Novel Insights into the Pathogenesis of Inflammatory Bowel Diseases
Abstract
1. Introduction
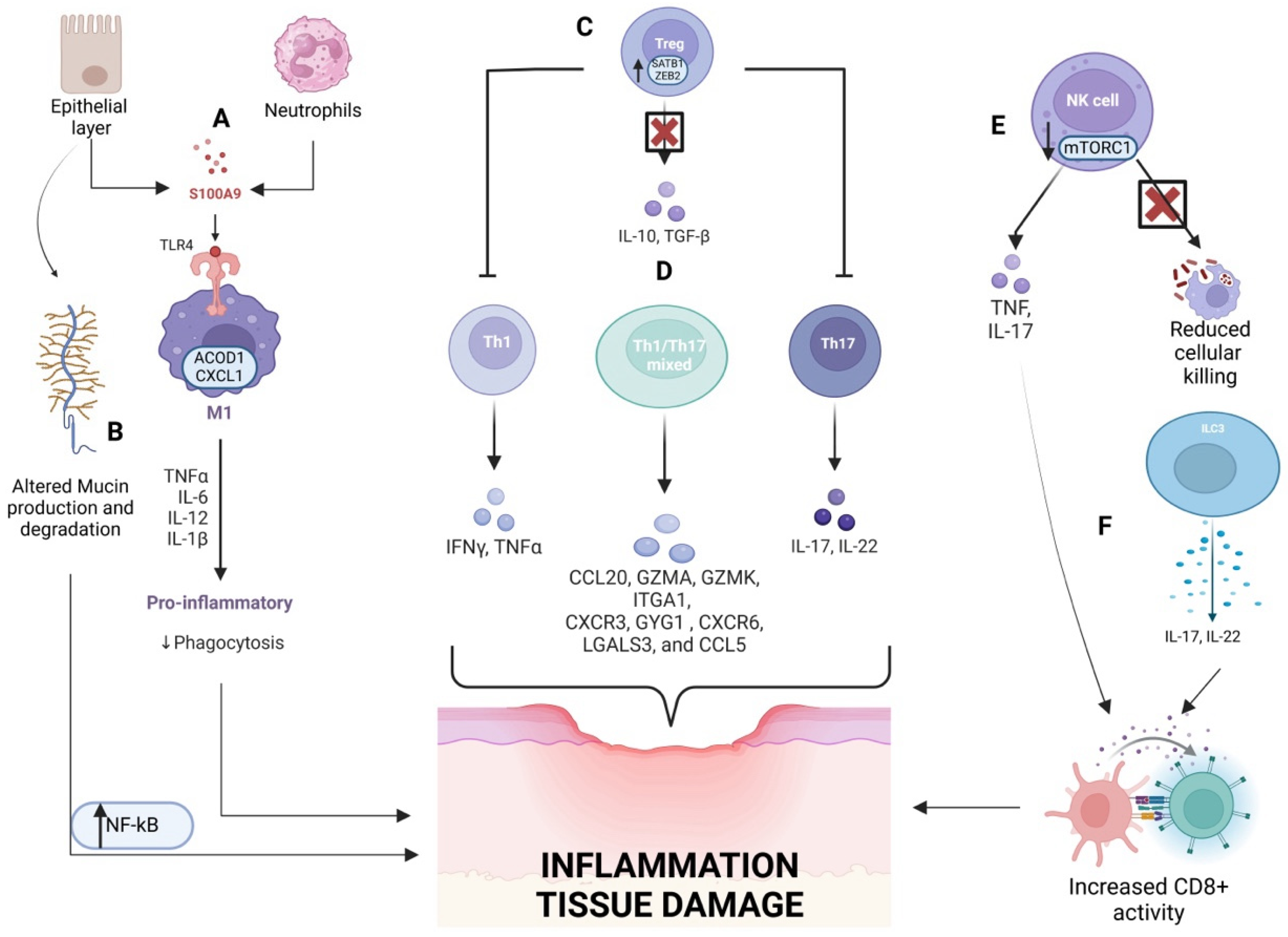
2. The Innate and Adaptive Immune System
2.1. Innate Immune System
2.2. Adaptive Immune System
3. Gut Microbiota
4. Genetics
4.1. Polygenic Contributions to IBD
4.2. Monogenic IBD
4.3. Genetics–Microbiota Interactions
5. The Exposome
5.1. Childhood Exposures
5.2. Adulthood Exposures
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.-Z. Inflammatory Bowel Disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, M.; Mohammed, N. A Review on Inflammatory Bowel Diseases: Recent Molecular Pathophysiology Advances. Biol. Targets Ther. 2022, 16, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Annese, V. Genetics and Epigenetics of IBD. Pharmacol. Res. 2020, 159, 104892. [Google Scholar] [CrossRef] [PubMed]
- Aden, K.; Reindl, W. The Gut Microbiome in Inflammatory Bowel Diseases: Diagnostic and Therapeutic Implications. Visc. Med. 2019, 35, 332–337. [Google Scholar] [CrossRef]
- Ho, S.-M.; Lewis, J.D.; Mayer, E.A.; Bernstein, C.N.; Plevy, S.E.; Chuang, E.; Rappaport, S.M.; Croitoru, K.; Korzenik, J.R.; Krischer, J.; et al. Challenges in IBD Research: Environmental Triggers. Inflamm. Bowel Dis. 2019, 25, S13–S23. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q. A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019, 2019, 7247238. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Carding, S.R. Inflammatory Bowel Disease: Cause and Immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Pokatayev, V.; Lefkovith, A.; Carter, G.T.; Creasey, E.A.; Krishna, C.; Subramanian, S.; Kochar, B.; Ashenberg, O.; Lau, H.; et al. The Landscape of Immune Dysregulation in Crohn’s Disease Revealed through Single-Cell Transcriptomic Profiling in the Ileum and Colon. Immunity 2023, 56, 444–458.e5. [Google Scholar] [CrossRef]
- Okumura, R.; Takeda, K. Roles of Intestinal Epithelial Cells in the Maintenance of Gut Homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ho, S.B. Intestinal Goblet Cells and Mucins in Health and Disease: Recent Insights and Progress. Curr. Gastroenterol. Rep. 2010, 12, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Chen, C.; Feng, J.; Zhou, S.; Feng, X.; Yang, Z.; Lu, H.; Tao, H.; Li, L.; Xv, H.; et al. Muc2 Mucin O-Glycosylation Interacts with Enteropathogenic Escherichia Coli to Influence the Development of Ulcerative Colitis Based on the NF-kB Signaling Pathway. J. Transl. Med. 2023, 21, 793. [Google Scholar] [CrossRef] [PubMed]
- Ashton, J.J.; Boukas, K.; Davies, J.; Stafford, I.S.; Vallejo, A.F.; Haggarty, R.; Coelho, T.A.F.; Batra, A.; Afzal, N.A.; Vadgama, B.; et al. Ileal Transcriptomic Analysis in Paediatric Crohn’s Disease Reveals IL17- and NOD-Signalling Expression Signatures in Treatment-Naïve Patients and Identifies Epithelial Cells Driving Differentially Expressed Genes. J. Crohns Colitis 2021, 15, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Bennstein, S.B.; Uhrberg, M. Circulating Innate Lymphoid Cells (cILCs): Unconventional Lymphocytes with Hidden Talents. J. Allergy Clin. Immunol. 2024, 154, 523–536. [Google Scholar] [CrossRef]
- Krämer, B.; Goeser, F.; Lutz, P.; Glässner, A.; Boesecke, C.; Schwarze-Zander, C.; Kaczmarek, D.; Nischalke, H.D.; Branchi, V.; Manekeller, S.; et al. Compartment-Specific Distribution of Human Intestinal Innate Lymphoid Cells Is Altered in HIV Patients under Effective Therapy. PLoS Pathog. 2017, 13, e1006373. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Sapra, L.; Bhardwaj, A.; Mishra, P.K.; Verma, B.; Baig, Z. Unravelling the Immunobiology of Innate Lymphoid Cells (ILCs): Implications in Health and Disease. Cytokine Growth Factor Rev. 2023, 74, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Wang, Y.; Boudreau, P.; Song, X.; Wu, M.; Chen, X.; Patik, I.; Tang, Y.; Ouahed, J.; Ringel, A.; et al. Bacterial Sphingolipids Exacerbate Colitis by Inhibiting ILC3-Derived IL-22 Production. Cell. Mol. Gastroenterol. Hepatol. 2024, 18, 101350. [Google Scholar] [CrossRef] [PubMed]
- Eken, A.; Singh, A.K.; Treuting, P.M.; Oukka, M. IL-23R+ Innate Lymphoid Cells Induce Colitis via Interleukin-22-Dependent Mechanism. Mucosal Immunol. 2014, 7, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Mazzurana, L.; Bonfiglio, F.; Forkel, M.; D’Amato, M.; Halfvarson, J.; Mjösberg, J. Crohn’s Disease Is Associated With Activation of Circulating Innate Lymphoid Cells. Inflamm. Bowel Dis. 2021, 27, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.M.; Wang, A.; Parhar, K.S.; Johnston, M.J.G.; Van Rooijen, N.; Beck, P.L.; McKay, D.M. In Vitro-Derived Alternatively Activated Macrophages Reduce Colonic Inflammation in Mice. Gastroenterology 2010, 138, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Guo, J.; Yan, W.; Xu, L. Macrophage Polarization in Inflammatory Bowel Disease. Cell Commun. Signal. 2023, 21, 367. [Google Scholar] [CrossRef]
- Lissner, D.; Schumann, M.; Batra, A.; Kredel, L.-I.; Kühl, A.A.; Erben, U.; May, C.; Schulzke, J.-D.; Siegmund, B. Monocyte and M1 Macrophage-Induced Barrier Defect Contributes to Chronic Intestinal Inflammation in IBD. Inflamm. Bowel Dis. 2015, 21, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhang, J.; Liu, H.; Li, S.; Wang, Q. The Role of Tissue-Resident Macrophages in the Development and Treatment of Inflammatory Bowel Disease. Front. Cell Dev. Biol. 2022, 10, 896591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, X.; Zhang, Q.; Yang, J.; Liu, G. Roles of Macrophages on Ulcerative Colitis and Colitis-Associated Colorectal Cancer. Front. Immunol. 2023, 14, 1103617. [Google Scholar] [CrossRef]
- Garrido-Trigo, A.; Corraliza, A.M.; Veny, M.; Dotti, I.; Melón-Ardanaz, E.; Rill, A.; Crowell, H.L.; Corbí, Á.; Gudiño, V.; Esteller, M.; et al. Macrophage and Neutrophil Heterogeneity at Single-Cell Spatial Resolution in Human Inflammatory Bowel Disease. Nat. Commun. 2023, 14, 4506. [Google Scholar] [CrossRef] [PubMed]
- Azramezani Kopi, T.; Amini Kadijani, A.; Parsian, H.; Shahrokh, S.; Asadzadeh Aghdaei, H.; Mirzaei, A.; Balaii, H.; Zali, M.R. The Value of mRNA Expression of S100A8 and S100A9 as Blood-Based Biomarkers of Inflammatory Bowel Disease. Arab J. Gastroenterol. 2019, 20, 135–140. [Google Scholar] [CrossRef]
- Okada, K.; Itoh, H.; Ikemoto, M. Circulating S100A8/A9 Is Potentially a Biomarker That Could Reflect the Severity of Experimental Colitis in Rats. Heliyon 2020, 6, e03470. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wang, R.; Zhang, X.; Wen, X.; Deng, S.; Xie, W. Identification CCL2,CXCR2,S100A9 of the Immune-Related Gene Markers and Immune Infiltration Characteristics of Inflammatory Bowel Disease and Heart Failure via Bioinformatics Analysis and Machine Learning. Front. Cardiovasc. Med. 2023, 10, 1268675. [Google Scholar] [CrossRef]
- Poggi, A.; Benelli, R.; Venè, R.; Costa, D.; Ferrari, N.; Tosetti, F.; Zocchi, M.R. Human Gut-Associated Natural Killer Cells in Health and Disease. Front. Immunol. 2019, 10, 961. [Google Scholar] [CrossRef] [PubMed]
- Zaiatz Bittencourt, V.; Jones, F.; Tosetto, M.; Doherty, G.A.; Ryan, E.J. Dysregulation of Metabolic Pathways in Circulating Natural Killer Cells Isolated from Inflammatory Bowel Disease Patients. J. Crohns Colitis 2021, 15, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and Adaptive Immunity in Inflammatory Bowel Disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef]
- Kałużna, A.; Olczyk, P.; Komosińska-Vassev, K. The Role of Innate and Adaptive Immune Cells in the Pathogenesis and Development of the Inflammatory Response in Ulcerative Colitis. J. Clin. Med. 2022, 11, 400. [Google Scholar] [CrossRef] [PubMed]
- Mitsialis, V.; Wall, S.; Liu, P.; Ordovas-Montanes, J.; Parmet, T.; Vukovic, M.; Spencer, D.; Field, M.; McCourt, C.; Toothaker, J.; et al. Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn’s Disease. Gastroenterology 2020, 159, 591–608.e10. [Google Scholar] [CrossRef] [PubMed]
- Medina, T.S.; Murison, A.; Smith, M.; Kinker, G.S.; Chakravarthy, A.; Vitiello, G.A.F.; Turpin, W.; Shen, S.Y.; Yau, H.L.; Sarmento, O.F.; et al. The Chromatin and Single-Cell Transcriptional Landscapes of CD4 T Cells in Inflammatory Bowel Disease Link Risk Loci with a Proinflammatory Th17 Cell Population. Front. Immunol. 2023, 14, 1161901. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Chen, P.; Peng, B.; Cheng, Y.; Xie, J.; Hou, Z.; Chen, H.; Ye, L.; Li, H.; Wang, H.; et al. The Transcription Factor ELF4 Alleviates Inflammatory Bowel Disease by Activating IL1RN Transcription, Suppressing Inflammatory TH17 Cell Activity, and Inducing Macrophage M2 Polarization. Front. Immunol. 2023, 14, 1270411. [Google Scholar] [CrossRef]
- Tanemoto, S.; Sujino, T.; Miyamoto, K.; Moody, J.; Yoshimatsu, Y.; Ando, Y.; Koya, I.; Harada, Y.; Tojo, A.O.; Ono, K.; et al. Single-Cell Transcriptomics of Human Gut T Cells Identifies Cytotoxic CD4+CD8A+ T Cells Related to Mouse CD4 Cytotoxic T Cells. Front. Immunol. 2022, 13, 977117. [Google Scholar] [CrossRef] [PubMed]
- Casalegno Garduño, R.; Däbritz, J. New Insights on CD8+ T Cells in Inflammatory Bowel Disease and Therapeutic Approaches. Front. Immunol. 2021, 12, 738762. [Google Scholar] [CrossRef] [PubMed]
- Corridoni, D.; Antanaviciute, A.; Gupta, T.; Fawkner-Corbett, D.; Aulicino, A.; Jagielowicz, M.; Parikh, K.; Repapi, E.; Taylor, S.; Ishikawa, D.; et al. Single-Cell Atlas of Colonic CD8+ T Cells in Ulcerative Colitis. Nat. Med. 2020, 26, 1480–1490. [Google Scholar] [CrossRef] [PubMed]
- Globig, A.-M.; Mayer, L.S.; Heeg, M.; Andrieux, G.; Ku, M.; Otto-Mora, P.; Hipp, A.V.; Zoldan, K.; Pattekar, A.; Rana, N.; et al. Exhaustion of CD39-Expressing CD8+ T Cells in Crohn’s Disease Is Linked to Clinical Outcome. Gastroenterology 2022, 163, 965–981.e31. [Google Scholar] [CrossRef] [PubMed]
- Mayne, C.G.; Williams, C.B. Induced and Natural Regulatory T Cells in the Development of Inflammatory Bowel Disease: Inflamm. Bowel Dis. 2013, 19, 1772–1788. [Google Scholar] [CrossRef]
- Yan, J.; Zeng, Y.; Guan, Z.; Li, Z.; Luo, S.; Niu, J.; Zhao, J.; Gong, H.; Huang, T.; Li, Z.; et al. Inherent Preference for Polyunsaturated Fatty Acids Instigates Ferroptosis of Treg Cells That Aggravates High-Fat-Diet-Related Colitis. Cell Rep. 2024, 43, 114636. [Google Scholar] [CrossRef] [PubMed]
- Boland, B.S.; He, Z.; Tsai, M.S.; Olvera, J.G.; Omilusik, K.D.; Duong, H.G.; Kim, E.S.; Limary, A.E.; Jin, W.; Milner, J.J.; et al. Heterogeneity and Clonal Relationships of Adaptive Immune Cells in Ulcerative Colitis Revealed by Single-Cell Analyses. Sci. Immunol. 2020, 5, eabb4432. [Google Scholar] [CrossRef]
- Zhang, Q.; Zeng, Z.; Wei, N.; Su, Y.; Wang, J.; Ni, Q.; Wang, Y.; Yang, J.; Liu, X.; Xu, H.; et al. Mesenteric Lymph Nodes: A Critical Site for the up-Regulatory Effect of hUC-MSCs on Treg Cells by Producing TGF-β1 in Colitis Treatment. Stem Cell Res. Ther. 2024, 15, 190. [Google Scholar] [CrossRef]
- Eiro, N.; Fraile, M.; González-Jubete, A.; González, L.O.; Vizoso, F.J. Mesenchymal (Stem) Stromal Cells Based as New Therapeutic Alternative in Inflammatory Bowel Disease: Basic Mechanisms, Experimental and Clinical Evidence, and Challenges. Int. J. Mol. Sci. 2022, 23, 8905. [Google Scholar] [CrossRef] [PubMed]
- Stavely, R.; Robinson, A.M.; Fraser, S.; Filippone, R.T.; Stojanovska, V.; Eri, R.; Apostolopoulos, V.; Sakkal, S.; Nurgali, K. Bone Marrow-Derived Mesenchymal Stem Cells Mitigate Chronic Colitis and Enteric Neuropathy via Anti-Inflammatory and Anti-Oxidative Mechanisms. Sci. Rep. 2024, 14, 6649. [Google Scholar] [CrossRef]
- Dave, M.; Dev, A.; Somoza, R.A.; Zhao, N.; Viswanath, S.; Mina, P.R.; Chirra, P.; Obmann, V.C.; Mahabeleshwar, G.H.; Menghini, P.; et al. MSCs Mediate Long-Term Efficacy in a Crohn’s Disease Model by Sustained Anti-Inflammatory Macrophage Programming via Efferocytosis. npj Regen. Med. 2024, 9, 6. [Google Scholar] [CrossRef]
- Scanu, M.; Toto, F.; Petito, V.; Masi, L.; Fidaleo, M.; Puca, P.; Baldelli, V.; Reddel, S.; Vernocchi, P.; Pani, G.; et al. An Integrative Multi-Omic Analysis Defines Gut Microbiota, Mycobiota, and Metabolic Fingerprints in Ulcerative Colitis Patients. Front. Cell. Infect. Microbiol. 2024, 14, 1366192. [Google Scholar] [CrossRef]
- Luchen, C.C.; Chibuye, M.; Spijker, R.; Simuyandi, M.; Chisenga, C.; Bosomprah, S.; Chilengi, R.; Schultsz, C.; Mende, D.R.; Harris, V.C. Impact of Antibiotics on Gut Microbiome Composition and Resistome in the First Years of Life in Low- to Middle-Income Countries: A Systematic Review. PLoS Med. 2023, 20, e1004235. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; He, Y.; Yuan, J.; Zhao, S.; Ma, M.; Chai, Z.; Ji, X.; Hu, X.; He, C.; Zhou, D.; et al. The Role of Antibiotic Exposure as Risk Factor for IBD Epidemic: An Updated Meta-Analysis. J. Gastroenterol. Hepatol. 2024, 39, 2561–2571. [Google Scholar] [CrossRef]
- Mårild, K.; Lerchova, T.; Östensson, M.; Imberg, H.; Størdal, K.; Ludvigsson, J. Early-Life Infections, Antibiotics and Later Risk of Childhood and Early Adult-Onset Inflammatory Bowel Disease: Pooled Analysis of Two Scandinavian Birth Cohorts. Aliment. Pharmacol. Ther. 2024, 61, 323–334. [Google Scholar] [CrossRef]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A Microbial Signature for Crohn’s Disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, L.I.H.; Morgan, X.C. Searching for a Consensus Among Inflammatory Bowel Disease Studies: A Systematic Meta-Analysis. Inflamm. Bowel Dis. 2023, 29, 125–139. [Google Scholar] [CrossRef]
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial Imbalance in Inflammatory Bowel Disease Patients at Different Taxonomic Levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.A.; Sergaki, C.; Logan, A.; Iriarte, R.; Bannaga, A.; Chandrapalan, S.; Wellington, E.M.H.; Rijpkema, S.; Arasaradnam, R.P. Exploring How Microbiome Signatures Change across Inflammatory Bowel Disease Conditions and Disease Locations. Sci. Rep. 2021, 11, 18699. [Google Scholar] [CrossRef]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt+ Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 2019, 50, 212–224.e4. [Google Scholar] [CrossRef]
- Sugihara, K.; Kamada, N. Metabolic Network of the Gut Microbiota in Inflammatory Bowel Disease. Inflamm. Regen. 2024, 44, 11. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Su, T.; Chen, W.; Wang, D.; Xue, Y.; Lu, Q.; Jiang, C.; Ni, Q.; Mao, E.; Peng, Y. Clostridioides Difficile Aggravates Dextran Sulfate Solution (DSS)-Induced Colitis by Shaping the Gut Microbiota and Promoting Neutrophil Recruitment. Gut Microbes 2023, 15, 2192478. [Google Scholar] [CrossRef] [PubMed]
- Leon-Coria, A.; Kumar, M.; Workentine, M.; Moreau, F.; Surette, M.; Chadee, K. Muc2 Mucin and Nonmucin Microbiota Confer Distinct Innate Host Defense in Disease Susceptibility and Colonic Injury. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 77–98. [Google Scholar] [CrossRef]
- Li, M.; Ding, Y.; Wei, J.; Dong, Y.; Wang, J.; Dai, X.; Yan, J.; Chu, F.; Zhang, K.; Meng, F.; et al. Gut Microbiota Metabolite Indole-3-Acetic Acid Maintains Intestinal Epithelial Homeostasis Through Mucin Sulfation. Gut Microbes 2024, 16, 2377576. [Google Scholar] [CrossRef]
- Leibovitzh, H.; Lee, S.-H.; Xue, M.; Raygoza Garay, J.A.; Hernandez-Rocha, C.; Madsen, K.L.; Meddings, J.B.; Guttman, D.S.; Espin-Garcia, O.; Smith, M.I.; et al. Altered Gut Microbiome Composition and Function Are Associated with Gut Barrier Dysfunction in Healthy Relatives of Patients With Crohn’s Disease. Gastroenterology 2022, 163, 1364–1376.e10. [Google Scholar] [CrossRef] [PubMed]
- Luchetti, M.M.; Ciccia, F.; Avellini, C.; Benfaremo, D.; Rizzo, A.; Spadoni, T.; Svegliati, S.; Marzioni, D.; Santinelli, A.; Costantini, A.; et al. Gut Epithelial Impairment, Microbial Translocation and Immune System Activation in Inflammatory Bowel Disease–Associated Spondyloarthritis. Rheumatology 2021, 60, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Alexander, K.L.; Zhao, Q.; Reif, M.; Rosenberg, A.F.; Mannon, P.J.; Duck, L.W.; Elson, C.O. Human Microbiota Flagellins Drive Adaptive Immune Responses in Crohn’s Disease. Gastroenterology 2021, 161, 522–535.e6. [Google Scholar] [CrossRef] [PubMed]
- Vujkovic-Cvijin, I.; Welles, H.C.; Ha, C.W.Y.; Huq, L.; Mistry, S.; Brenchley, J.M.; Trinchieri, G.; Devkota, S.; Belkaid, Y. The Systemic Anti-Microbiota IgG Repertoire Can Identify Gut Bacteria That Translocate Across Gut Barrier Surfaces. Sci. Transl. Med. 2022, 14, eabl3927. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, K.; Catesson, A.; Griffin, J.L.; Holmes, E.; Williams, H.R.T. Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review. J. Crohns Colitis 2021, 15, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Zhou, Y.-L.; Sun, H.; Zhang, Y.; Shen, C.; Wang, Z.; Xuan, B.; Zhao, Y.; Ma, Y.; Yan, Y.; et al. Microbiome and Metabolome Features in Inflammatory Bowel Disease via Multi-Omics Integration Analyses across Cohorts. Nat. Commun. 2023, 14, 7135. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, Y.; Xu, J.; Zhang, Y.; Wang, H.; Zhao, C.; Huang, H.; Yang, J.; Huang, C.; Li, Y.; et al. Short-Chain Fatty Acid-Producing Bacterial Strains Attenuate Experimental Ulcerative Colitis by Promoting M2 Macrophage Polarization via JAK/STAT3/FOXO3 Axis Inactivation. J. Transl. Med. 2024, 22, 369. [Google Scholar] [CrossRef]
- Tian, X.; Li, S.; Wang, C.; Zhang, Y.; Feng, X.; Yan, Q.; Guo, R.; Wu, F.; Wu, C.; Wang, Y.; et al. Gut Virome-Wide Association Analysis Identifies Cross-Population Viral Signatures for Inflammatory Bowel Disease. Microbiome 2024, 12, 130. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, F.; Massimino, L.; Furfaro, F.; Rimoldi, V.; Peyrin-Biroulet, L.; D’Alessio, S.; Danese, S. Metagenomic Analysis of Intestinal Mucosa Revealed a Specific Eukaryotic Gut Virome Signature in Early-Diagnosed Inflammatory Bowel Disease. Gut Microbes 2019, 10, 149–158. [Google Scholar] [CrossRef]
- Bernardi, F.; Ungaro, F.; D’Amico, F.; Zilli, A.; Parigi, T.L.; Massimino, L.; Allocca, M.; Danese, S.; Furfaro, F. The Role of Viruses in the Pathogenesis of Immune-Mediated Gastro-Intestinal Diseases. Int. J. Mol. Sci. 2024, 25, 8301. [Google Scholar] [CrossRef] [PubMed]
- Carasso, S.; Zaatry, R.; Hajjo, H.; Kadosh-Kariti, D.; Ben-Assa, N.; Naddaf, R.; Mandelbaum, N.; Pressman, S.; Chowers, Y.; Gefen, T.; et al. Inflammation and Bacteriophages Affect DNA Inversion States and Functionality of the Gut Microbiota. Cell Host Microbe 2024, 32, 322–334.e9. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Ding, H.; Gong, S.; Luo, Y.; Lin, H.; Mu, Y.; Li, H.; Li, X.; Zhong, M. Fungal Dysbiosis Facilitates Inflammatory Bowel Disease by Enhancing CD4+ T Cell Glutaminolysis. Front. Cell. Infect. Microbiol. 2023, 13, 1140757. [Google Scholar] [CrossRef]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef]
- Zhang, W.; Lyu, M.; Bessman, N.J.; Xie, Z.; Arifuzzaman, M.; Yano, H.; Parkhurst, C.N.; Chu, C.; Zhou, L.; Putzel, G.G.; et al. Gut-Innervating Nociceptors Regulate the Intestinal Microbiota to Promote Tissue Protection. Cell 2022, 185, 4170–4189.e20. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Deleu, S.; Godny, L.; Petito, V.; Puca, P.; Facciotti, F.; Sokol, H.; Ianiro, G.; Masucci, L.; Abreu, M.; et al. The First International Rome Consensus Conference on Gut Microbiota and Faecal Microbiota Transplantation in Inflammatory Bowel Disease. Gut 2023, 72, 1642–1650. [Google Scholar] [CrossRef] [PubMed]
- Costello, S.P.; Soo, W.; Bryant, R.V.; Jairath, V.; Hart, A.L.; Andrews, J.M. Systematic Review with Meta-Analysis: Faecal Microbiota Transplantation for the Induction of Remission for Active Ulcerative Colitis. Aliment. Pharmacol. Ther. 2017, 46, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Caldeira, L.d.F.; Borba, H.H.; Tonin, F.S.; Wiens, A.; Fernandez-Llimos, F.; Pontarolo, R. Fecal Microbiota Transplantation in Inflammatory Bowel Disease Patients: A Systematic Review and Meta-Analysis. PLoS ONE 2020, 15, e0238910. [Google Scholar] [CrossRef] [PubMed]
- Loddo, I.; Romano, C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front. Immunol. 2015, 6, 551. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-C.; Stappenbeck, T.S. Genetics and Pathogenesis of Inflammatory Bowel Disease. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 127–148. [Google Scholar] [CrossRef]
- Palmieri, O.; Latiano, A.; Valvano, R.; D’Incà, R.; Vecchi, M.; Sturniolo, G.C.; Saibeni, S.; Peyvandi, F.; Bossa, F.; Zagaria, C.; et al. Variants of OCTN1–2 Cation Transporter Genes Are Associated with Both Crohn’s Disease and Ulcerative Colitis. Aliment. Pharmacol. Ther. 2006, 23, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Vich Vila, A.; Gacesa, R.; Collij, V.; Stevens, C.; Fu, J.M.; Wong, I.; Talkowski, M.E.; Rivas, M.A.; Imhann, F.; et al. Whole Exome Sequencing Analyses Reveal Gene–Microbiota Interactions in the Context of IBD. Gut 2020, 70, 285–296. [Google Scholar] [CrossRef]
- Galluccio, M.; Tripicchio, M.; Pochini, L. The Human OCTN Sub-Family: Gene and Protein Structure, Expression, and Regulation. Int. J. Mol. Sci. 2024, 25, 8743. [Google Scholar] [CrossRef]
- Del Chierico, F.; Masi, L.; Petito, V.; Baldelli, V.; Puca, P.; Benvenuto, R.; Fidaleo, M.; Palucci, I.; Lopetuso, L.R.; Caristo, M.E.; et al. Solute Transporter OCTN1/Slc22a4 Affects Disease Severity and Response to Infliximab in Experimental Colitis: Role of Gut Microbiota and Immune Modulation. Inflamm. Bowel Dis. 2024, 30, 2259–2270. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Yang, Z.; Chen, B.; Zhong, X. The Multifaceted Role of CARD9 in Inflammatory Bowel Disease. J. Cell. Mol. Med. 2020, 24, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Michaudel, C.; Skerniskyte, J.; Planchais, J.; Magniez, A.; Agus, A.; Michel, M.-L.; Lamas, B.; Da Costa, G.; Spatz, M.; et al. CARD9 in Neutrophils Protects from Colitis and Controls Mitochondrial Metabolism and Cell Survival. Gut 2023, 72, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Lamas, B.; Lavelle, A.; Michel, M.-L.; Da Costa, G.; Pham, H.-P.; Lefevre, A.; Bridonneau, C.; Bredon, M.; Planchais, J.; et al. Dissecting the Respective Roles of Microbiota and Host Genetics in the Susceptibility of Card9−/− Mice to Colitis. Microbiome 2024, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Valatas, V.; Kolios, G.; Bamias, G. TL1A (TNFSF15) and DR3 (TNFRSF25): A Co-Stimulatory System of Cytokines With Diverse Functions in Gut Mucosal Immunity. Front. Immunol. 2019, 10, 583. [Google Scholar] [CrossRef]
- Sun, R.; Hedl, M.; Abraham, C. TNFSF15 Promotes Antimicrobial Pathways in Human Macrophages and These Are Modulated by TNFSF15 Disease-Risk Variants. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 249–272. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, Y.; Ichikawa, R.; Fuyuno, Y.; Hirano, A.; Umeno, J.; Torisu, T.; Watanabe, K.; Asakura, A.; Nakano, T.; Izumiyama, Y.; et al. An Integrated Genomic and Transcriptomic Analysis Reveals Candidates of Susceptibility Genes for Crohn’s Disease in Japanese Populations. Sci. Rep. 2020, 10, 10236. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, Y.; Jiang, H.; Chen, Z.; Lu, B.; Li, J.; Shen, X. Polymorphism Rs6478109 in the TNFSF15 Gene Contributes to the Susceptibility to Crohn’s Disease but Not Ulcerative Colitis: A Meta-Analysis. J. Int. Med. Res. 2020, 48, 0300060520961675. [Google Scholar] [CrossRef] [PubMed]
- Muise, A.M.; Snapper, S.B.; Kugathasan, S. The Age of Gene Discovery in Very Early Onset Inflammatory Bowel Disease. Gastroenterology 2012, 143, 285–288. [Google Scholar] [CrossRef]
- Uhlig, H.H.; Schwerd, T.; Koletzko, S.; Shah, N.; Kammermeier, J.; Elkadri, A.; Ouahed, J.; Wilson, D.C.; Travis, S.P.; Turner, D.; et al. The Diagnostic Approach to Monogenic Very Early Onset Inflammatory Bowel Disease. Gastroenterology 2014, 147, 990–1007.e3. [Google Scholar] [CrossRef]
- Nambu, R.; Warner, N.; Mulder, D.J.; Kotlarz, D.; McGovern, D.P.B.; Cho, J.; Klein, C.; Snapper, S.B.; Griffiths, A.M.; Iwama, I.; et al. A Systematic Review of Monogenic Inflammatory Bowel Disease. Clin. Gastroenterol. Hepatol. 2022, 20, e653–e663. [Google Scholar] [CrossRef] [PubMed]
- Arnadottir, G.A.; Norddahl, G.L.; Gudmundsdottir, S.; Agustsdottir, A.B.; Sigurdsson, S.; Jensson, B.O.; Bjarnadottir, K.; Theodors, F.; Benonisdottir, S.; Ivarsdottir, E.V.; et al. A Homozygous Loss-of-Function Mutation Leading to CYBC1 Deficiency Causes Chronic Granulomatous Disease. Nat. Commun. 2018, 9, 4447. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Motal, U.M.; Hubrack, S.Z.; Bullock, A.N.; Al-Marri, A.A.; Agrebi, N.; Al-Subaiey, A.A.; Ibrahim, N.A.; Charles, A.K.; Al-Kaabi, S.R.; Al-Mohannadi, M.J.; et al. Human AGR2 Deficiency Causes Mucus Barrier Dysfunction and Infantile Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1809–1830. [Google Scholar] [CrossRef]
- Neehus, A.-L.; Moriya, K.; Nieto-Patlán, A.; Le Voyer, T.; Lévy, R.; Özen, A.; Karakoc-Aydiner, E.; Baris, S.; Yildiran, A.; Altundag, E.; et al. Impaired Respiratory Burst Contributes to Infections in PKCδ-Deficient Patients. J. Exp. Med. 2021, 218, e20210501. [Google Scholar] [CrossRef] [PubMed]
- Ozen, A.; Comrie, W.A.; Ardy, R.C.; Conde, C.D.; Dalgic, B.; Beser, F.; Morawski, A.R.; Karakoc-Aydiner, E.; Tutar, E.; Baris, S.; et al. CD55 Deficiency, Early-Onset Protein-Losing Enteropathy, and Thrombosis. N. Engl. J. Med. 2017, 377, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Azabdaftari, A.; Jones, K.D.J.; Kammermeier, J.; Uhlig, H.H. Monogenic Inflammatory Bowel Disease-Genetic Variants, Functional Mechanisms and Personalised Medicine in Clinical Practice. Hum. Genet. 2023, 142, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, P.; Andrighetti, T.; Verstockt, S.; Caenepeel, C.; Ferrante, M.; Sabino, J.; Verstockt, B.; Vermeire, S. Integrated Analysis of Microbe-Host Interactions in Crohn’s Disease Reveals Potential Mechanisms of Microbial Proteins on Host Gene Expression. iScience 2022, 25, 103963. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Bernstein, C.N.; Vatn, M.H.; Lakatos, P.L.; Loftus, E.V.; Tysk, C.; O’Morain, C.; Moum, B.; Colombel, J.-F. Epidemiology and Natural History Task Force of the International Organization of Inflammatory Bowel Disease (IOIBD) Geographical Variability and Environmental Risk Factors in Inflammatory Bowel Disease. Gut 2013, 62, 630–649. [Google Scholar] [CrossRef] [PubMed]
- Mahid, S.S.; Minor, K.S.; Soto, R.E.; Hornung, C.A.; Galandiuk, S. Smoking and Inflammatory Bowel Disease: A Meta-Analysis. Mayo Clin. Proc. 2006, 81, 1462–1471. [Google Scholar] [CrossRef] [PubMed]
- Dunford, A.R.; Sangster, J.M. Maternal and Paternal Periconceptional Nutrition as an Indicator of Offspring Metabolic Syndrome Risk in Later Life through Epigenetic Imprinting: A Systematic Review. Diabetes Metab. Syndr. 2017, 11 (Suppl. S2), S655–S662. [Google Scholar] [CrossRef]
- Zong, D.; Liu, X.; Li, J.; Ouyang, R.; Chen, P. The Role of Cigarette Smoke-Induced Epigenetic Alterations in Inflammation. Epigenet. Chromatin 2019, 12, 65. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, A.; Patro-Małysza, J.; Kimber-Trojnar, Ż.; Marciniak, B.; Oleszczuk, J.; Leszczyńska-Gorzelak, B. Fetal Programming of the Metabolic Syndrome. Taiwan. J. Obstet. Gynecol. 2017, 56, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Grossniklaus, U.; Kelly, W.G.; Kelly, B.; Ferguson-Smith, A.C.; Pembrey, M.; Lindquist, S. Transgenerational Epigenetic Inheritance: How Important Is It? Nat. Rev. Genet. 2013, 14, 228–235. [Google Scholar] [CrossRef]
- Ventham, N.T.; Kennedy, N.A.; Nimmo, E.R.; Satsangi, J. Beyond Gene Discovery in Inflammatory Bowel Disease: The Emerging Role of Epigenetics. Gastroenterology 2013, 145, 293–308. [Google Scholar] [CrossRef]
- Schaible, T.D.; Harris, R.A.; Dowd, S.E.; Smith, C.W.; Kellermayer, R. Maternal Methyl-Donor Supplementation Induces Prolonged Murine Offspring Colitis Susceptibility in Association with Mucosal Epigenetic and Microbiomic Changes. Hum. Mol. Genet. 2011, 20, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.I.; McFadden, T.; Link, V.M.; Han, S.-J.; Karlsson, R.-M.; Stacy, A.; Farley, T.K.; Lima-Junior, D.S.; Harrison, O.J.; Desai, J.V.; et al. Prenatal Maternal Infection Promotes Tissue-Specific Immunity and Inflammation in Offspring. Science 2021, 373, eabf3002. [Google Scholar] [CrossRef] [PubMed]
- Van Der Sloot, K.W.J.; Weersma, R.K.; Alizadeh, B.Z.; Dijkstra, G. Identification of Environmental Risk Factors Associated With the Development of Inflammatory Bowel Disease. J. Crohns Colitis 2020, 14, 1662–1671. [Google Scholar] [CrossRef]
- Wiklund, P.; Karhunen, V.; Richmond, R.C.; Parmar, P.; Rodriguez, A.; De Silva, M.; Wielscher, M.; Rezwan, F.I.; Richardson, T.G.; Veijola, J.; et al. DNA Methylation Links Prenatal Smoking Exposure to Later Life Health Outcomes in Offspring. Clin. Epigenet. 2019, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Fragou, D.; Pakkidi, E.; Aschner, M.; Samanidou, V.; Kovatsi, L. Smoking and DNA Methylation: Correlation of Methylation with Smoking Behavior and Association with Diseases and Fetus Development Following Prenatal Exposure. Food Chem. Toxicol. 2019, 129, 312–327. [Google Scholar] [CrossRef]
- McLean, C.; Jun, S.; Kozyrskyj, A. Impact of Maternal Smoking on the Infant Gut Microbiota and Its Association with Child Overweight: A Scoping Review. World J. Pediatr. 2019, 15, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lochhead, P.; Ko, Y.; Claggett, B.; Leong, R.W.; Ananthakrishnan, A.N. Systematic Review with Meta-Analysis: Breastfeeding and the Risk of Crohn’s Disease and Ulcerative Colitis. Aliment. Pharmacol. Ther. 2017, 46, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Klement, E.; Cohen, R.V.; Boxman, J.; Joseph, A.; Reif, S. Breastfeeding and Risk of Inflammatory Bowel Disease: A Systematic Review with Meta-Analysis. Am. J. Clin. Nutr. 2004, 80, 1342–1352. [Google Scholar] [CrossRef]
- Gordon, H.; Blad, W.; Trier Møller, F.; Orchard, T.; Steel, A.; Trevelyan, G.; Ng, S.; Harbord, M. UK IBD Twin Registry: Concordance and Environmental Risk Factors of Twins with IBD. Dig. Dis. Sci. 2022, 67, 2444–2450. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; van den Brandt, P.A.; Kummeling, I.; Snijders, B.; Stelma, F.; Adams, H.; van Ree, R.; Stobberingh, E.E. Gut Microbiota Composition and Development of Atopic Manifestations in Infancy: The KOALA Birth Cohort Study. Gut 2007, 56, 661–667. [Google Scholar] [CrossRef]
- M’Rabet, L.; Vos, A.P.; Boehm, G.; Garssen, J. Breast-Feeding and Its Role in Early Development of the Immune System in Infants: Consequences for Health Later in Life1. J. Nutr. 2008, 138, 1782S–1790S. [Google Scholar] [CrossRef]
- Keag, O.E.; Norman, J.E.; Stock, S.J. Long-Term Risks and Benefits Associated with Cesarean Delivery for Mother, Baby, and Subsequent Pregnancies: Systematic Review and Meta-Analysis. PLoS Med. 2018, 15, e1002494. [Google Scholar] [CrossRef]
- Li, Y.; Tian, Y.; Zhu, W.; Gong, J.; Gu, L.; Zhang, W.; Guo, Z.; Li, N.; Li, J. Cesarean Delivery and Risk of Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Scand. J. Gastroenterol. 2014, 49, 834–844. [Google Scholar] [CrossRef]
- Bruce, A.; Black, M.; Bhattacharya, S. Mode of Delivery and Risk of Inflammatory Bowel Disease in the Offspring: Systematic Review and Meta-Analysis of Observational Studies. Inflamm. Bowel Dis. 2014, 20, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Gosalbes, M.J.; Friedrichs, A.; Knecht, H.; Artacho, A.; Eismann, K.; Otto, W.; Rojo, D.; Bargiela, R.; von Bergen, M.; et al. Gut Microbiota Disturbance during Antibiotic Therapy: A Multi-Omic Approach. Gut 2013, 62, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, R.; Bernstein, C.N.; Gearry, R.; Hviid, A.; Kolho, K.-L.; Kronman, M.P.; Shaw, S.; Van Kruiningen, H.; Colombel, J.-F.; Atreja, A. Antibiotics Associated with Increased Risk of New-Onset Crohn’s Disease but Not Ulcerative Colitis: A Meta-Analysis. Am. J. Gastroenterol. 2014, 109, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The Gut Microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Cholapranee, A.; Ananthakrishnan, A.N. Environmental Hygiene and Risk of Inflammatory Bowel Diseases: A Systematic Review and Meta-Analysis. Inflamm. Bowel Dis. 2016, 22, 2191–2199. [Google Scholar] [CrossRef]
- Michaux, M.; Chan, J.M.; Bergmann, L.; Chaves, L.F.; Klinkenberg, B.; Jacobson, K. Spatial Cluster Mapping and Environmental Modeling in Pediatric Inflammatory Bowel Disease. World J. Gastroenterol. 2023, 29, 3688–3702. [Google Scholar] [CrossRef]
- Song, C.; Yang, J.; Ye, W.; Zhang, Y.; Tang, C.; Li, X.; Zhou, X.; Xie, Y. Urban-Rural Environmental Exposure during Childhood and Subsequent Risk of Inflammatory Bowel Disease: A Meta-Analysis. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 591–602. [Google Scholar] [CrossRef]
- Wu, X.-W.; Ji, H.-Z.; Yang, M.-F.; Wu, L.; Wang, F.-Y. Helicobacter Pylori Infection and Inflammatory Bowel Disease in Asians: A Meta-Analysis. World J. Gastroenterol. 2015, 21, 4750–4756. [Google Scholar] [CrossRef]
- Castaño-Rodríguez, N.; Kaakoush, N.O.; Lee, W.S.; Mitchell, H.M. Dual Role of Helicobacter and Campylobacter Species in IBD: A Systematic Review and Meta-Analysis. Gut 2017, 66, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Luther, J.; Dave, M.; Higgins, P.D.R.; Kao, J.Y. Association between Helicobacter Pylori Infection and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Inflamm. Bowel Dis. 2010, 16, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Rokkas, T.; Gisbert, J.P.; Niv, Y.; O’Morain, C. The Association between Helicobacter Pylori Infection and Inflammatory Bowel Disease Based on Meta-Analysis. United Eur. Gastroenterol. J. 2015, 3, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Codolo, G.; Mazzi, P.; Amedei, A.; Del Prete, G.; Berton, G.; D’Elios, M.M.; de Bernard, M. The Neutrophil-Activating Protein of Helicobacter Pylori down-Modulates Th2 Inflammation in Ovalbumin-Induced Allergic Asthma. Cell. Microbiol. 2008, 10, 2355–2363. [Google Scholar] [CrossRef]
- Luther, J.; Owyang, S.Y.; Takeuchi, T.; Cole, T.S.; Zhang, M.; Liu, M.; Erb-Downward, J.; Rubenstein, J.H.; Chen, C.-C.; Pierzchala, A.V.; et al. Helicobacter Pylori DNA Decreases Pro-Inflammatory Cytokine Production by Dendritic Cells and Attenuates Dextran Sodium Sulphate-Induced Colitis. Gut 2011, 60, 1479–1486. [Google Scholar] [CrossRef]
- Arnold, I.C.; Hitzler, I.; Müller, A. The Immunomodulatory Properties of Helicobacter Pylori Confer Protection against Allergic and Chronic Inflammatory Disorders. Front. Cell. Infect. Microbiol. 2012, 2, 10. [Google Scholar] [CrossRef]
- Okada, H.; Kuhn, C.; Feillet, H.; Bach, J.-F. The “hygiene Hypothesis” for Autoimmune and Allergic Diseases: An Update. Clin. Exp. Immunol. 2010, 160, 1–9. [Google Scholar] [CrossRef]
- Schaub, B.; Liu, J.; Höppler, S.; Schleich, I.; Huehn, J.; Olek, S.; Wieczorek, G.; Illi, S.; von Mutius, E. Maternal Farm Exposure Modulates Neonatal Immune Mechanisms through Regulatory T Cells. J. Allergy Clin. Immunol. 2009, 123, 774–782.e5. [Google Scholar] [CrossRef]
- Hasosah, M.; Alhashmi, W.; Abualsaud, R.; Alamoudi, A.; Aljawad, A.; Tunkar, M.; Felemban, N.; Basalim, A.; Khan, M.; Alanazi, A.; et al. Environmental Risk Factors for Childhood Inflammatory Bowel Diseases: A Multicenter Case-Control Study. Children 2022, 9, 438. [Google Scholar] [CrossRef]
- Strisciuglio, C.; Giugliano, F.; Martinelli, M.; Cenni, S.; Greco, L.; Staiano, A.; Miele, E. Impact of Environmental and Familial Factors in a Cohort of Pediatric Patients With Inflammatory Bowel Disease. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Piovani, D.; Danese, S.; Peyrin-Biroulet, L.; Nikolopoulos, G.K.; Lytras, T.; Bonovas, S. Environmental Risk Factors for Inflammatory Bowel Diseases: An Umbrella Review of Meta-Analyses. Gastroenterology 2019, 157, 647–659.e4. [Google Scholar] [CrossRef]
- Jakobsen, C.; Paerregaard, A.; Munkholm, P.; Wewer, V. Environmental Factors and Risk of Developing Paediatric Inflammatory Bowel Disease—A Population Based Study 2007–2009. J. Crohns Colitis 2013, 7, 79–88. [Google Scholar] [CrossRef]
- Sidik, S. Chronic Stress Can Inflame the Gut—Now Scientists Know Why. Nature 2023, 618, 221–222. [Google Scholar] [CrossRef]
- Schneider, K.M.; Blank, N.; Alvarez, Y.; Thum, K.; Lundgren, P.; Litichevskiy, L.; Sleeman, M.; Bahnsen, K.; Kim, J.; Kardo, S.; et al. The Enteric Nervous System Relays Psychological Stress to Intestinal Inflammation. Cell 2023, 186, 2823–2838.e20. [Google Scholar] [CrossRef]
- Bayrer, J.R.; Castro, J.; Venkataraman, A.; Touhara, K.K.; Rossen, N.D.; Morrie, R.D.; Maddern, J.; Hendry, A.; Braverman, K.N.; Garcia-Caraballo, S.; et al. Gut Enterochromaffin Cells Drive Visceral Pain and Anxiety. Nature 2023, 616, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Karl, J.P.; Hatch, A.M.; Arcidiacono, S.M.; Pearce, S.C.; Pantoja-Feliciano, I.G.; Doherty, L.A.; Soares, J.W. Effects of Psychological, Environmental and Physical Stressors on the Gut Microbiota. Front. Microbiol. 2018, 9, 2013. [Google Scholar] [CrossRef] [PubMed]
- Oligschlaeger, Y.; Yadati, T.; Houben, T.; Condello Oliván, C.M.; Shiri-Sverdlov, R. Inflammatory Bowel Disease: A Stressed “Gut/Feeling”. Cells 2019, 8, 659. [Google Scholar] [CrossRef] [PubMed]
- Konturek, P.C.; Brzozowski, T.; Konturek, S.J. Stress and the Gut: Pathophysiology, Clinical Consequences, Diagnostic Approach and Treatment Options. J. Physiol. Pharmacol. 2011, 62, 591–599. [Google Scholar] [PubMed]
- Hu, D.; Ren, J.; Wang, G.; Gu, G.; Liu, S.; Wu, X.; Chen, J.; Ren, H.; Hong, Z.; Li, J. Geographic Mapping of Crohn’s Disease and Its Relation to Affluence in Jiangsu Province, an Eastern Coastal Province of China. Gastroenterol. Res. Pract. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.G.; Hubbard, J.; Korzenik, J.; Sands, B.E.; Panaccione, R.; Ghosh, S.; Wheeler, A.J.; Villeneuve, P.J. The Inflammatory Bowel Diseases and Ambient Air Pollution: A Novel Association. Am. J. Gastroenterol. 2010, 105, 2412–2419. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, D.K.; Heilmann, R.M.; Paital, B.; Patel, A.; Yadav, V.K.; Wong, D.; Jergens, A.E. Oxidative Stress, Hormones, and Effects of Natural Antioxidants on Intestinal Inflammation in Inflammatory Bowel Disease. Front. Endocrinol. 2023, 14, 1217165. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, Z.; Zhang, J. Pathomechanisms of Oxidative Stress in Inflammatory Bowel Disease and Potential Antioxidant Therapies. Oxid. Med. Cell. Longev. 2017, 2017, 4535194. [Google Scholar] [CrossRef]
- Qin, J.; Xia, W.; Liang, G.; Xu, S.; Zhao, X.; Wang, D.; Sun, X.; Li, Y.; Liu, H. Association of Fine Particulate Matter with Glucose and Lipid Metabolism: A Longitudinal Study in Young Adults. Occup. Environ. Med. 2021, 78, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Arancibia, S.; Miranda-Martínez, A.; Rodríguez-Martínez, E.; Hernández-Orozco, E.; Valdés-Fuentes, M.; De La Rosa-Sierra, R. Ozone Environmental Pollution: Relationship between the Intestine and Neurodegenerative Diseases. Antioxidants 2023, 12, 1323. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; McGinley, E.L.; Binion, D.G.; Saeian, K. Ambient Air Pollution Correlates with Hospitalizations for Inflammatory Bowel Disease: An Ecologic Analysis. Inflamm. Bowel Dis. 2011, 17, 1138–1145. [Google Scholar] [CrossRef]
- Adami, G.; Pontalti, M.; Cattani, G.; Rossini, M.; Viapiana, O.; Orsolini, G.; Benini, C.; Bertoldo, E.; Fracassi, E.; Gatti, D.; et al. Association between Long-Term Exposure to Air Pollution and Immune-Mediated Diseases: A Population-Based Cohort Study. RMD Open 2022, 8, e002055. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, T.; Si, B.; Du, H.; Liu, Y.; Waqas, A.; Huang, S.; Zhao, G.; Chen, S.; Xu, A. Intratracheally Instillated Diesel PM2.5 Significantly Altered the Structure and Composition of Indigenous Murine Gut Microbiota. Ecotoxicol. Environ. Saf. 2021, 210, 111903. [Google Scholar] [CrossRef]
- Okafor, P.N.; Dahlen, A.; Youssef, M.; Olayode, A.; Sonu, I.; Neshatian, L.; Nguyen, L.; Charu, V. Environmental Pollutants Are Associated With Irritable Bowel Syndrome in a Commercially Insured Cohort of California Residents. Clin. Gastroenterol. Hepatol. 2023, 21, 1617–1626.e9. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Guieysse, B.; Jefferson, B.; Cartmell, E.; Lester, J.N. Nonylphenol in the Environment: A Critical Review on Occurrence, Fate, Toxicity and Treatment in Wastewaters. Environ. Int. 2008, 34, 1033–1049. [Google Scholar] [CrossRef]
- Wagner, M.; Schlüsener, M.P.; Ternes, T.A.; Oehlmann, J. Identification of Putative Steroid Receptor Antagonists in Bottled Water: Combining Bioassays and High-Resolution Mass Spectrometry. PLoS ONE 2013, 8, e72472. [Google Scholar] [CrossRef]
- Perl, D.P.; Fogarty, U.; Harpaz, N.; Sachar, D.B. Bacterial-Metal Interactions: The Potential Role of Aluminum and Other Trace Elements in the Etiology of Chrohn’s Disease. Inflamm. Bowel Dis. 2004, 10, 881–883. [Google Scholar] [CrossRef]
- Jowett, S.L. Influence of Dietary Factors on the Clinical Course of Ulcerative Colitis: A Prospective Cohort Study. Gut 2004, 53, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Aamodt, G.; Bukholm, G.; Jahnsen, J.; Moum, B.; Vatn, M.H.; The IBSEN Study Group. The Association Between Water Supply and Inflammatory Bowel Disease Based on a 1990–1993 Cohort Study in Southeastern Norway. Am. J. Epidemiol. 2008, 168, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Jädert, C.; Petersson, J.; Massena, S.; Ahl, D.; Grapensparr, L.; Holm, L.; Lundberg, J.O.; Phillipson, M. Decreased Leukocyte Recruitment by Inorganic Nitrate and Nitrite in Microvascular Inflammation and NSAID-Induced Intestinal Injury. Free Radic. Biol. Med. 2012, 52, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Chai, P.; Dong, X.; Liang, Z.; Yang, Z.; Li, J.; Teng, G.; Sun, S.; Xu, M.; Zheng, Z.-J.; et al. Drinking Water Quality and Inflammatory Bowel Disease: A Prospective Cohort Study. Environ. Sci. Pollut. Res. 2023, 30, 71171–71183. [Google Scholar] [CrossRef]
- Johnson, G.J.; Cosnes, J.; Mansfield, J.C. Review Article: Smoking Cessation as Primary Therapy to Modify the Course of Crohn’s Disease. Aliment. Pharmacol. Ther. 2005, 21, 921–931. [Google Scholar] [CrossRef]
- Rozich, J.J.; Holmer, A.; Singh, S. Effect of Lifestyle Factors on Outcomes in Patients With Inflammatory Bowel Diseases. Am. J. Gastroenterol. 2020, 115, 832–840. [Google Scholar] [CrossRef]
- Higuchi, L.M.; Khalili, H.; Chan, A.T.; Richter, J.M.; Bousvaros, A.; Fuchs, C.S. A Prospective Study of Cigarette Smoking and the Risk of Inflammatory Bowel Disease in Women. Am. J. Gastroenterol. 2012, 107, 1399–1406. [Google Scholar] [CrossRef]
- Nasr, S.; Nsiri, I.; Fredj, M.B. Effectiveness of Smoking Cessation Interventions for Smokers with Crohn’s Disease: A Systematic Review. Future Sci. OA 2023, 9, FSO870. [Google Scholar] [CrossRef] [PubMed]
- Nunes, T.; Etchevers, M.J.; Merino, O.; Gallego, S.; García-Sánchez, V.; Marín-Jiménez, I.; Menchén, L.; Barreiro-de Acosta, M.; Bastida, G.; García, S.; et al. High Smoking Cessation Rate in Crohn’s Disease Patients after Physician Advice—The TABACROHN Study. J. Crohns Colitis 2013, 7, 202–207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cosnes, J.; Beaugerie, L.; Carbonnel, F.; Gendre, J. Smoking Cessation and the Course of Crohn’s Disease: An Intervention Study. Gastroenterology 2001, 120, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.A.; Lipowska, A.M.; Zmeter, N.; Perez, E.; Kavitt, R.; Rubin, D.T. Poor Sleep Quality in Crohn’s Disease Is Associated With Disease Activity and Risk for Hospitalization or Surgery. Inflamm. Bowel Dis. 2020, 26, 1251–1259. [Google Scholar] [CrossRef]
- Zhang, J.-Z.; Song, X.-Z.; Song, X.-N.; Shen, Y.-L.; Tang, H.; Li, H. Prevalence and Risk Factors of Sleep Disorders in Inflammatory Bowel Disease: A Cross-Sectional Study. Int. J. Colorectal Dis. 2024, 39, 140. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Orr, W.C. Sleep Disturbances and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2014, 20, 1986–1995. [Google Scholar] [CrossRef]
- Vieujean, S.; Caron, B.; Haghnejad, V.; Jouzeau, J.-Y.; Netter, P.; Heba, A.-C.; Ndiaye, N.C.; Moulin, D.; Barreto, G.; Danese, S.; et al. Impact of the Exposome on the Epigenome in Inflammatory Bowel Disease Patients and Animal Models. Int. J. Mol. Sci. 2022, 23, 7611. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, U.; Mustafi, R.; Zhu, H.; Zhu, X.; Deb, D.; Meredith, S.C.; Ayaloglu-Butun, F.; Fletcher, M.; Sanchez, A.; Pekow, J.; et al. Upregulation of Polycistronic microRNA-143 and microRNA-145 in Colonocytes Suppresses Colitis and Inflammation-Associated Colon Cancer. Epigenetics 2021, 16, 1317–1334. [Google Scholar] [CrossRef]
- Wei, M.; Gao, X.; Liu, L.; Li, Z.; Wan, Z.; Dong, Y.; Chen, X.; Niu, Y.; Zhang, J.; Yang, G. Visceral Adipose Tissue Derived Exosomes Exacerbate Colitis Severity via Pro-Inflammatory MiRNAs in High Fat Diet Fed Mice. ACS Nano 2020, 14, 5099–5110. [Google Scholar] [CrossRef] [PubMed]
- Timms, V.J.; Daskalopoulos, G.; Mitchell, H.M.; Neilan, B.A. The Association of Mycobacterium Avium Subsp. Paratuberculosis with Inflammatory Bowel Disease. PLoS ONE 2016, 11, e0148731. [Google Scholar] [CrossRef]
- Guo, Z.; Cai, X.; Guo, X.; Xu, Y.; Gong, J.; Li, Y.; Zhu, W. Let-7b Ameliorates Crohn’s Disease-Associated Adherent-Invasive E Coli Induced Intestinal Inflammation via Modulating Toll-Like Receptor 4 Expression in Intestinal Epithelial Cells. Biochem. Pharmacol. 2018, 156, 196–203. [Google Scholar] [CrossRef]
- Melhem, H.; Hansmannel, F.; Bressenot, A.; Battaglia-Hsu, S.-F.; Billioud, V.; Alberto, J.M.; Gueant, J.L.; Peyrin-Biroulet, L. Methyl-Deficient Diet Promotes Colitis and SIRT1-Mediated Endoplasmic Reticulum Stress. Gut 2016, 65, 595–606. [Google Scholar] [CrossRef]
- Trakman, G.L.; Lin, W.Y.Y.; Hamilton, A.L.; Wilson-O’Brien, A.L.; Stanley, A.; Ching, J.Y.; Yu, J.; Mak, J.W.Y.; Sun, Y.; Niu, J.; et al. Processed Food as a Risk Factor for the Development and Perpetuation of Crohn’s Disease—The ENIGMA Study. Nutrients 2022, 14, 3627. [Google Scholar] [CrossRef]
- De Chambrun, G.P.; Body-Malapel, M.; Frey-Wagner, I.; Djouina, M.; Deknuydt, F.; Atrott, K.; Esquerre, N.; Altare, F.; Neut, C.; Arrieta, M.C.; et al. Aluminum Enhances Inflammation and Decreases Mucosal Healing in Experimental Colitis in Mice. Mucosal Immunol. 2014, 7, 589–601. [Google Scholar] [CrossRef]
- Becker, H.M.; Bertschinger, M.M.; Rogler, G. Microparticles and Their Impact on Intestinal Immunity. Dig. Dis. 2012, 30, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, S.M. Implications of the Exposome for Exposure Science. J. Expo. Sci. Environ. Epidemiol. 2011, 21, 5–9. [Google Scholar] [CrossRef]
- Vel Szic, K.S.; Declerck, K.; Vidaković, M.; Vanden Berghe, W. From Inflammaging to Healthy Aging by Dietary Lifestyle Choices: Is Epigenetics the Key to Personalized Nutrition? Clin. Epigenet. 2015, 7, 33. [Google Scholar] [CrossRef]
- Lippai, D.; Bala, S.; Catalano, D.; Kodys, K.; Szabo, G. Micro-RNA-155 Deficiency Prevents Alcohol-Induced Serum Endotoxin Increase and Small Bowel Inflammation in Mice. Alcohol. Clin. Exp. Res. 2014, 38, 2217–2224. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Xing, Y.; Liu, J.; Dong, D.; Liu, Y.; Qiao, H.; Zhang, Y.; Hu, L. Lonicerin Targets EZH2 to Alleviate Ulcerative Colitis by Autophagy-Mediated NLRP3 Inflammasome Inactivation. Acta Pharm. Sin. B 2021, 11, 2880–2899. [Google Scholar] [CrossRef]
- Galleggiante, V.; De Santis, S.; Liso, M.; Verna, G.; Sommella, E.; Mastronardi, M.; Campiglia, P.; Chieppa, M.; Serino, G. Quercetin-Induced miR-369-3p Suppresses Chronic Inflammatory Response Targeting C/EBP-β. Mol. Nutr. Food Res. 2019, 63, 1801390. [Google Scholar] [CrossRef]
- James, S.; Aparna, J.S.; Babu, A.; Paul, A.M.; Lankadasari, M.B.; Athira, S.R.; Kumar, S.S.; Vijayan, Y.; Namitha, N.N.; Mohammed, S.; et al. Cardamonin Attenuates Experimental Colitis and Associated Colorectal Cancer. Biomolecules 2021, 11, 661. [Google Scholar] [CrossRef]
- Qiao, C.-X.; Xu, S.; Wang, D.-D.; Gao, S.-Y.; Zhao, S.-F.; Zhang, M.-L.; Yu, B.; Yin, Q.; Zhao, G. MicroRNA-19b Alleviates Lipopolysaccharide-Induced Inflammatory Injury in Human Intestinal Cells by up-Regulation of Runx3. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5284–5294. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Khalili, H.; Konijeti, G.G.; Higuchi, L.M.; de Silva, P.; Korzenik, J.R.; Fuchs, C.S.; Willett, W.C.; Richter, J.M.; Chan, A.T. A Prospective Study of Long-Term Intake of Dietary Fiber and Risk of Crohn’s Disease and Ulcerative Colitis. Gastroenterology 2013, 145, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.K.; Abraham, B.; El-Serag, H. Dietary Intake and Risk of Developing Inflammatory Bowel Disease: A Systematic Review of the Literature. Am. J. Gastroenterol. 2011, 106, 563–573. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvez, V.; Puca, P.; Di Vincenzo, F.; Del Gaudio, A.; Bartocci, B.; Murgiano, M.; Iaccarino, J.; Parand, E.; Napolitano, D.; Pugliese, D.; et al. Novel Insights into the Pathogenesis of Inflammatory Bowel Diseases. Biomedicines 2025, 13, 305. https://doi.org/10.3390/biomedicines13020305
Calvez V, Puca P, Di Vincenzo F, Del Gaudio A, Bartocci B, Murgiano M, Iaccarino J, Parand E, Napolitano D, Pugliese D, et al. Novel Insights into the Pathogenesis of Inflammatory Bowel Diseases. Biomedicines. 2025; 13(2):305. https://doi.org/10.3390/biomedicines13020305
Chicago/Turabian StyleCalvez, Valentin, Pierluigi Puca, Federica Di Vincenzo, Angelo Del Gaudio, Bianca Bartocci, Marco Murgiano, Jacopo Iaccarino, Erfan Parand, Daniele Napolitano, Daniela Pugliese, and et al. 2025. "Novel Insights into the Pathogenesis of Inflammatory Bowel Diseases" Biomedicines 13, no. 2: 305. https://doi.org/10.3390/biomedicines13020305
APA StyleCalvez, V., Puca, P., Di Vincenzo, F., Del Gaudio, A., Bartocci, B., Murgiano, M., Iaccarino, J., Parand, E., Napolitano, D., Pugliese, D., Gasbarrini, A., & Scaldaferri, F. (2025). Novel Insights into the Pathogenesis of Inflammatory Bowel Diseases. Biomedicines, 13(2), 305. https://doi.org/10.3390/biomedicines13020305






