Mutations in CREBBP and EP300 HAT and Bromo Domains Drive Hypermutation and Predict Survival in GI Cancers Treated with Immunotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohorts and Data Acquisition
2.2. TMB and MSI Assessment
2.3. Mutation Classification and Sample Grouping
2.4. Analysis of Mutation Types and Structural Mapping
2.5. Cohorts and Datasets for Prognostic and Predictive Biomarker Evaluation
2.6. Raw Data Processing and Bioinformatics Analysis
2.7. Statistical Analysis
2.8. Gene-Enrichment Analysis
3. Results
3.1. Cohort Overview and Study Group Characteristics
3.2. Analysis of Differences in TMB and MSI Values Between CREBBP and/or EP300 Mutant and WT Samples
3.3. Correlation Analysis of Genes Carrying Coding Mutations Associated with TMB-High
3.4. Landscape of Genes Co-Mutated with CREBBP and EP300
3.5. CREBBP and EP300 as Key Co-Mutants of Genes Associated with TMB-High
3.6. Pathway Enrichment Analysis Among Significant Co-Mutated Genes with CREBBP and EP300
3.7. Analysis of Mutation Types and Their Domain Localization Effects in CREBBP and EP300 Genes
3.8. Prognostic and Predictive Significance of CREBBP and EP300 Mutational Status
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BER | Base Excision Repair |
| CI | Confidence Interval |
| CREBBP | CREB-binding protein |
| dMMR | Deficient Mismatch Repair |
| DSBR | Double-Strand Break Repair |
| EBV | Epstein–Barr Virus |
| EP300 | E1A Binding Protein P300 |
| FDR | False Discovery Rate |
| GEJ | Gastroesophageal Junction |
| GSEA | Gene Set Enrichment Analysis |
| HAT | Histone Acetyltransferase |
| HR | Hazard Ratio |
| HRR | Homologous Recombination Repair |
| ICIs | Immune Checkpoint Inhibitors |
| IQR | Interquartile Range |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MMR | Mismatch Repair |
| MSI | Microsatellite Instability |
| NER | Nucleotide Excision Repair |
| NGS | Next-Generation Sequencing |
| NHEJ | Non-Homologous End Joining |
| OR | Odds Ratio |
| OS | Overall Survival |
| PD | Progressive Disease |
| PR | Partial Response |
| PTV | Protein-Truncating Variant |
| SD | Stable Disease |
| SNV | Single Nucleotide Variant |
| TMB | Tumor Mutational Burden |
| WES | Whole-Exome Sequencing |
| WT | Wild-Type |
References
- Yuan, J.; Hegde, P.S.; Clynes, R.; Foukas, P.G.; Harari, A.; Kleen, T.O.; Kvistborg, P.; Maccalli, C.; Maecker, H.T.; Page, D.B.; et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J. Immunother. Cancer 2016, 4, 10. [Google Scholar] [CrossRef]
- Kovács, S.A.; Fekete, J.T.; Győrffy, B. Predictive biomarkers of immunotherapy response with pharmacological applications in solid tumors. Acta Pharmacol. Sin. 2023, 44, 1879–1889. [Google Scholar] [CrossRef]
- Bhamidipati, D.; Subbiah, V. Impact of tissue-agnostic approvals for patients with gastrointestinal malignancies. Trends Cancer 2023, 9, 237–249. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Z.; Zhang, W.; Zhang, W.; Buzdin, A.; Mu, X.; Yan, Q.; Zhao, X.; Chang, H.-H.; Duhon, M.; et al. FDA-approved and emerging next generation predictive biomarkers for immune checkpoint inhibitors in cancer patients. Front. Oncol. 2021, 11, 683419. [Google Scholar] [CrossRef]
- Prasad, V.; Addeo, A. The FDA approval of pembrolizumab for patients with TMB >10 mut/Mb: Was it a wise decision? No. Ann. Oncol. 2020, 31, 1112–1114. [Google Scholar] [CrossRef]
- Gandara, D.R.; Agarwal, N.; Gupta, S.; Klempner, S.J.; Andrews, M.C.; Mahipal, A.; Subbiah, V.; Eskander, R.N.; Carbone, D.P.; Riess, J.W.; et al. Tumor mutational burden and survival on immune checkpoint inhibition in >8000 patients across 24 cancer types. J. Immunother. Cancer 2025, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- McGrail, D.J.; Pilié, P.G.; Rashid, N.U.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.B.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 10. [Google Scholar] [CrossRef]
- Venetis, K.; Frascarelli, C.; Boscolo Bielo, L.; Cursano, G.; Adorisio, R.; Ivanova, M.; Mane, E.; Peruzzo, V.; Concardi, A.; Negrelli, M.; et al. Mismatch repair (MMR) and microsatellite instability (MSI) phenotypes across solid tumors: A comprehensive cBioPortal study on prevalence and prognostic impact. Eur. J. Cancer 2025, 217, 115233. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.J.; Huang, Y.S.; An, N.; Hu, J.J.; Wu, C.Y.; Chen, Y.X.; Chen, J.Y.; Zhao, Q.; Xu, R.H.; Yuan, S.Q.; et al. Pan-cancer analysis of heterogeneity of tumor mutational burden and genomic mutation under treatment pressure. ESMO Open 2024, 9, 103494. [Google Scholar] [CrossRef] [PubMed]
- Taban, H.; Ergun, Y. Challenges in using tumor mutational burden as a post-treatment biomarker. ESMO Open 2024, 9, 103999. [Google Scholar] [CrossRef]
- Heydt, C.; Rehker, J.; Pappesch, R.; Buhl, T.; Ball, M.; Siebolts, U.; Haak, A.; Lohneis, P.; Büttner, R.; Hillmer, A.M.; et al. Analysis of tumor mutational burden: Correlation of five large gene panels with whole exome sequencing. Sci. Rep. 2020, 10, 11387. [Google Scholar] [CrossRef] [PubMed]
- Swat, W.; Kumar, D.; Boyle, T.A.; Dalton, E.; Comer, S.; Solomon, J.; Golem, S.; Eyring, K.R.; Baskovich, B.; Vashisht, A.; et al. Distributional range of tumor mutational burden (TMB) across multiple cancer types emphasizing the importance of different cutoff values in different tumor types for clinical interpretation: A consensus statement from the SEQUOIA consortium. J. Clin. Oncol. 2023, 41, 2615. [Google Scholar] [CrossRef]
- Vickram, S.; Infant, S.S.; Manikandan, S.; Rani, D.J.; Muthu, C.M.M.; Chopra, H. Immune biomarkers and predictive signatures in gastric cancer: Optimizing immunotherapy responses. Pathol. Res. Pract. 2025, 265, 155743. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhu, W.; Xu, Y.; Ma, J.; He, C.; Wang, F. SWI/SNF complex gene variations are associated with a higher tumor mutational burden and a better response to immune checkpoint inhibitor treatment: A pan-cancer analysis of next-generation sequencing data corresponding to 4591 cases. Cancer Cell Int. 2022, 22, 347. [Google Scholar] [CrossRef]
- Jia, Y.; Yuan, H.; Xin, Y.; Xing, X.; Li, Z.; Zhu, H. Genomic alteration in chromatin remodeling genes as a potential predictive biomarker for immunotherapy in gastric cancer. J. Clin. Oncol. 2022, 40, e16083. [Google Scholar] [CrossRef]
- Liu, C.; Jin, Y.; Zhang, H.; Yan, J.; Guo, Y.; Bao, X.; Zhao, P. Effects of KMT2D mutation and its exon 39 mutation on the immune microenvironment and drug sensitivity in colorectal adenocarcinoma. Heliyon 2023, 9, e13629. [Google Scholar] [CrossRef]
- Li, Q.; Wu, R.; Wu, F.; Chen, Q. KMT2D promotes proliferation of gastric cancer cells: Evidence from ctDNA sequencing. J. Clin. Lab. Anal. 2021, 35, e23721. [Google Scholar] [CrossRef]
- Liu, R.; Niu, Y.; Zhang, X.; Ma, T. Association of KMT2C/D loss-of-function mutations with tumor infiltrating lymphocytes and response to immune checkpoint inhibitors in solid tumors. J. Clin. Oncol. 2021, 39, 2587. [Google Scholar] [CrossRef]
- Ni, C.; Wang, X.; Liu, S.; Zhang, J.; Luo, Z.; Xu, B. KMT2C mutation as a predictor of immunotherapeutic efficacy in colorectal cancer. Sci. Rep. 2024, 14, 8284. [Google Scholar] [CrossRef]
- Wang, J.; Xiu, J.; Baca, Y.; Goldberg, R.M.; Philip, P.A.; Seeber, A.; Battaglin, F.; Arai, H.; Soni, S.; Tokunaga, R.; et al. Molecular landscape of gastric cancer (GC) harboring mutations of histone methyltransferases. J. Clin. Oncol. 2020, 38, 418. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, C.; Li, L.; Zhang, T.; Wang, X. Pan-cancer analysis reveals that E1A binding protein p300 mutations increase genome instability and antitumor immunity. Front. Cell Dev. Biol. 2021, 9, 729927. [Google Scholar] [CrossRef]
- Wang, N.; Li, D.; Zhang, T.; Pachai, M.R.; Cho, W.H.; Khudoynazarova, M.N.; Schoeps, D.M.; Bao, Y.; Liu, M.; Tang, L.; et al. Loss of Kmt2c/d promotes gastric cancer initiation and confers vulnerability to mTORC1 inhibition and anti-PD1 immunotherapy. bioRxiv 2025. [Google Scholar] [CrossRef]
- Blümli, S.; Wiechens, N.; Wu, M.-Y.; Singh, V.; Gierlinski, M.; Schweikert, G.; Gilbert, N.; Naughton, C.; Sundaramoorthy, R.; Varghese, J.; et al. Acute depletion of the ARID1A subunit of SWI/SNF complexes reveals distinct pathways for activation and repression of transcription. Cell Rep. 2021, 37, 109943. [Google Scholar] [CrossRef] [PubMed]
- Gronkowska, K.; Robaszkiewicz, A. Genetic dysregulation of EP300 in cancers in light of cancer epigenome control—Targeting of p300-proficient and -deficient cancers. Mol. Ther. Oncol. 2024, 32, 200871. [Google Scholar] [CrossRef] [PubMed]
- Duex, J.E.; Swain, K.E.; Dancik, G.M.; Paucek, R.D.; Owens, C.; Churchill, M.E.A.; Theodorescu, D. Functional impact of chromatin remodeling gene mutations and predictive signature for therapeutic response in bladder cancer. Mol. Cancer Res. 2018, 16, 69–77. [Google Scholar] [CrossRef]
- Arany, Z.; Sellers, W.R.; Livingston, D.M.; Eckner, R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 1994, 77, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, X.; Tang, S.; Wang, Y. The important role of the histone acetyltransferases p300/CBP in cancer and the promising anticancer effects of p300/CBP inhibitors. Cell Biol. Toxicol. 2025, 41, 32. [Google Scholar] [CrossRef]
- Huang, Y.H.; Cai, K.; Xu, P.P.; Wang, L.; Huang, C.X.; Fang, Y.; Cheng, S.; Sun, X.J.; Liu, F.; Huang, J.Y.; et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct. Target. Ther. 2021, 6, 10. [Google Scholar] [CrossRef]
- Korfi, K.; Ali, S.; Heward, J.A.; Fitzgibbon, J. Follicular lymphoma, a B cell malignancy addicted to epigenetic mutations. Epigenetics 2017, 12, 23–35. [Google Scholar] [CrossRef]
- Sermer, D.; Pasqualucci, L.; Wendel, H.-G.; Melnick, A.; Younes, A. Emerging epigenetic-modulating therapies in lymphoma. Nat. Rev. Clin. Oncol. 2019, 16, 494–507. [Google Scholar] [CrossRef]
- Bakhshi, T.J.; Georgel, P.T. Genetic and epigenetic determinants of diffuse large B-cell lymphoma. Blood Cancer J. 2020, 10, 123. [Google Scholar] [CrossRef]
- Chen, J.; Hu, X.; Zheng, Y.; Huang, M. Investigation of CREBBP mutation and correlation with immunotherapy biomarker in Chinese bladder cancer patients. J. Clin. Oncol. 2022, 40, e16537. [Google Scholar] [CrossRef]
- Rao, A.; McGrath, J.E.; Xiu, J.; De Souza, A.L.; Gulati, S.; Abuali, I.; Sagaram, S.; Nabhan, C.; Korn, W.M.; Ryan, C.J.; et al. Characterization of microsatellite instability (dMMR/MSI-H) and mutational landscape in a large contemporary cohort of upper tract urothelial cancer (UTUC) patients. J. Clin. Oncol. 2021, 39 Suppl. 6, 465. [Google Scholar] [CrossRef]
- Zhu, G.; Pei, L.; Li, Y.; Gou, X. EP300 mutation is associated with tumor mutation burden and promotes antitumor immunity in bladder cancer patients. Aging 2020, 12, 2132–2147. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Yoo, N.J.; Lee, S.H. Expressional and mutational analysis of CREBBP gene in gastric and colorectal cancers with microsatellite instability. Pathol. Oncol. Res. 2014, 20, 769–777. [Google Scholar] [CrossRef]
- Liu, K.; Wang, J.F.; Zhan, Y.; Kong, D.L.; Wang, C. Prognosis model of colorectal cancer patients based on NOTCH3, KMT2C, and CREBBP mutations. J. Gastrointest. Oncol. 2021, 12, 987–999. [Google Scholar] [CrossRef]
- Lumish, M.A.; Walch, H.; Maron, S.B.; Chatila, W.; Kemel, Y.; Maio, A.; Ku, G.Y.; Ilson, D.H.; Won, E.; Li, J.; et al. Clinical and molecular characteristics of early-onset vs average-onset esophagogastric cancer. J. Natl. Cancer Inst. 2024, 116, djad186. [Google Scholar] [CrossRef] [PubMed]
- Janjigian, Y.Y.; Maron, S.B.; Chatila, W.K.; Millang, B.; Chavan, S.S.; Alterman, C.; Chou, J.F.; Segal, M.F.; Simmons, M.Z.; Momtaz, P.; et al. First-line pembrolizumab and trastuzumab in HER2-positive oesophageal, gastric, or gastro-oesophageal junction cancer: An open-label, single-arm, phase 2 trial. Lancet Oncol. 2020, 21, 1061–1071. [Google Scholar] [CrossRef]
- Maron, S.B.; Chatila, W.; Walch, H.; Chou, J.F.; Ceglia, N.; Ptashkin, R.; Do, R.K.G.; Paroder, V.; Pandit-Taskar, N.; Lewis, J.S.; et al. Determinants of survival with combined HER2 and PD-1 blockade in metastatic esophagogastric cancer. Clin. Cancer Res. 2023, 29, 1234–1247. [Google Scholar] [CrossRef]
- Sihag, S.; Nussenzweig, S.C.; Walch, H.S.; Hsu, M.; Tan, K.S.; De La Torre, S.; Janjigian, Y.Y.; Maron, S.B.; Ku, G.Y.; Tang, L.H.; et al. The role of the TP53 pathway in predicting response to neoadjuvant therapy in esophageal adenocarcinoma. Clin. Cancer Res. 2022, 28, 5678–5689. [Google Scholar] [CrossRef]
- Chen, K.; Yang, D.; Li, X.; Sun, B.; Song, F.; Cao, W.; Brat, D.J.; Gao, Z.; Li, H.; Liang, H.; et al. Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 1107–1112. [Google Scholar] [CrossRef]
- Janigian, Y.Y.; Sanchez-Vega, F.; Jonsson, P.; Chatila, W.K.; Hechtman, J.F.; Ku, G.Y.; Riches, J.C.; Tuvy, Y.; Kundra, R.; Bouvier, N.; et al. Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov. 2018, 8, 49–58. [Google Scholar] [CrossRef]
- Dulak, A.M.; Stojanov, P.; Peng, S.; Lawrence, M.S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S.E.; Shefler, E.; et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013, 45, 478–486. [Google Scholar] [CrossRef] [PubMed]
- GDAC Broad Institute. Index of /runs/stddata__2016_01_28/data/ESCA/20160128. Available online: https://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/ESCA/20160128/ (accessed on 8 June 2025).
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.N.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef]
- GDAC Broad Institute. Index of /runs/stddata__2016_01_28/data/STAD/20160128. Available online: https://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/STAD/20160128/ (accessed on 8 June 2025).
- Kakiuchi, M.; Nishizawa, T.; Ueda, H.; Gotoh, K.; Tanaka, A.; Hayashi, A.; Yamamoto, S.; Tatsuno, K.; Katoh, H.; Watanabe, Y.; et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet. 2014, 46, 583–587. [Google Scholar] [CrossRef]
- Wang, K.; Kan, J.; Yuen, S.T.; Shi, S.T.; Chu, K.M.; Law, S.; Chan, T.L.; Kan, Z.; Chan, A.S.Y.; Tsui, W.Y.; et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 2011, 43, 1219–1223. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Ye, K.; Zhang, Q.; Lu, C.; Xie, M.; McLellan, M.D.; Wendl, M.C.; Ding, L. MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014, 30, 1015–1016. [Google Scholar] [CrossRef]
- Johansen, A.F.B.; Kassentoft, C.G.; Knudsen, M.; Laursen, M.B.; Madsen, A.H.; Iversen, L.H.; Sunesen, K.G.; Rasmussen, M.H.; Andersen, C.L. Validation of computational determination of microsatellite status using whole exome sequencing data from colorectal cancer patients. BMC Cancer 2019, 19, 971. [Google Scholar] [CrossRef] [PubMed]
- Anthony, H.; Seoighe, C. Performance assessment of computational tools to detect microsatellite instability. Brief. Bioinform. 2024, 25, bbae390. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.-M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- The Genomic Data Commons. Available online: https://docs.gdc.cancer.gov/Data/Introduction/ (accessed on 28 August 2025).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- gnomAD. Available online: https://gnomad.broadinstitute.org/ (accessed on 28 August 2025).
- Jia, P.; Yang, X.; Guo, L.; Liu, B.; Lin, J.; Liang, H.; Sun, J.; Zhang, C.; Ye, K. MSIsensor-pro: Fast, accurate, and matched-normal-sample-free detection of microsatellite instability. Genom. Proteom. Bioinform. 2020, 18, 65–71. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Fujibuchi, W.; Kanehisa, M. Computation with the KEGG pathway database. Biosystems 1998, 47, 119–128. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.N.; Scuoppo, C.; Vlasevska, S.; Bal, E.; Holmes, A.B.; Holloman, M.; Garcia-Ibanez, L.; Nataraj, S.; Duval, R.; Vantrimpont, T.; et al. Unique and shared epigenetic programs of the CREBBP and EP300 acetyltransferases in germinal center B cells reveal targetable dependencies in lymphoma. Immunity 2019, 51, 535–547.e9. [Google Scholar] [CrossRef]
- Dutto, I.; Scalera, C.; Tillhon, M.; Ticli, G.; Passaniti, G.; Cazzalini, O.; Savio, M.; Stivala, L.A.; Gervasini, C.; Larizza, L.; et al. Mutations in CREBBP and EP300 genes affect DNA repair of oxidative damage in Rubinstein-Taybi syndrome cells. Carcinogenesis 2020, 41, 257–266. [Google Scholar] [CrossRef]
- Dutto, I.; Scalera, C.; Prosperi, E. CREBBP and p300 lysine acetyl transferases in the DNA damage response. Cell. Mol. Life Sci. 2018, 75, 1325–1338. [Google Scholar] [CrossRef]
- Gujar, V.; Li, H.; Paull, T.T.; Neumann, C.A.; Weyemi, U. Unraveling the nexus: Genomic instability and metabolism in cancer. Cell Rep. 2025, 44, 115540. [Google Scholar] [CrossRef]
- Kumar, M.; Molkentine, D.; Molkentine, J.; Bridges, K.; Xie, T.; Yang, L.; Hefner, A.; Gao, M.; Bahri, R.; Dhawan, A.; et al. Inhibition of histone acetyltransferase function radiosensitizes CREBBP/EP300 mutants via repression of homologous recombination, potentially targeting a gain of function. Nat. Commun. 2021, 12, 6340. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, J.; Qu, X.; Yang, Y.; Gong, Z.; Yang, Y.; Wu, Z.; Guo, W. Microsatellite instability-related ACVR2A mutations partially account for decreased lymph node metastasis in MSI-H gastric cancers. Onco Targets Ther. 2020, 13, 3809–3821. [Google Scholar] [CrossRef]
- Weinstein, H.N.W.; Hu, K.; Fish, L.; Chen, Y.-A.; Allegakoen, P.; Pham, J.H.; Hui, K.S.F.; Chang, C.-H.; Tutar, M.; Benitez-Rivera, L.; et al. RPL22 is a tumor suppressor in MSI-high cancers and a splicing regulator of MDM4. Cell Rep. 2024, 43, 114622. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, J.; Luo, H.; Meng, X.; Chen, M.; Zhu, D. Wnt signaling pathway in cancer immunotherapy. Cancer Lett. 2022, 525, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Zhou, X. Targeting Hippo signaling in cancer: Novel perspectives and therapeutic potential. MedComm 2023, 4, e375. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef]
- Li, J.; Chin, C.R.; Ying, H.-Y.; Meydan, C.; Teater, M.R.; Xia, M.; Farinha, P.; Takata, K.; Chu, C.-S.; Jiang, Y.; et al. Loss of CREBBP and KMT2D cooperate to accelerate lymphomagenesis and shape the lymphoma immune microenvironment. Nat. Commun. 2024, 15, 47012. [Google Scholar] [CrossRef]
- Mondello, P.; Ansell, S.M.; Nowakowski, G.S. Immune epigenetic crosstalk between malignant B cells and the tumor microenvironment in B cell lymphoma. Front. Genet. 2022, 13, 826594. [Google Scholar] [CrossRef]
- Krupar, R.; Watermann, C.; Idel, C.; Ribbat-Idel, J.; Offermann, A.; Pasternack, H.; Kirfel, J.; Sikora, A.G.; Perner, S. In silico analysis reveals EP300 as a panCancer inhibitor of anti-tumor immune response via metabolic modulation. Sci. Rep. 2020, 10, 9389. [Google Scholar] [CrossRef] [PubMed]
- Kögl, J.; Pan, T.L.; Marth, C.; Zeimet, A.G. The game-changing impact of POLE mutations in oncology—A review from a gynecologic oncology perspective. Front. Oncol. 2024, 14, 1369189. [Google Scholar] [CrossRef] [PubMed]
- Garmezy, B.; Gheeya, J.; Lin, H.Y.; Huang, Y.; Kim, T.; Jiang, X.; Thein, K.Z.; Pilié, P.G.; Zeineddine, F.; Wang, W.; et al. Clinical and molecular characterization of POLE mutations as predictive biomarkers of response to immune checkpoint inhibitors in advanced cancers. JCO Precis. Oncol. 2022, 6, e2100267. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Dong, L.; Liu, X.; Ou, K.; Yang, L. POLE/POLD1 mutation and tumor immunotherapy. J. Exp. Clin. Cancer Res. 2022, 41, 216. [Google Scholar] [CrossRef]
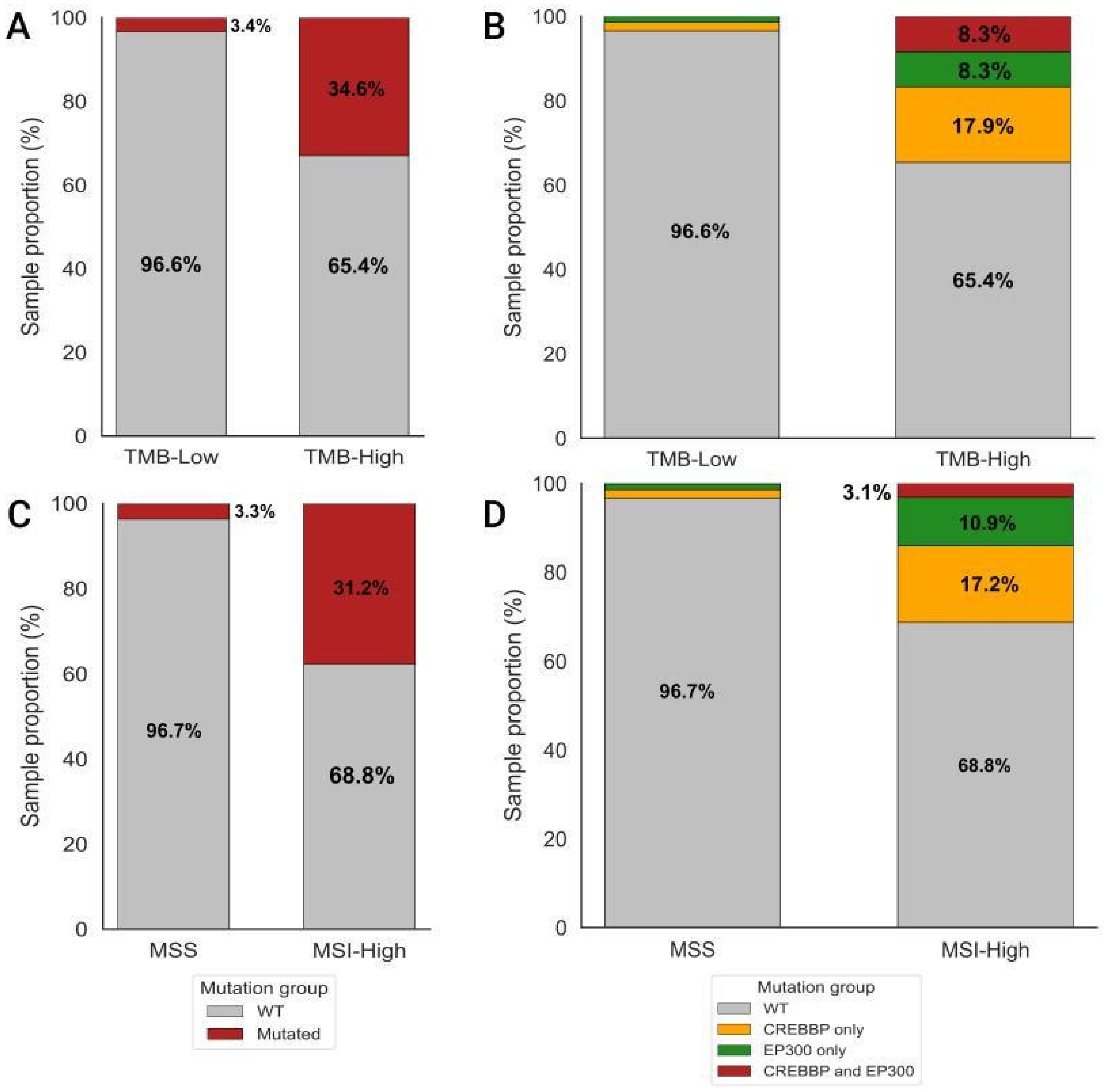
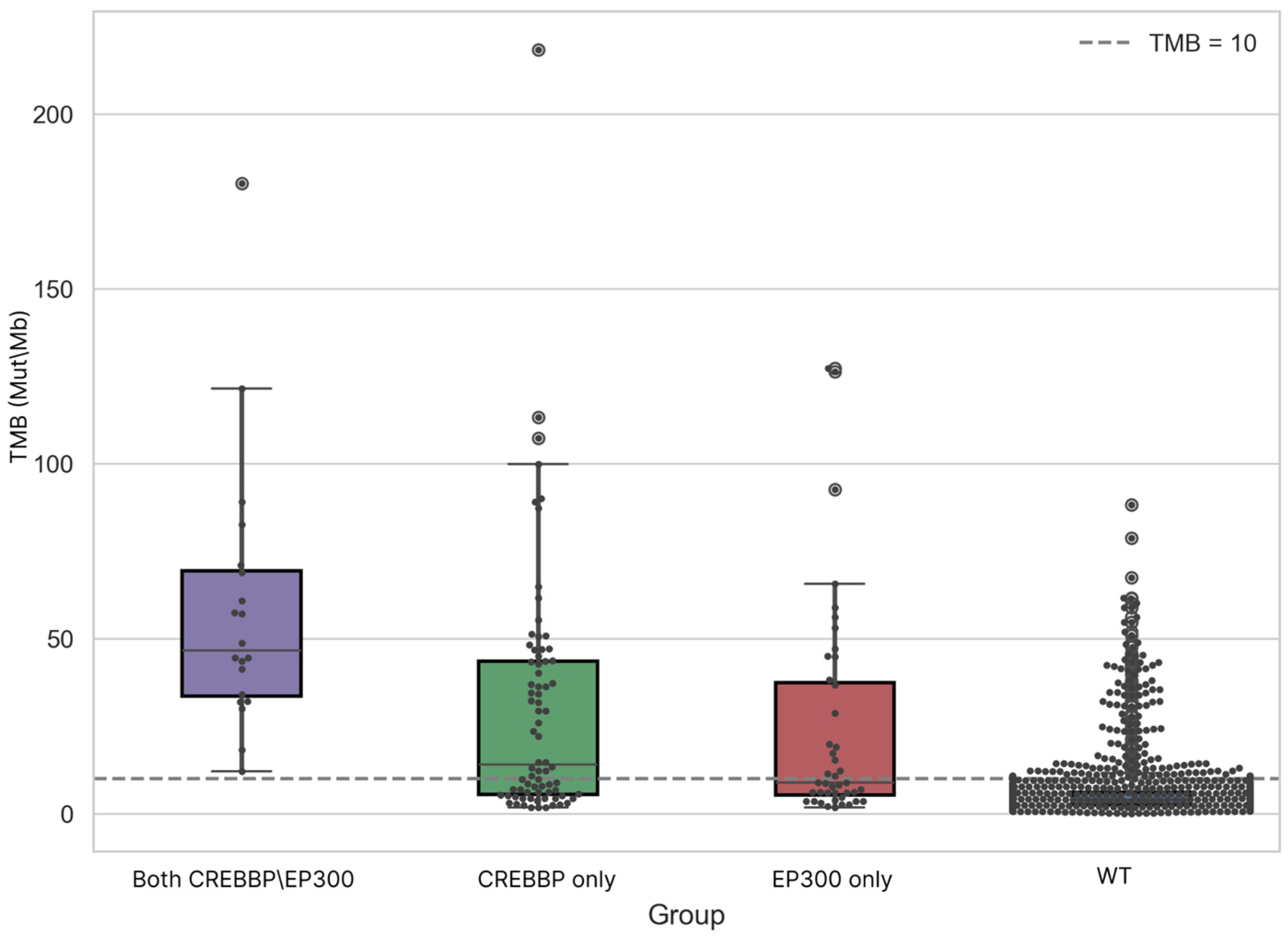
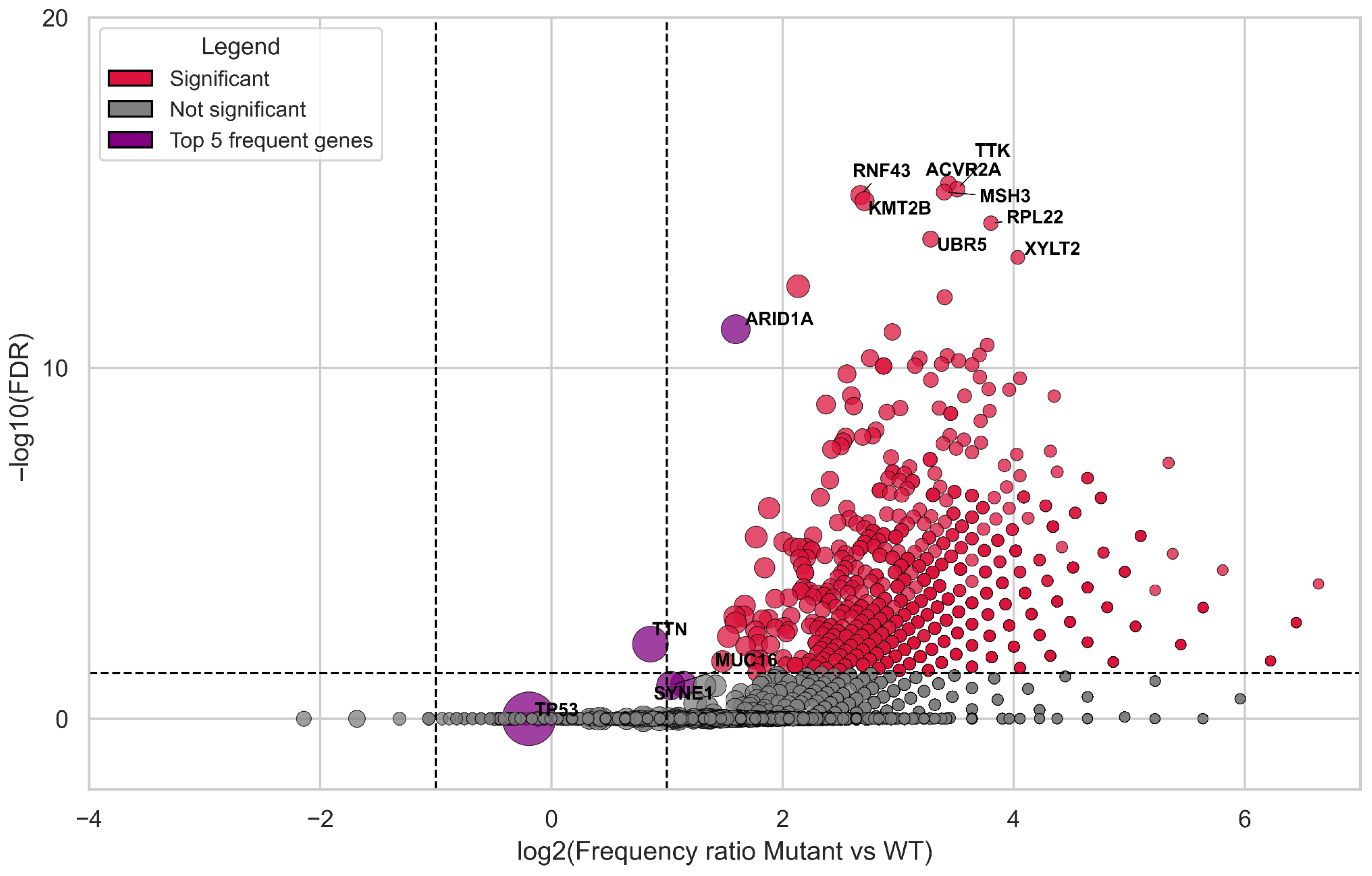
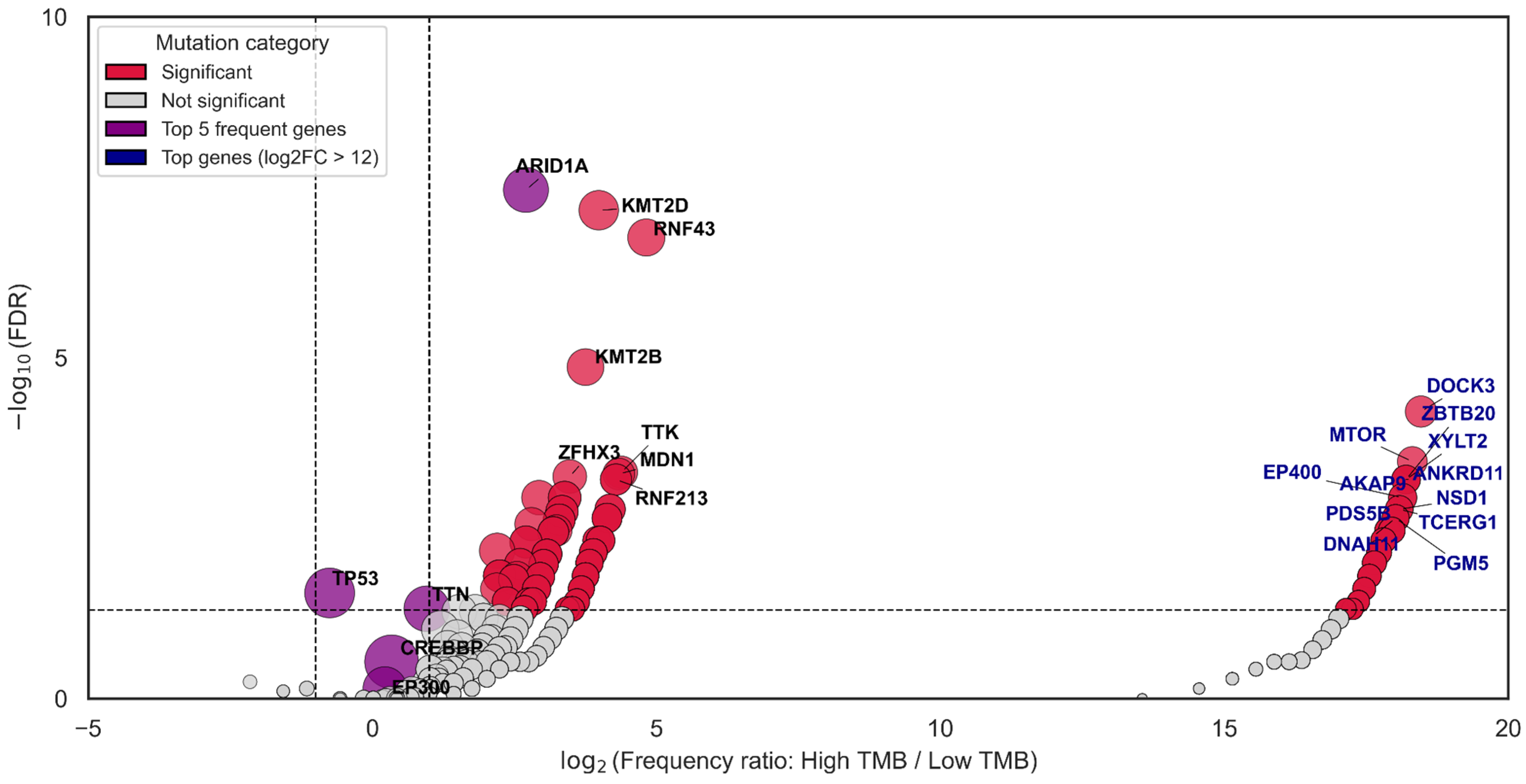
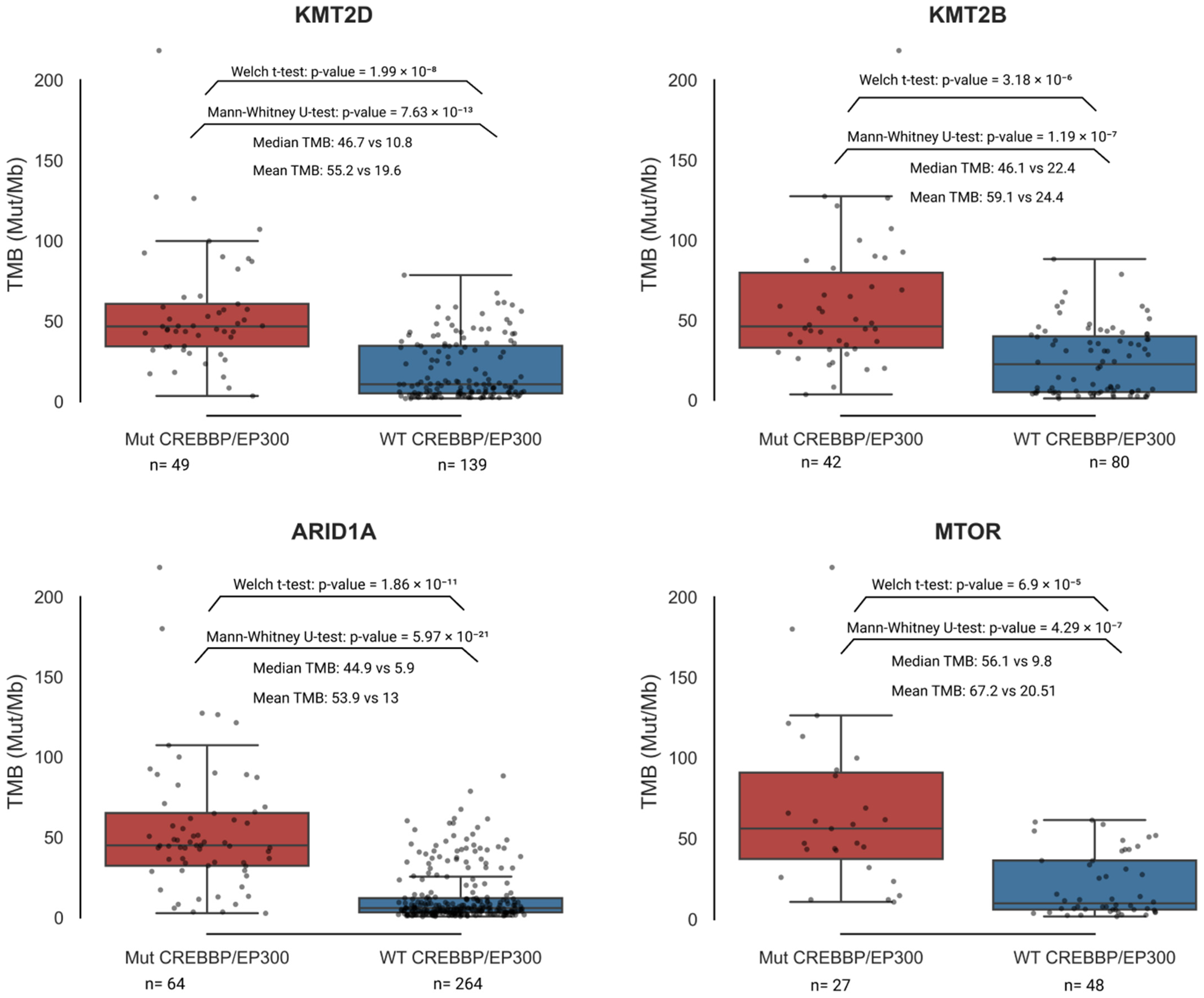
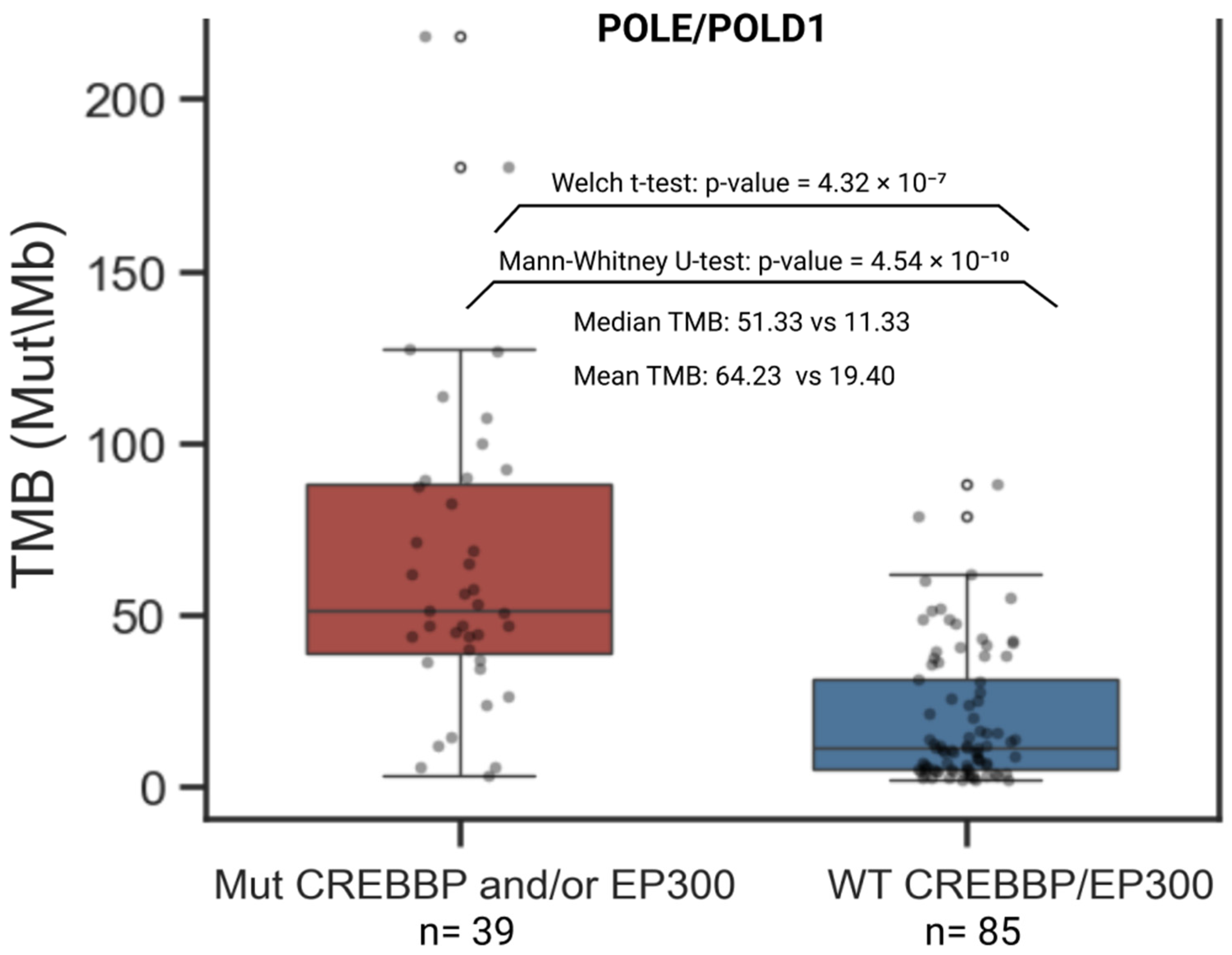
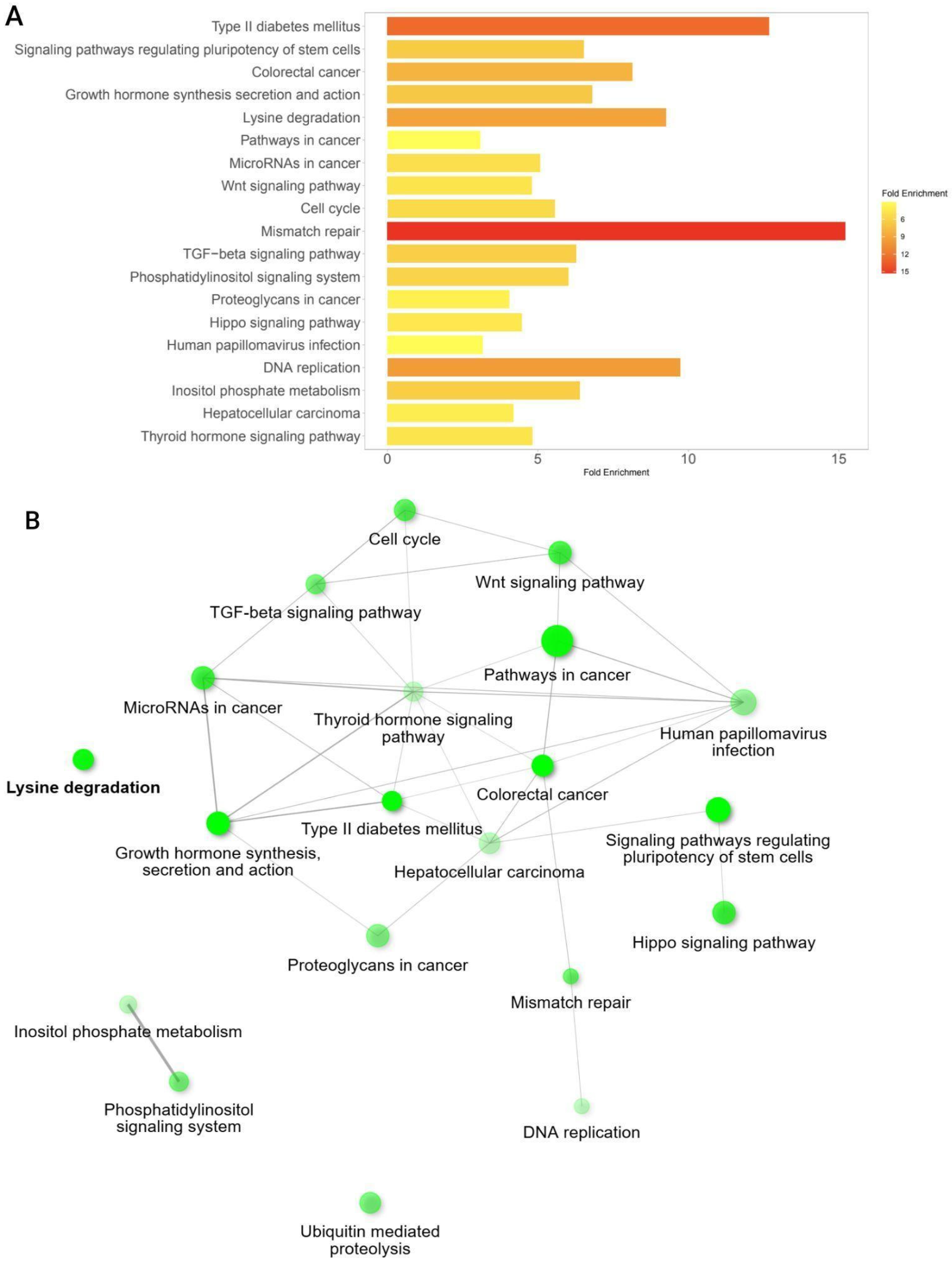
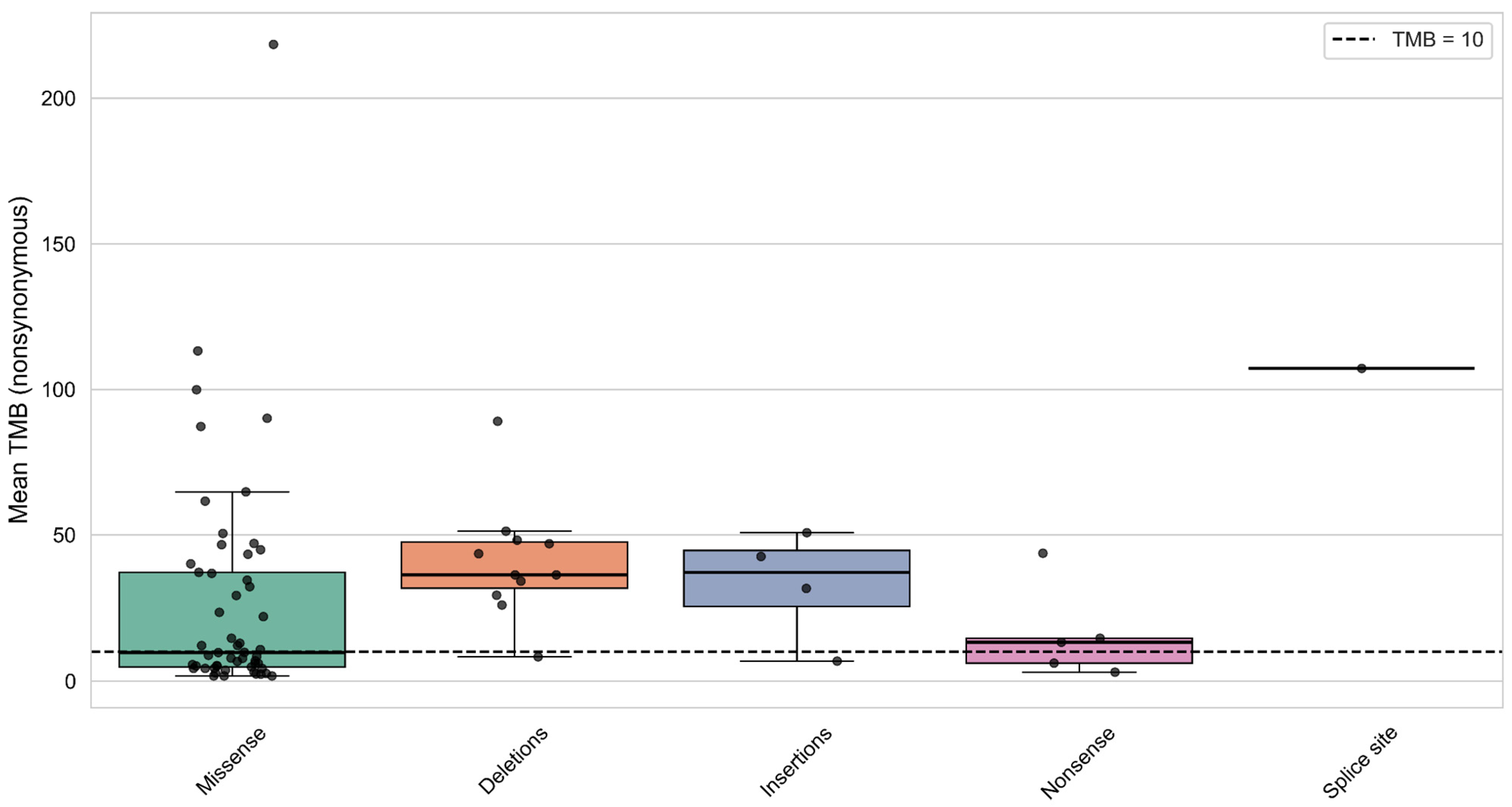
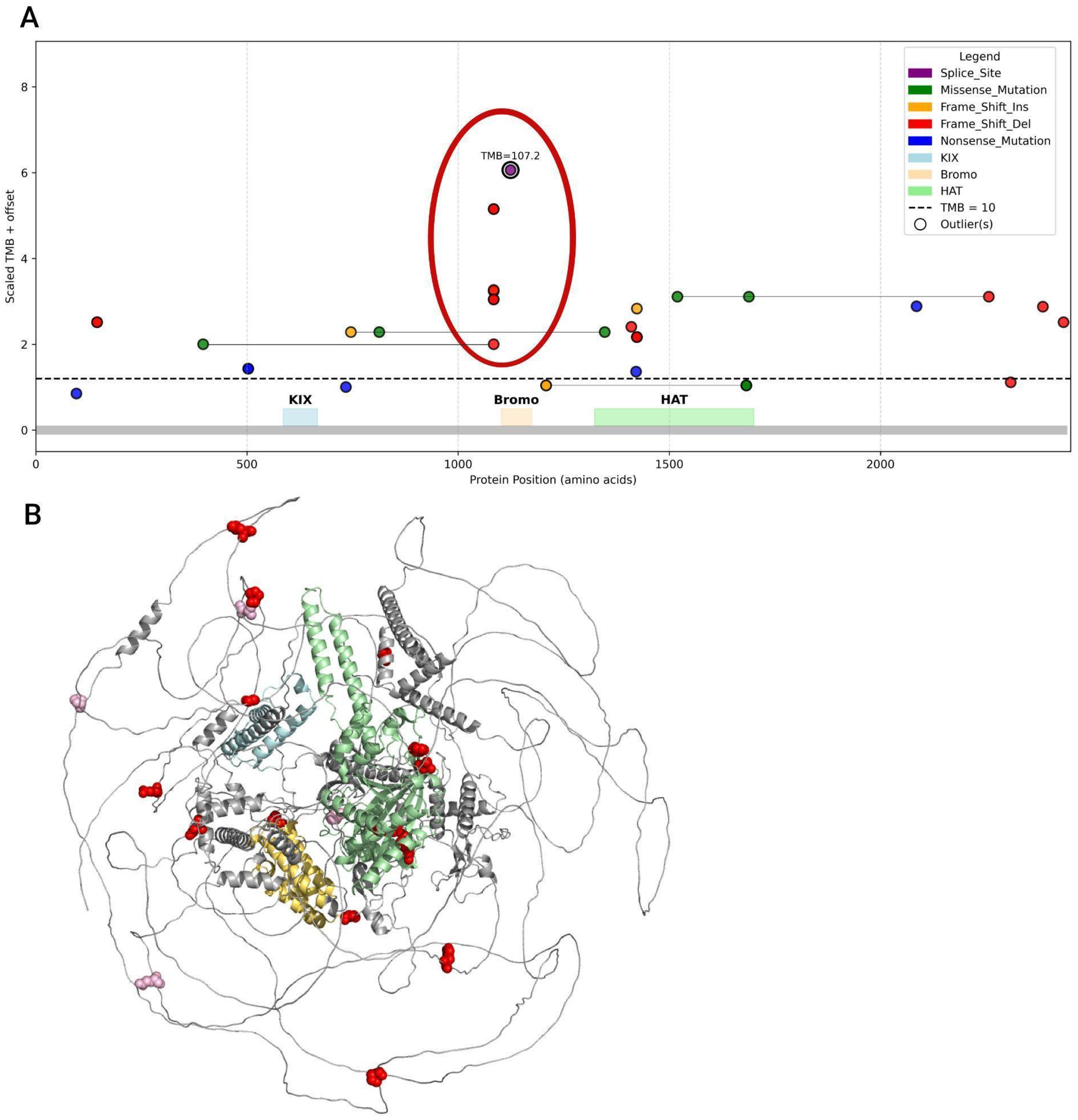
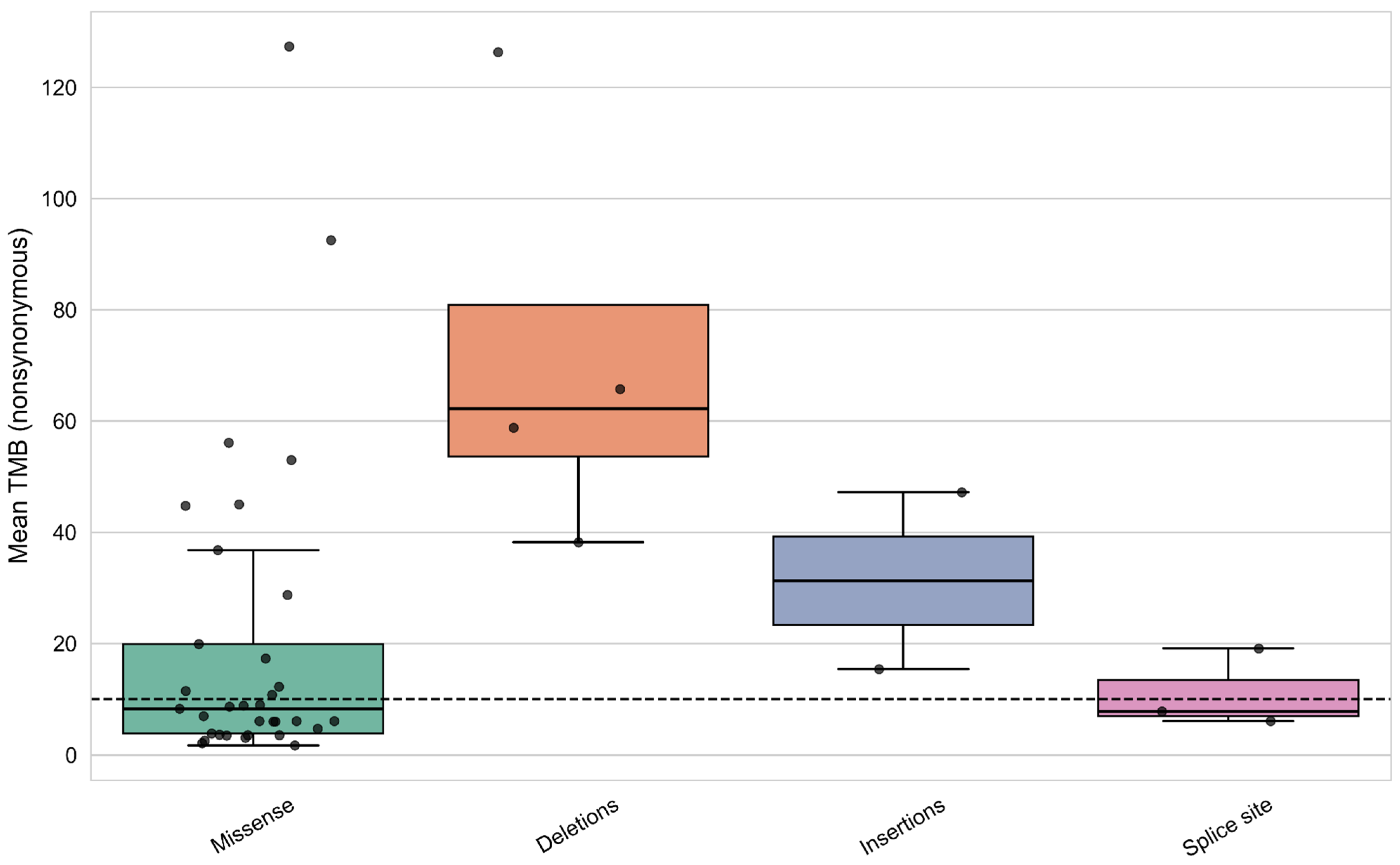
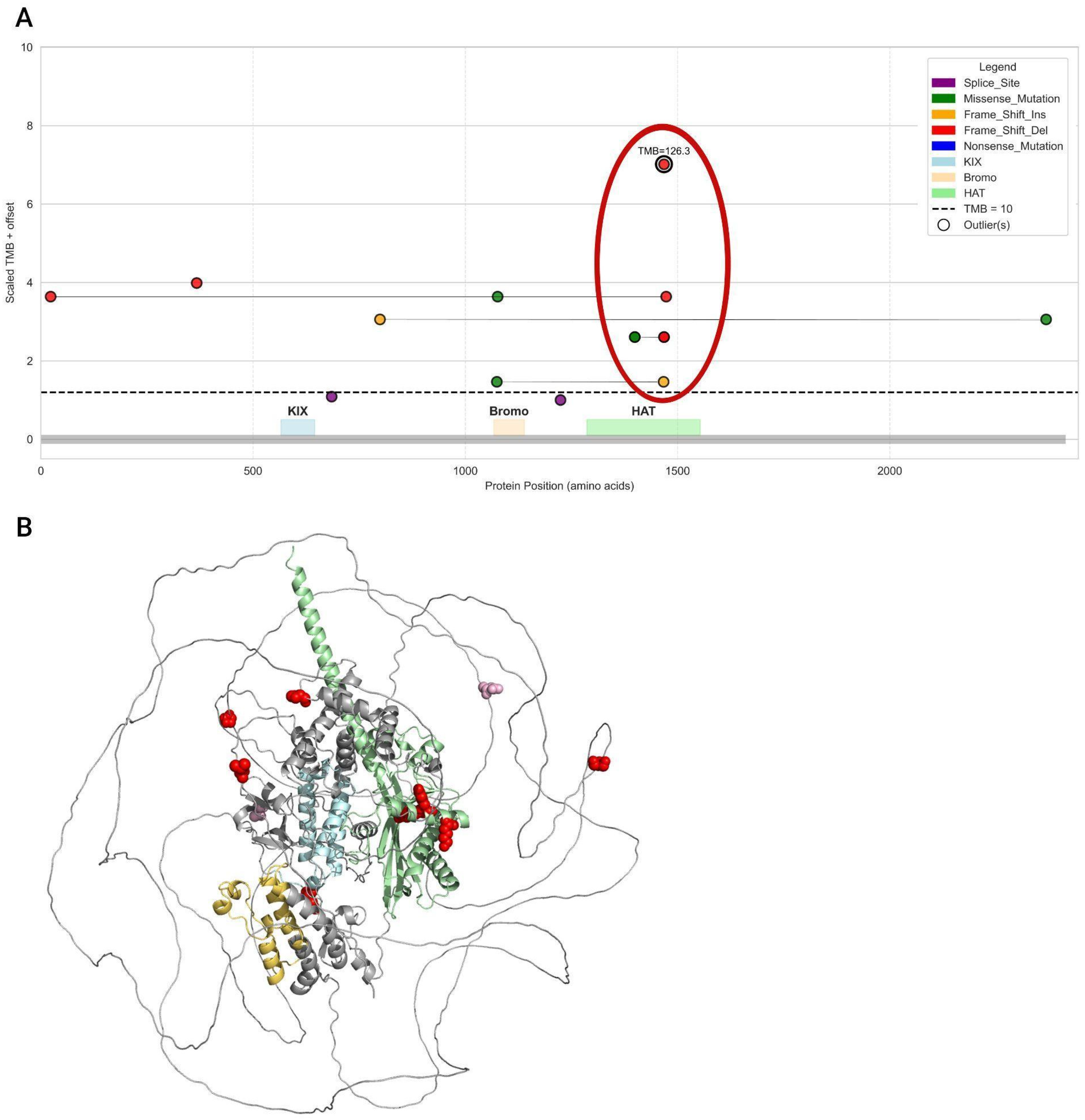
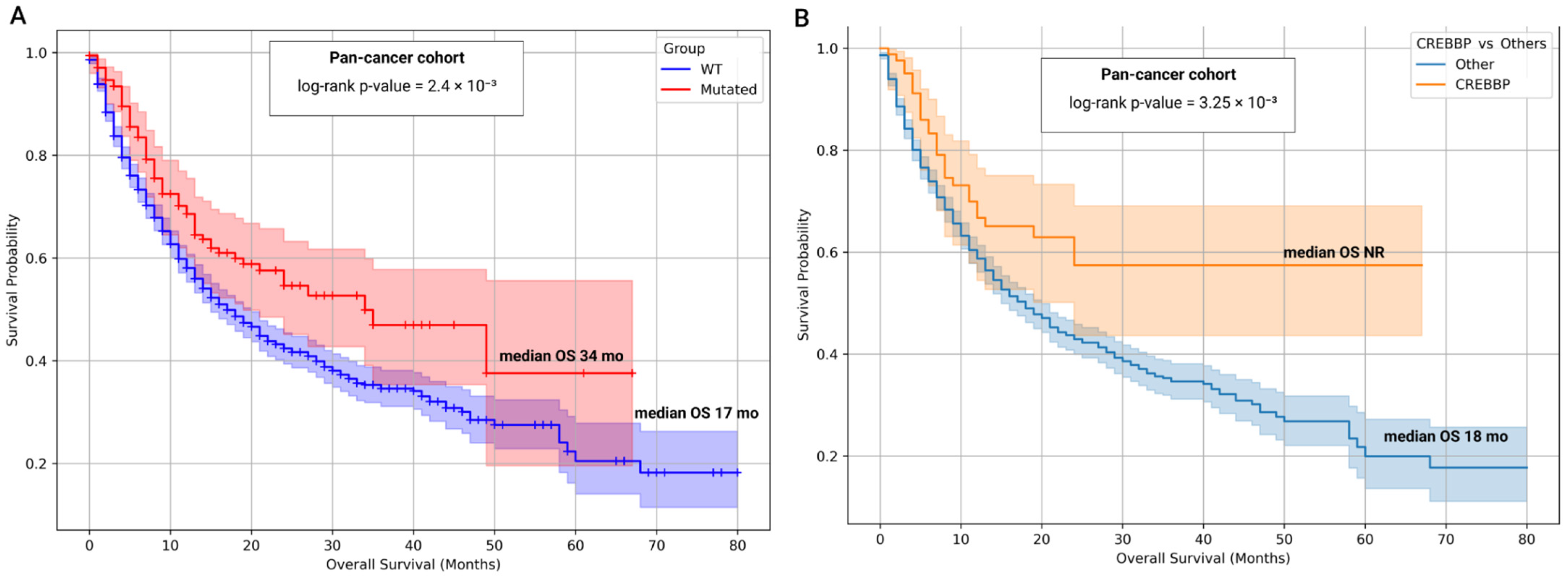
| Group | Samples Analyzed for TMB | TMB ≥ 10 (n, %) | TMB ≥ 20 (n, %) | TMB ≥ 50 (n, %) | TMB ≥ 100 (n, %) | TMB ≥ 200 (n, %) | Samples Analyzed for MSI | MSI-High (Score ≥ 3.5) (n, %) |
|---|---|---|---|---|---|---|---|---|
| WT | 1732 | 157 (9.1%) | 74 (4.3%) | 13 (0.8%) | 0 | 0 | 744 | 44 (5.9%) |
| CREBBP | 76 | 43 (56.6%) | 36 (47.4%) | 13 (17.1%) | 3 (3.9%) | 1 (1.3%) | 25 | 11 (44.0%) |
| EP300 | 43 | 20 (46.5%) | 13 (30.2%) | 7 (16.3%) | 2 (4.7%) | 0 | 16 | 7 (43.8%) |
| CREBBP/EP300 | 20 | 20 (100%) | 18 (90.0%) | 9 (45.0%) | 2 (10.0%) | 0 | 3 | 2 (66.7%) |
| Group | Median | Mean | Min. | Max. | Q1 | Q3 | IQR |
|---|---|---|---|---|---|---|---|
| CREBBP/EP300 | 46.55 | 58.44 | 12.11 | 180.13 | 33.50 | 69.40 | 35.90 |
| CREBBP | 13.99 | 29.74 | 1.73 | 218.47 | 5.55 | 43.59 | 38.04 |
| EP300 | 8.81 | 24.34 | 1.73 | 127.33 | 5.30 | 37.47 | 32.17 |
| WT | 3.97 | 5.95 | 0.03 | 88.20 | 2.53 | 6.18 | 3.64 |
| Gene | Mutated | WT | Correlation, r1 (TMB-High, Cut-Off = 10 Mut\Mb) | Correlation, r2 (TMB-High, Cut-Off = 20 Mut\Mb) |
|---|---|---|---|---|
| KMT2D | 181 | 1705 | 0.51 | 0.56 |
| ACVR2A | 77 | 1809 | 0.46 | 0.55 |
| KMT2B | 120 | 1766 | 0.44 | 0.57 |
| MSH3 | 74 | 1812 | 0.44 | 0.54 |
| RNF43 | 125 | 1761 | 0.42 | 0.56 |
| ARID1A | 332 | 1554 | 0.41 | 0.46 |
| RPL22 | 56 | 1830 | 0.41 | 0.52 |
| TTK | 69 | 1817 | 0.41 | 0.49 |
| DOCK3 | 77 | 1809 | 0.40 | 0.46 |
| XYLT2 | 47 | 1839 | 0.39 | 0.51 |
| BRCA2 | 96 | 1790 | 0.37 | 0.41 |
| ZFHX3 | 111 | 1775 | 0.37 | 0.45 |
| IRS1 | 58 | 1828 | 0.36 | 0.48 |
| UBR5 | 72 | 1814 | 0.36 | 0.45 |
| MTOR | 75 | 1811 | 0.35 | 0.39 |
| UPF3A | 39 | 1847 | 0.35 | 0.42 |
| MBD6 | 51 | 1835 | 0.35 | 0.41 |
| PGM5 | 45 | 1841 | 0.35 | 0.46 |
| CREBBP | 92 | 1794 | 0.35 | 0.40 |
| RGS12 | 58 | 1828 | 0.35 | 0.42 |
| HLA-B | 52 | 1834 | 0.35 | 0.41 |
| KMT2C | 124 | 1762 | 0.35 | 0.38 |
| PDS5B | 46 | 1840 | 0.35 | 0.38 |
| RNF213 | 71 | 1815 | 0.34 | 0.40 |
| TRIO | 53 | 1833 | 0.34 | 0.43 |
| ZBTB20 | 49 | 1837 | 0.34 | 0.44 |
| PHF2 | 45 | 1841 | 0.34 | 0.45 |
| POLE | 65 | 1821 | 0.34 | 0.39 |
| TCERG1 | 43 | 1843 | 0.33 | 0.43 |
| LARP4B | 43 | 1843 | 0.33 | 0.38 |
| Cancer Type | WT (n) | Mutated (n) | Median OS WT (mo) | Median OS Mut (mo) | Log-Rank p-Value | Cox HR | 95% CI | Cox p-Value |
|---|---|---|---|---|---|---|---|---|
| Pan-Cancer | 1438 | 172 | 17.0 | 34.0 | 0.0024 | 0.68 | 0.52–0.87 | 0.0026 |
| Bladder Cancer | 166 | 45 | 16.0 | NR | 0.0306 | 0.55 | 0.31–0.95 | 0.0337 |
| Colorectal Cancer | 87 | 22 | 13.0 | NR | 0.0048 | 0.25 | 0.09–0.71 | 0.0089 |
| Gastrointestinal Cancers | 195 | 32 | 13.0 | NR | 0.0012 | 0.31 | 0.14–0.65 | 0.0021 |
| Esophagogastric Cancer | 108 | 10 | 15.0 | NR | 0.2125 | 0.48 | 0.15–1.58 | 0.2291 |
| Cancer of Unknown Primary | 73 | 12 | 9.0 | NR | 0.0800 | 0.30 | 0.07–1.26 | 0.1004 |
| Glioma | 108 | 8 | 13.0 | 13.0 | 0.5154 | 0.72 | 0.26–1.97 | 0.5206 |
| Head and Neck Cancer | 118 | 11 | 11.0 | 9.0 | 0.5012 | 0.74 | 0.32–1.74 | 0.4944 |
| Melanoma | 276 | 37 | 42.0 | 49.0 | 0.9580 | 1.02 | 0.57–1.81 | 0.9595 |
| Non-Small Cell Lung Cancer | 319 | 25 | 11.0 | 14.0 | 0.4410 | 0.81 | 0.48–1.37 | 0.4314 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gusakova, M.; Sharko, F.; Mamchur, A.; Boulygina, E.; Mochalova, A.; Bullikh, A.; Patrushev, M. Mutations in CREBBP and EP300 HAT and Bromo Domains Drive Hypermutation and Predict Survival in GI Cancers Treated with Immunotherapy. Biomedicines 2025, 13, 2592. https://doi.org/10.3390/biomedicines13112592
Gusakova M, Sharko F, Mamchur A, Boulygina E, Mochalova A, Bullikh A, Patrushev M. Mutations in CREBBP and EP300 HAT and Bromo Domains Drive Hypermutation and Predict Survival in GI Cancers Treated with Immunotherapy. Biomedicines. 2025; 13(11):2592. https://doi.org/10.3390/biomedicines13112592
Chicago/Turabian StyleGusakova, Mariia, Fedor Sharko, Aleksandra Mamchur, Eugenia Boulygina, Anastasia Mochalova, Artem Bullikh, and Maxim Patrushev. 2025. "Mutations in CREBBP and EP300 HAT and Bromo Domains Drive Hypermutation and Predict Survival in GI Cancers Treated with Immunotherapy" Biomedicines 13, no. 11: 2592. https://doi.org/10.3390/biomedicines13112592
APA StyleGusakova, M., Sharko, F., Mamchur, A., Boulygina, E., Mochalova, A., Bullikh, A., & Patrushev, M. (2025). Mutations in CREBBP and EP300 HAT and Bromo Domains Drive Hypermutation and Predict Survival in GI Cancers Treated with Immunotherapy. Biomedicines, 13(11), 2592. https://doi.org/10.3390/biomedicines13112592






