Drug Repurposing for AML: Structure-Based Virtual Screening and Molecular Simulations of FDA-Approved Compounds with Polypharmacological Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Structures Preparation
2.2. Ligand Dataset Preparation
2.3. Molecular Docking
2.4. ADMET Prediction
2.5. Molecular Dynamics Simulations
3. Results
3.1. Docking Protocol Validation
3.2. Structure-Based Virtual Screening Results
3.3. ADMET Analysis
3.4. Molecular Dynamics Trajectory Analysis
3.4.1. LSD1 Complexes Analysis
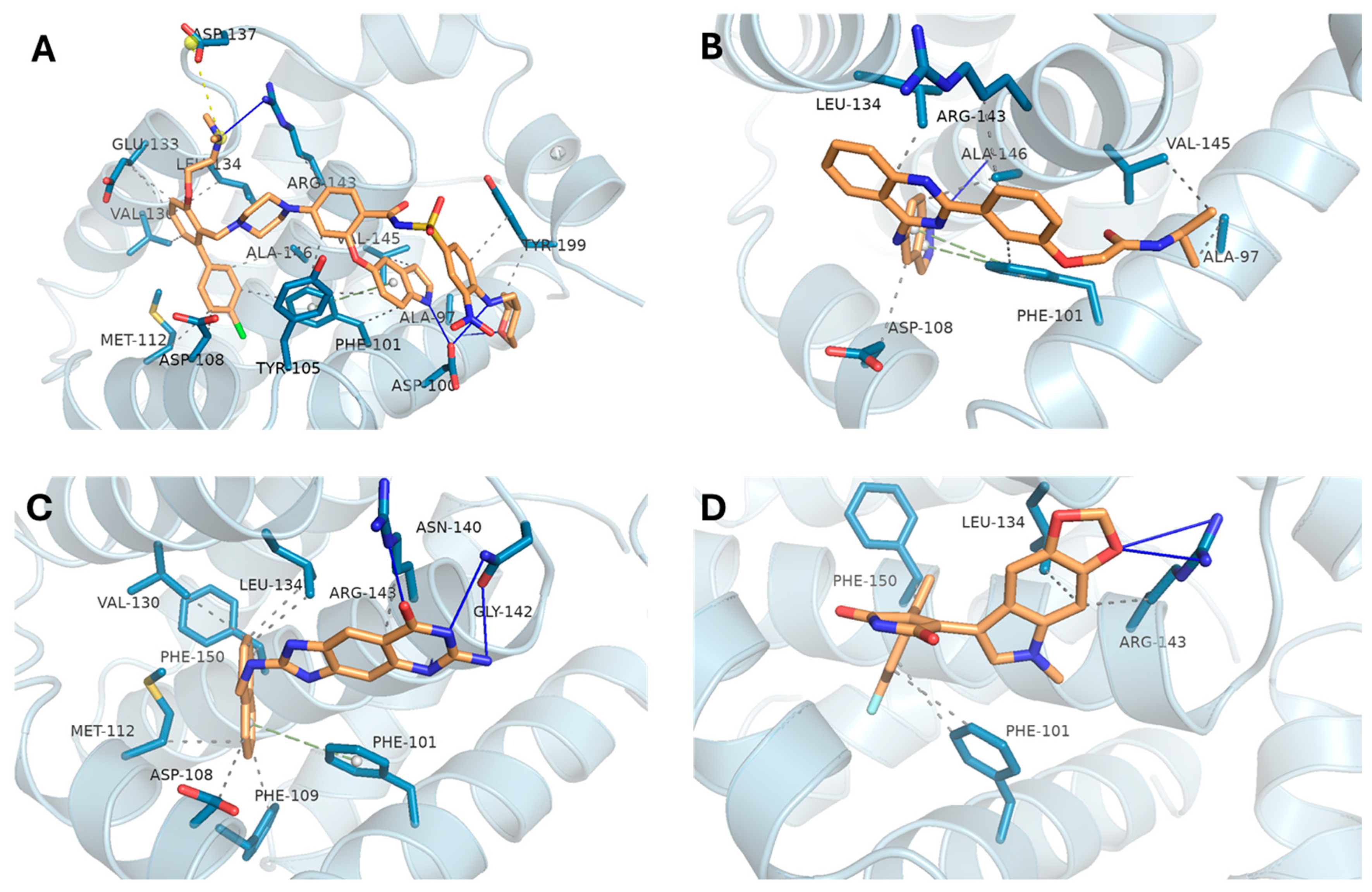
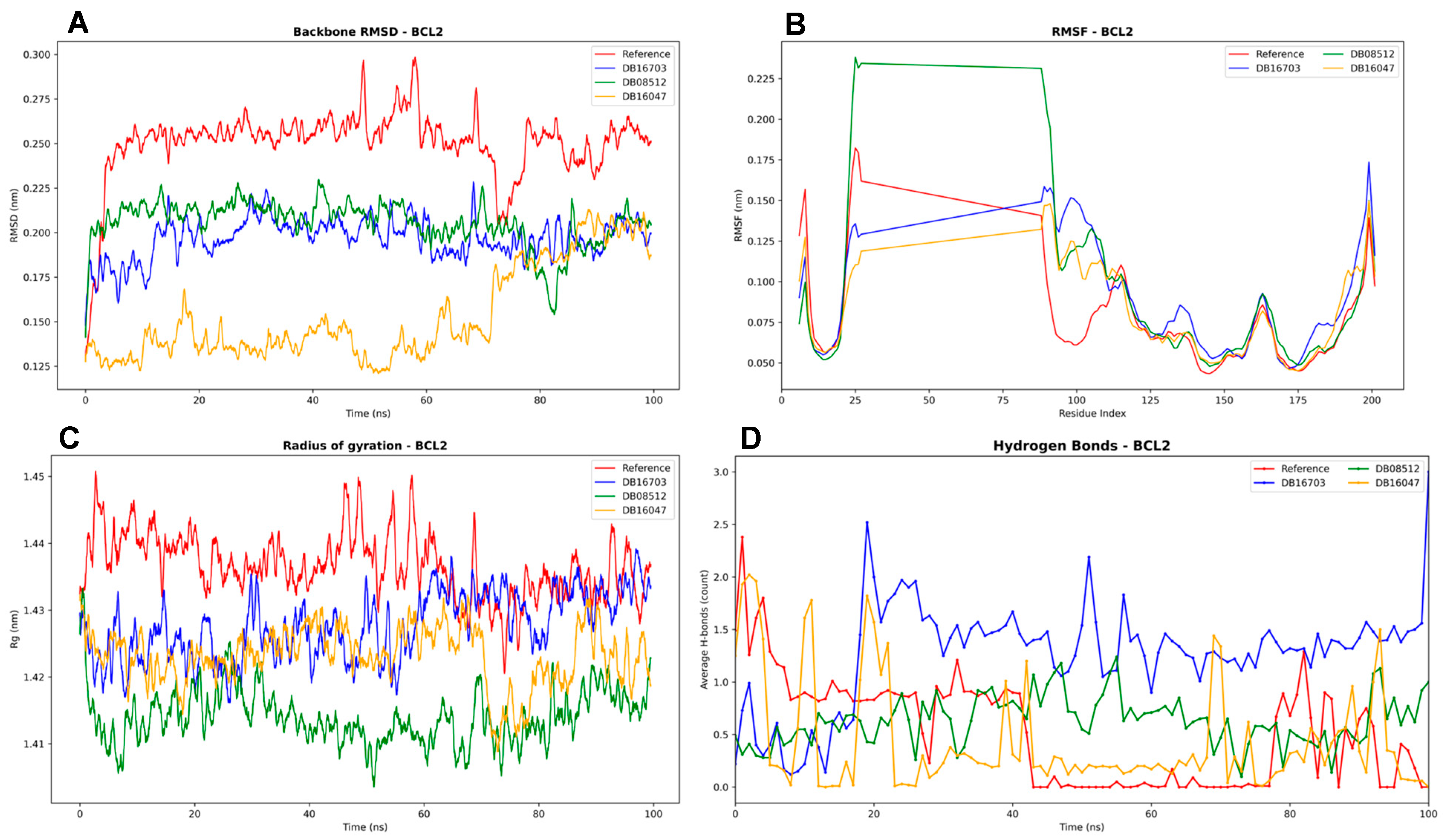
3.4.2. BCL2 Complexes Analysis
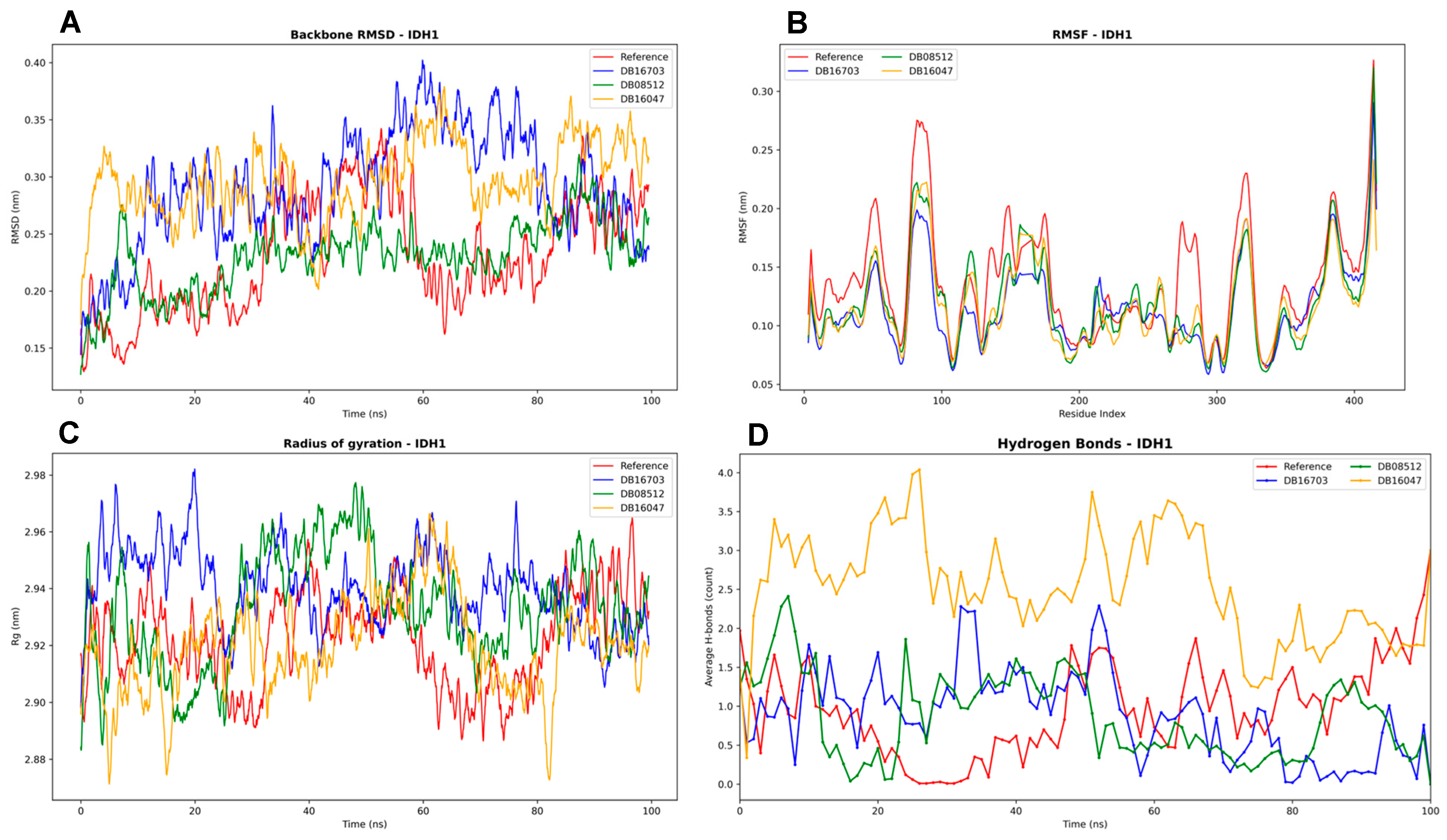
3.4.3. IDH1 Complexes Analysis
4. Discussion
4.1. Docking and Molecular Dynamics Insights
4.2. Polypharmacology in AML: A Multitarget Strategy
4.3. Rationale for Concurrent LSD1, BCL2, and IDH1 Inhibition
5. Limitations and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Yashar, W.M.; Curtiss, B.M.; Coleman, D.J.; VanCampen, J.; Kong, G.; Macaraeg, J.; Estabrook, J.; Demir, E.; Long, N.; Bottomly, D.; et al. Disruption of the MYC Superenhancer Complex by Dual Targeting of FLT3 and LSD1 in Acute Myeloid Leukemia. Mol. Cancer Res. 2023, 21, 631–647. [Google Scholar] [CrossRef]
- Madan, V.; Koeffler, H.P. Differentiation therapy of myeloid leukemia: Four decades of development. Haematologica 2021, 106, 26–38. [Google Scholar] [CrossRef]
- Yu, P.-H.; Zhu, C.-Y.; Kang, Y.-Y.; Naranmandura, H.; Yang, C. Mutation in the Unrearranged PML Allele Confers Resistance to Arsenic Trioxide in Acute Promyelocytic Leukemia. Research 2025, 8, 0696. [Google Scholar] [CrossRef]
- Bercier, P.; Wang, Q.Q.; Zang, N.; Zhang, J.; Yang, C.; Maimaitiyiming, Y.; Abou-Ghali, M.; Berthier, C.; Wu, C.; Niwa-Kawakita, M.; et al. Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function. Cancer Discov. 2023, 13, 2548–2565. [Google Scholar] [CrossRef]
- Ravasio, R.; Ceccacci, E.; Nicosia, L.; Hosseini, A.; Rossi, P.L.; Barozzi, I.; Fornasari, L.; Zuffo, R.D.; Valente, S.; Fioravanti, R.; et al. Targeting the scaffolding role of LSD1 (KDM1A) poises acute myeloid leukemia cells for retinoic acid–induced differentiation. Sci. Adv. 2020, 6, eaax2746. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.R.; Wang, E.S. Full article: What potential is there for LSD1 inhibitors to reach approval for AML? Expert Opin. Emerg. Drugs 2019, 24, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Waclawiczek, A.; Leppä, A.-M.; Renders, S.; Stumpf, K.; Reyneri, C.; Betz, B.; Janssen, M.; Shahswar, R.; Donato, E.; Karpova, D.; et al. Combinatorial BCL2 Family Expression in Acute Myeloid Leukemia Stem Cells Predicts Clinical Response to Azacitidine/Venetoclax. Cancer Discov. 2023, 13, 1408–1427. [Google Scholar] [CrossRef]
- Watts, J.M.; Shaw, S.J.; Jonas, B.A. Looking Beyond the Surface: Olutasidenib and Ivosidenib for Treatment of mIDH1 Acute Myeloid Leukemia. Curr. Treat. Options Oncol. 2024, 25, 1345–1353. [Google Scholar] [CrossRef]
- Kanouni, T.; Severin, C.; Cho, R.W.; Yuen, N.Y.Y.; Xu, J.; Shi, L.; Lai, C.; Del Rosario, J.R.; Stansfield, R.K.; Lawton, L.N.; et al. Discovery of CC-90011: A Potent and Selective Reversible Inhibitor of Lysine Specific Demethylase 1 (LSD1). J. Med. Chem. 2020, 63, 14522–14529. [Google Scholar] [CrossRef] [PubMed]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Pusch, S.; Krausert, S.; Fischer, V.; Balss, J.; Ott, M.; Schrimpf, D.; Capper, D.; Sahm, F.; Eisel, J.; Beck, A.C.; et al. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 2017, 133, 629–644. [Google Scholar] [CrossRef]
- Kouranov, A.; Xie, L.; de la Cruz, J.; Chen, L.; Westbrook, J.; Bourne, P.E.; Berman, H.M. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006, 34 (Suppl. 1), D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Boulaamane, Y.; Bolivar Avila, S.; Hurtado, J.R.; Touati, I.; Sadoq, B.-E.; Al-Mutairi, A.A.; Irfan, A.; Al-Hussain, S.A.; Maurady, A.; Zaki, M.E.A. Computational screening of natural products as tryptophan 2,3-dioxygenase inhibitors: Insights from CNN-based QSAR, molecular docking, ADMET, and molecular dynamics simulations. Comput. Biol. Med. 2025, 191, 110199. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Lee, Y.; Lee, H.J.; Kwon, Y.-S.; Chun, W. In silico screening of GABA aminotransferase inhibitors from the constituents of Valeriana officinalis by molecular docking and molecular dynamics simulation study. J. Mol. Model. 2020, 26, 228. [Google Scholar] [CrossRef]
- Bjelkmar, P.; Larsson, P.; Cuendet, M.A.; Hess, B.; Lindahl, E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J. Chem. Theory Comput. 2010, 6, 459–466. [Google Scholar] [CrossRef]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Al-Khafaji, K.; Taskin Tok, T. Molecular dynamics simulation, free energy landscape and binding free energy computations in exploration the anti-invasive activity of amygdalin against metastasis. Comput. Methods Programs Biomed. 2020, 195, 105660. [Google Scholar] [CrossRef] [PubMed]
- Boulaamane, Y.; Jangid, K.; Britel, M.R.; Maurady, A. Probing the molecular mechanisms of α-synuclein inhibitors unveils promising natural candidates through machine-learning QSAR, pharmacophore modeling, and molecular dynamics simulations. Mol. Divers. 2023, 28, 2495–2511. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Cutler, C.; Lee, S.J.; Arai, S.; Rotta, M.; Zoghi, B.; Lazaryan, A.; Ramakrishnan, A.; DeFilipp, Z.; Salhotra, A.; Chai-Ho, W.; et al. Belumosudil for chronic graft-versus-host disease after 2 or more prior lines of therapy: The ROCKstar Study. Blood 2021, 138, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Birkinshaw, R.W.; Gong, J.; Luo, C.S.; Lio, D.; White, C.A.; Anderson, M.A.; Blombery, P.; Lessene, G.; Majewski, I.J.; Thijssen, R.; et al. Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations. Nat. Commun. 2019, 10, 2385. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.; Cavalcante, L.; Giles, F.J.; Taylor, A. Elraglusib (9-ING-41), a selective small-molecule inhibitor of glycogen synthase kinase-3 beta, reduces expression of immune checkpoint molecules PD-1, TIGIT and LAG-3 and enhances CD8+ T cell cytolytic killing of melanoma cells. J. Hematol. Oncol. 2022, 15, 134. [Google Scholar] [CrossRef]
- Mahalingam, D.; Saeed, A.; Powell, S.F.; Huerta, M.; Sahai, V.; Coveler, A.L.; Davis, E.J.; Steeghs, N.; Mulcahy, M.; Raufi, A.G.; et al. Phase II study of elraglusib (9-ING-41), a GSK-3β inhibitor, in combination with gemcitabine plus nab-paclitaxel in previously untreated metastatic pancreatic cancer. ESMO Open 2025, 10, 105122. [Google Scholar] [CrossRef]
- Andresen, V.; Gjertsen, B.T. Drug Repurposing for the Treatment of Acute Myeloid Leukemia. Front. Med. 2017, 4, 211. [Google Scholar] [CrossRef]
- Abdelsayed, M. AI-Driven Polypharmacology in Small-Molecule Drug Discovery. Int. J. Mol. Sci. 2025, 26, 6996. [Google Scholar] [CrossRef]
- Deb, G.; Wingelhofer, B.; Amaral, F.M.R.; Maiques-Diaz, A.; Chadwick, J.A.; Spencer, G.J.; Williams, E.L.; Leong, H.-S.; Maes, T.; Somervaille, T.C.P. Pre-clinical activity of combined LSD1 and mTORC1 inhibition in MLL-translocated acute myeloid leukaemia. Leukemia 2020, 34, 1266–1277. [Google Scholar] [CrossRef]
- Chan, S.M.; Thomas, D.; Corces-Zimmerman, M.R.; Xavy, S.; Rastogi, S.; Hong, W.-J.; Zhao, F.; Medeiros, B.C.; Tyvoll, D.A.; Majeti, R. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 2015, 21, 178–184. [Google Scholar] [CrossRef]
- National Cancer Institute (NCI). Phase 1 Trial of Iadademstat in Combination with Venetoclax and Azacitidine in Patients with Treatment Naive AML; NCI: Bethesda, MD, USA, 2025. Available online: https://clinicaltrials.gov/study/NCT06514261 (accessed on 28 August 2025).
- Hosseini, A.; Dhall, A.; Ikonen, N.; Sikora, N.; Nguyen, S.; Shen, Y.; Amaral, M.L.J.; Jiao, A.; Wallner, F.; Sergeev, P.; et al. Perturbing LSD1 and WNT rewires transcription to synergistically induce AML differentiation. Nature 2025, 642, 508–518. [Google Scholar] [CrossRef] [PubMed]





| Protein | Gene | PDB ID | Reference Drug | Resolution |
|---|---|---|---|---|
| Lysine-specific histone demethylase 1A | LSD1 | 6W4K | Pulrodemstat | 2.93 Å |
| B-cell lymphoma 2 | BCL2 | 4MAN | BDBM178570 | 2.07 Å |
| Isocitrate Dehydrogenase 1 | IDH1 mutant (R132H) | 5LGE | BDBM389289 | 2.70 Å |
| Target | Ligand | Docking Score (kcal/mol) | H Bonds | Hydrophobic |
|---|---|---|---|---|
| LSD1 | Pulrodemstat | −8.42 | ASP-555 | PHE-538, ALA-539 |
| DB16703 | −10.7 | THR-335, ALA-539, PHE-560, HIS-564 | ILE-356, PHE-538, GLU-559, TYR-761, ALA-809, THR-810 | |
| DB08512 | −10.28 | ALA-539, TRP-552, VAL-764 | VAL-333, PHE-538, TRP-695, ALA-809 | |
| DB16047 | −10.54 | THR-335, PHE-560, HIS-564 | GLU-559 | |
| BCL2 | BDBM178570 | −10.84 | ALA-97, ASP-100, ARG-143 | PHE-101, TYR-105, ASP-108, MET-112, VAL-130, GLU-133, LEU-134, ARG-143, VAL-145, ALA-146, TYR-199 |
| DB16703 | −9.63 | ALA-146 | ALA-97A, PHE-101A, ASP-108A, LEU-134A, ARG-143A, VAL-145A, ALA-146A | |
| DB08512 | −9.84 | ASN-140, GLY-142, ARG-143 | ASP-108, PHE-109, MET-112, VAL-130, LEU-134, ARG-143, PHE-150 | |
| DB16047 | −9.39 | ARG-143 | PHE-101, LEU-134, ARG-143, PHE-150 | |
| IDH1 R132H mutant | BDBM389289 | −11.09 | ASN-271B, SER-280B, VAL-281B | LEU-120A, TRP-124B, ILE-128B, ILE-130B, VAL-255A, ALA-258A, ALA-258B, VAL-276A, GLN-277A, VAL-281A, VAL-281B |
| DB16703 | −10.45 | HIS-132B, LYS-212A, VAL-255A, GLN-277A, SER-278B, SER-280B | LEU-120B, TRP-124B, ILE-130B, VAL-255A, ALA-258B, TRP-267B, ASN-271B, VAL-276A, VAL-276B | |
| DB08512 | −10.71 | LYS-212A, SER-280B | TRP-124B, ILE-128B, TRP-267B, VAL-276A, TYR-285B | |
| DB16047 | −10.47 | GLN-277A, SER-278B, SER-280B, GLY-284B | TRP-124B, ILE-128B, ALA-258B, VAL-276A |
| Ligand | Water Solubility | Caco2 Permeability | HIA | BBB Permeability | CYP1A2 Inhibitior | CYP2D6 Inhibitior | Total Clearance | Renal OCT2 Substrate | AMES Toxicity |
|---|---|---|---|---|---|---|---|---|---|
| Ref-LSD1 | −5.07 | 0.74 | 91.34 | −0.93 | No | No | 0.20 | No | No |
| Ref-BCL2 | −3.84 | 0.34 | 96.05 | −2.15 | No | No | 0.09 | No | Yes |
| Ref-IDH1 | −6.75 | 0.81 | 87.67 | 0.01 | No | No | 0.19 | No | No |
| DB16703 | −5.56 | 0.67 | 92.50 | −0.70 | No | No | 0.95 | No | No |
| DB08512 | −4.58 | 0.53 | 91.26 | −0.50 | No | No | 1.15 | No | No |
| DB16047 | −4.95 | 1.01 | 92.31 | −0.53 | No | No | 1.06 | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelsayed, M.; Boulaamane, Y. Drug Repurposing for AML: Structure-Based Virtual Screening and Molecular Simulations of FDA-Approved Compounds with Polypharmacological Potential. Biomedicines 2025, 13, 2605. https://doi.org/10.3390/biomedicines13112605
Abdelsayed M, Boulaamane Y. Drug Repurposing for AML: Structure-Based Virtual Screening and Molecular Simulations of FDA-Approved Compounds with Polypharmacological Potential. Biomedicines. 2025; 13(11):2605. https://doi.org/10.3390/biomedicines13112605
Chicago/Turabian StyleAbdelsayed, Mena, and Yassir Boulaamane. 2025. "Drug Repurposing for AML: Structure-Based Virtual Screening and Molecular Simulations of FDA-Approved Compounds with Polypharmacological Potential" Biomedicines 13, no. 11: 2605. https://doi.org/10.3390/biomedicines13112605
APA StyleAbdelsayed, M., & Boulaamane, Y. (2025). Drug Repurposing for AML: Structure-Based Virtual Screening and Molecular Simulations of FDA-Approved Compounds with Polypharmacological Potential. Biomedicines, 13(11), 2605. https://doi.org/10.3390/biomedicines13112605





