-Autophagy Activities of Probiotics for Colorectal Cancer Prevention and Treatment: A Narrative Review" Biomedicines 13, no. 7: 1554. https://doi.org/10.3390/biomedicines13071554
APA StyleGaravaglia, B., Vallino, L., Ferraresi, A., Visciglia, A., Amoruso, A., Pane, M., Munteanu, C., & Isidoro, C. (2025). The Anti-Inflammatory, Immunomodulatory, and Pro-Autophagy Activities of Probiotics for Colorectal Cancer Prevention and Treatment: A Narrative Review. Biomedicines, 13(7), 1554. https://doi.org/10.3390/biomedicines13071554










