Abstract
Acute leukemias are highly aggressive hematologic malignancies that demand intensive chemotherapy regimens. However, drug toxicity remains a major barrier to treatment success and patient survival. In this context, pharmacogenomics offers a promising strategy by identifying single-nucleotide variants (SNVs) that influence drug metabolism, efficacy, and toxicity, ultimately impacting treatment outcomes. This study analyzed data from the ClinPGx/PharmGKB database to identify clinically annotated variants related to chemotherapy response in Acute Myeloid Leukemia (AML) and Acute Lymphoblastic Leukemia (ALL). A total of 24 variants were curated for AML and 57 for ALL. Among these, nonsynonymous variants were most frequent in ALL (31.6%), while synonymous variants predominated in AML (33.3%). Although traditionally considered neutral, synonymous and intronic variants may influence gene expression through regulatory or splicing mechanisms. The analysis revealed clinically significant variants associated with chemotherapy response, particularly in the ABCB1 gene, observed in 12.5% of AML and 10.5% of ALL cases. Several variants, particularly TPMT, NUDT15, ABCC1, SLC28A3, and RARG, were associated with severe adverse effects such as myelotoxicity, mucositis, cardiotoxicity, and hepatotoxicity. This study reinforces the importance of genetic variants in modulating the therapeutic response and toxicity to chemotherapy drugs in acute leukemias. Analysis of ClinPGx/PharmGKB data emphasizes ABCB1 as a potential resistance marker and supports pre-treatment genotyping of genes like TPMT and NUDT15 to prevent severe toxicities. Future advances should include the expansion of pharmacogenetic studies in underrepresented populations and the clinical validation of new markers in prospective trials, aiming to consolidate precision medicine as a routine part of the therapeutic management of acute leukemias.
1. Introduction
Acute leukemias represent a group of hematological malignancies of rapid progression and high lethality, which are characterized by the uncontrolled proliferation of immature hematopoietic cells [1,2]. Despite advances in diagnosis and clinical management, conventional chemotherapy remains the main therapeutic modality for these diseases. However, the chemotherapy regimens used are often associated with severe side effects, with toxicity being one of the main obstacles in treatment. Many patients progress to severe complications resulting from chemotherapy, which can compromise the continuity of treatment and, in more extreme cases, lead to death [3,4,5].
Faced with this challenging scenario, there is a global effort in the search for more effective and safer therapies, such as target-directed drugs, which promise a more personalized and less aggressive approach [6]. However, while these treatments are still in the consolidation phase for many forms of acute leukemia, variability in response to conventional chemotherapy remains a critical factor to consider.
Within this context, pharmacogenomics plays a key role in investigating how genetic variations affect drug response. Through the analysis of single-nucleotide variants (SNVs) it is possible to predict drug effectiveness and make dose adjustments in a personalized way. Genetic variants can be classified as synonymous, which do not alter the amino acid sequence of the encoded protein, or non-synonymous, which include missense mutations, nonsense mutations which introduce a premature stop codon, and frameshift mutations. Among these, non-synonymous variants, particularly nonsense and frameshift mutations, are generally more deleterious, as they tend to disrupt protein structure and function more severely [7,8].
As genomic technology advances and sequencing costs decreases, the integration of pharmacogenomics into clinical practice becomes increasingly feasible, enabling more precise and individualized care for patients [9,10,11]. Despite its potential, pharmacogenomic studies in acute leukemia are still scarce, especially in diverse populations, which limits the clinical application of this knowledge.
Therefore, this study aimed to compile all variants described in the ClinPGx/PharmGKB database for acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL), identifying the main effects of the presence of each variant in relation to the efficacy of treatment with chemotherapy and the adverse effects resulting from drug toxicity.
2. Methodology
Search Strategy
Secondary data were obtained from the ClinPGx/PharmGKB, a public database that curate information on how genetic variation affects drug response. This resource integrates data from the scientific literature and clinical studies to support pharmacogenomic research. [12]. In this study, the ClinPGx/PharmGKB was used to identify all genetic variants previously reported to influence chemotherapy response in acute leukemias, without restriction by publication period. Variants related to acute myeloid or lymphoblastic leukemia were retrieved from the “Clinical Annotations” section, which compiles evidence of their clinical relevance, and further explored through the “Variant Annotations” section to review supporting studies. For each variant, data were collected on genomic location, functional impact, and associations with drug efficacy and toxicity, excluding those related solely to metabolism or dosage adjustment. Population allele frequencies were sourced from the 1000 Genomes Project (Phase 3), using the five major superpopulations: African (AFR), Admixed American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS) [13]. The data were curated manually and with the aid of Microsoft Excel spreadsheets, where the variants were initially organized by gene, mutation type, population, and described clinical association. Functional annotations were complemented using literature references indicated by the database itself. After filtering, the variants were reorganized considering the associated gene, type of drug involved, and reported clinical impact.
The level of evidence (LOE) for gene variant–drug associations is a classification made by ClinPGx/PharmGKB that ranges from 1A to 4, based on the strength of available data. Level 1A represents the highest level of evidence, describing variant–drug combinations with specific guidelines in clinical practice guidelines or Food and Drug Administration (FDA) drug labels, requiring at least one additional supporting scientific publication. Level 1B also reflects strong evidence but does not include specific recommendations, requiring two independent publications.
Levels 2A and 2B include associations with moderate evidence, differentiated by the presence (2A) or absence (2B) of variants in ClinPGx/PharmGKB’s Very Important Pharmacogenes (VIPs), both requiring two supporting publications. Level 3 includes associations with limited evidence, which may be based on single studies or preliminary data. Notably, Level 4 is assigned when evidence is insufficient or contradicts the association, resulting in a negative score. This hierarchical classification system enables systematic evaluation of the reliability of gene variant-drug response associations.
3. Results
Initial screening of clinical notes available in the ClinPGx/PharmGKB database resulted in a total of 25 records of drug response-associated variants related to acute myeloid leukemia (AML) and 106 records associated with acute lymphoblastic leukemia (ALL). After applying the eligibility criteria, such as the presence of specific information on genetic variants (SNVs) and their association with therapeutic response or toxicity to drugs used in the treatment of acute leukemias, 24 annotations were selected for AML (Table 1) and 57 for ALL (Table 2).

Table 1.
Acute Myeloid Leukemia Drug Response-Associated Variants Annotations.

Table 2.
Acute Lymphoblastic Leukemia Drug Response-Associated Variants Annotations.
In total, all 24 variants identified in AML related to chemotherapy had a level of evidence (LOE) of 3. These variants are distributed in 17 genes, and the genes with the highest frequency of variants were ABCB1 (12.5%) and RRM2 (12.5%). Among the findings, most of the variants found are synonymous (33.33%) and non-synonymous (20.83%). They have a variety of associations with several drugs, with cytarabine (79.16%) and idarubicin (54.16%) being the drugs that were most present in correlation with the variants.
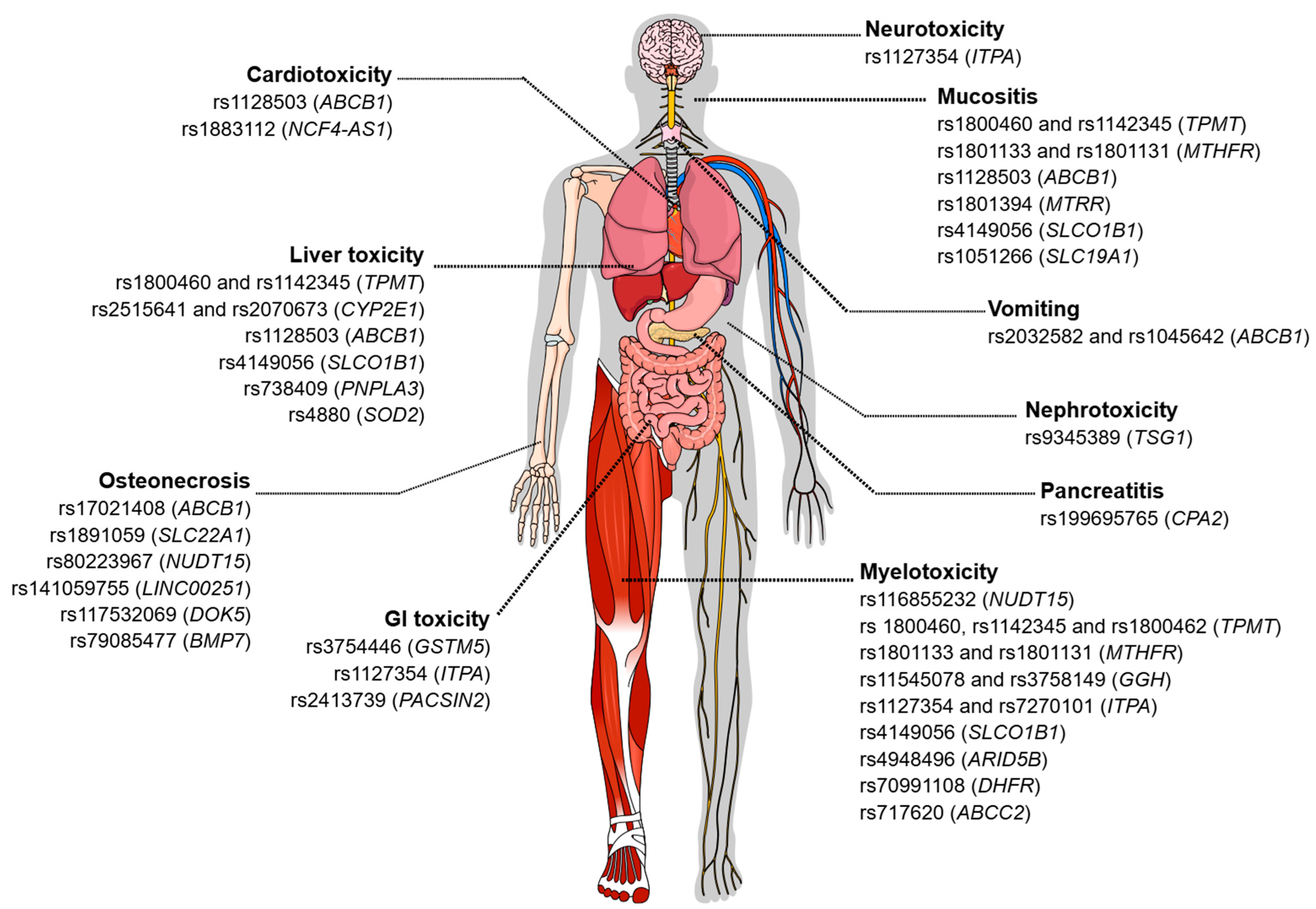
Depending on the genotype (heterozygous or homozygous mutated), an association was observed with the increase or reduction in the intensity of these adverse effects, with 75% of these effects being associated with the efficacy of the drugs and 50% associated with toxicity. The associations regarding the efficacy of chemotherapy drugs are still controversial. About 50% of the reported associations show that the presence of certain variants is related to a higher probability of overall survival (OS), event-free survival (EFS), complete remission (CR), and response to treatment. However, the presence of other specific variants confers a worse response. The main symptomatic characteristics observed regarding toxicity were vomiting, toxic liver disease, and cardiotoxicity (Figure 1).
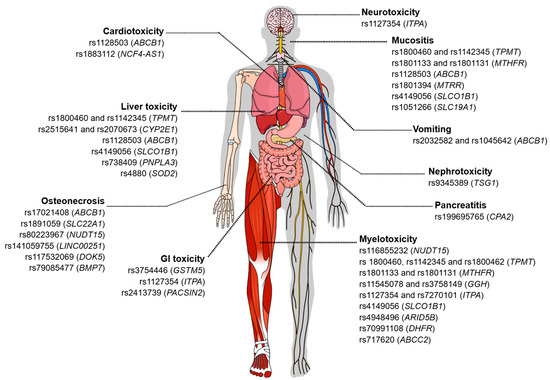
Figure 1.
Genetic variants associated with toxicity related to the treatment of acute leukemias. The figure illustrates different types of chemotherapy-induced toxicities and the genetic variants described in the literature that are associated with each adverse manifestation. Among the toxicities represented are neurotoxicity, cardiotoxicity, hepatotoxicity, pancreatitis, gastrointestinal toxicity, nephrotoxicity, myelotoxicity, mucositis, vomiting, and osteonecrosis. For each type of toxicity, single-nucleotide polymorphisms (SNPs) and the corresponding genes potentially involved in individual susceptibility to these adverse events are indicated. Figure created in the Mind the Graph platform (www.mindthegraph.com (accessed on 3 September 2025)).
In general, altered alleles are associated with a worse response/reaction to chemotherapy drugs. However, in AML, some variants showed positive effects related to the presence of the alternative allele or the homozygous genotype for this allele. This is the case of the rs80143932 and rs2306744 variants, located in the DCK gene, whose altered alleles are associated with a greater response to treatment using cytarabine and idarubicin. In addition, the rs1042919 (RRM1) variant, in turn, demonstrates that the presence of alleles—reference and altered—in heterozygosis, may be associated with a decrease in the response to treatment using cladribine and cytarabine in children.
In contrast to most cases, the absence of the rs1130609 (RRM2) and rs1561876 (STIM1) variants, i.e., the presence of only the reference alleles, is associated with decreased therapeutic response in children. Similarly, the rs11231825 (SLC22A12) variant demonstrates that the homozygous genotype of the reference allele may be related to a higher probability of fever occurrence.
Overall, the 57 variants in ALL are listed in 37 genes that are associated with the response to chemotherapy, of which 4 were level 1A, 1 level 2A, 49 level 3 and 3 level 4. The most frequently listed genes were ABCB1 (10.5%), GGH (7%), NUDT15, TPMT, and SLCO1B1 (5.2% each). The types of variants that occurred most frequently were variants and non-synonyms (31.6%) and intronics (28%). The drugs with the greatest relations with variants were methotrexate (80.7%), followed by asparaginase (21%), vincristine (19.3%) and mercaptopurine (15.7%), mostly being associated with a poorer response and/or increased toxicity.
The presence of variants, whether homozygous or heterozygous, has been associated with the modulation of the intensity of adverse effects, which may result in an increase or reduction in these events. Approximately 38.6% of these associations refer to therapeutic efficacy, in which the presence of at least one allele altered with the variant is associated with minimal residual disease (MRD) detection after induction, decreased event-free survival (EFS) and response to treatment, and increased likelihood of relapse and resistance. Meanwhile, about 84.2% of variants are related to drug toxicity, with the main reported toxic effects being myelosuppression, leukopenia, neutropenia/febrile neutropenia, thrombocytopenia, mucositis, toxic liver disease, osteonecrosis, and gastrointestinal toxicity (Figure 1).
Among the four variants classified with LOE 1A, one occurs in the NUDT15 gene (non-synonymous) and the other 3 were located in the TPMT gene (2 non-synonymous and 1 exonic/splicing). Notably, none of these variants were synonymous, which is consistent with their well-established functional impact on protein activity and, consequently, on drug metabolism. All were associated with toxicity related to chemotherapy agents mercaptopurine, thioguanine and methotrexate. The presence of mutated alleles generally increased the likelihood of myelosuppression, severe pancytopenia, and drug-related toxicity in patients.
The non-synonymous rs1801133 variant that occurs in the MTHFR gene has evidence level 2A and showed associations with the use of methotrexate. According to studies, the presence of the altered allele could indicate a greater probability of relapses and a higher risk of drug toxicity, mucositis, thrombocytopenia, leukopenia, neutropenia, and myelosuppression.
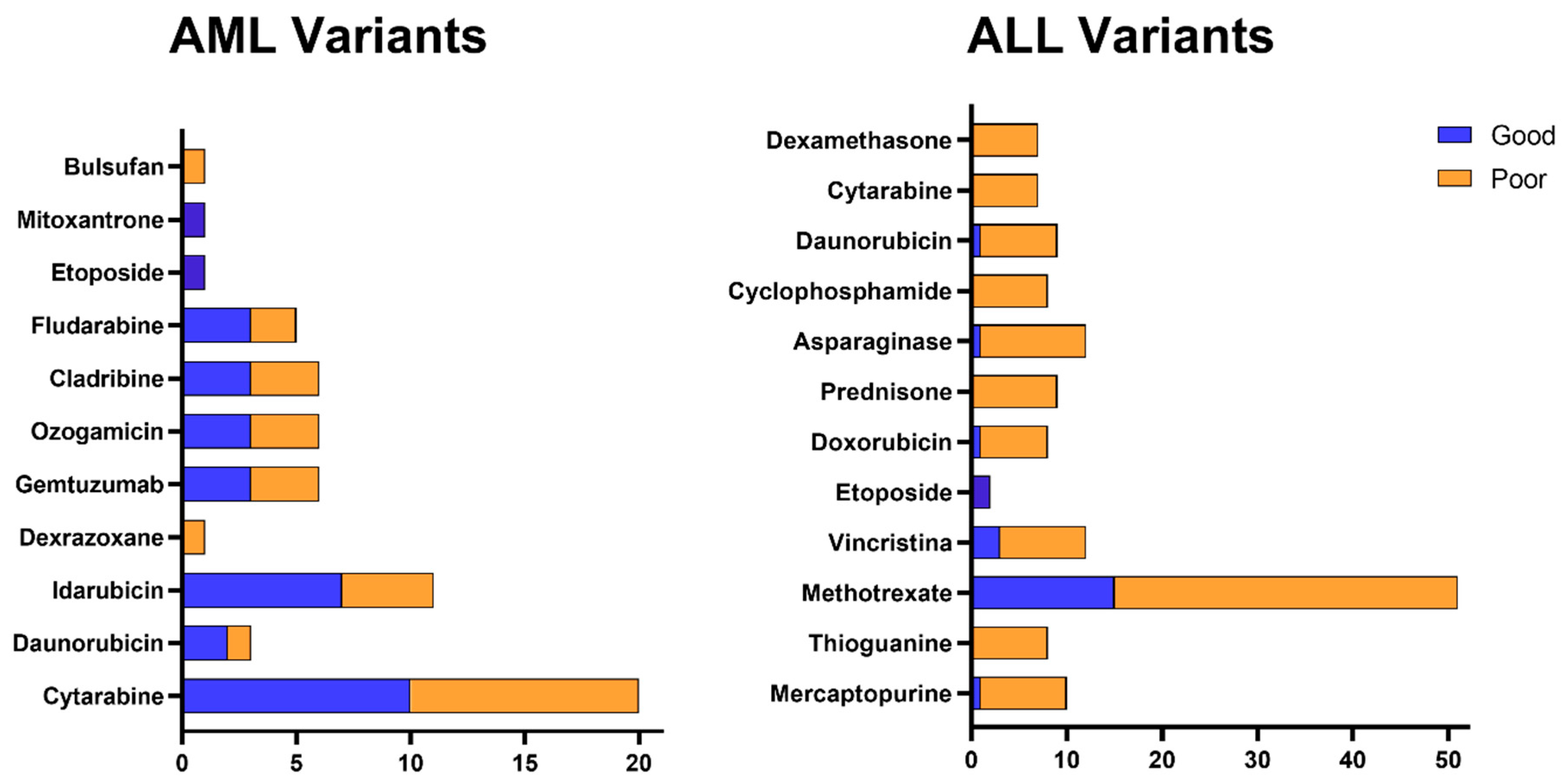
Figure 2 shows that, among the genetic variants described in the previous tables, the relationship with unfavorable outcomes predominates, either due to reduced therapeutic efficacy or increased risk of toxicity. Although variants related to good response are also present, they are less frequently distributed. It is also observed that certain drugs, such as cytarabine, daunorubicin, L-asparaginase, and methotrexate, concentrate a greater number of variants associated with a negative prognosis, while others, such as mercaptopurine and cyclophosphamide, have a more balanced profile between beneficial and adverse effects. Cytarabine stands out in particular, which not only brings together several variants, but is also related to genetic alterations with a relevant clinical impact on the therapeutic response.
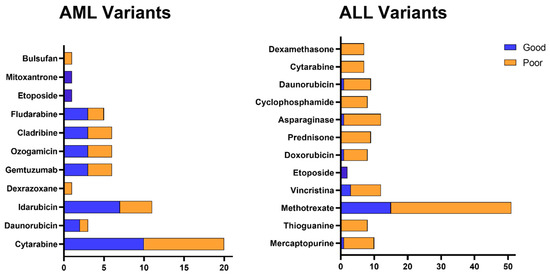
Figure 2.
Distribution of genetic variants associated with the response to drugs used in the treatment of acute leukemias. Association between genetic variants and response to the main chemotherapeutic agents used in the treatment of AML and ALL. The figure shows the prognosis associated with the genetic variants previously described in the tables, classified as related to good response (blue) or poor response (orange) for each therapeutic agent. Drugs depicted include alkylating agents, anthracyclines, topoisomerase II inhibitors, antimetabolites, enzymes, and conjugated antibodies.
Some variants of the ABCB1 gene are common between AML and ALL, such as rs1045642 and rs1128503. The rs1045642 in AML patients treated with cytarabine is associated with higher chances of complete remission and 3-year EFS, in addition to also increasing the probability of vomiting and increased liver enzymes [15,16,17,22]. In ALL patients treated with vincristine, methotrexate and etoposide, the absence of this same variant was associated with a higher probability of developing leukopenia, neutropenia, mucositis, hepatotoxicity, anemia, and thrombocytopenia [45,47,53,54,55,113,114].
The presence of SNV rs1128503 in AML patients treated with cytarabine is associated with lower overall survival (OS) and higher probability of cardiotoxicity. While in methotrexate-treated ALL patients, the presence of the variant is related to a higher risk of drug-induced liver injury and increased severity of mucositis.
In general, the presence of genetic variants is commonly related to a worse response or greater toxicity to chemotherapy treatment; however, in some cases the opposite is observed. The absence of the rs1127354 (ITPA), rs408626, and rs442767 (MSH3) variants is associated with a higher risk of leukopenia. The presence of rs1051266 (SLC19A1) variant in homozygosis is related to greater severity in mucositis, whereas its absence is associated with a higher probability of toxic liver disease, neutropenia, pancreatitis, vomiting, and myelosuppression.
4. Discussion
The investigation of genetic variants associated with therapeutic response in acute leukemia has been shown to be of great relevance in view of the high interindividual variability in the response to chemotherapy drugs and the significant frequency of serious adverse events related to drug toxicity. Understanding these changes allows the personalization of the treatment, contributing to the choice of safer and more effective therapeutic regimens, in addition to minimizing potentially lethal adverse reactions.
The survival rate in AML (32.9%) is generally lower than in ALL (92% for children and 40–60% for adults). This is due to the fact that AML is more prevalent in adults and the elderly, who generally tolerate intensive chemotherapy poorly. It is a disease with lower sensitivity to conventional chemotherapy, with fewer target-directed therapeutic options, and with more common and rapid relapses. ALL, on the other hand, is more prevalent in children, who usually respond and tolerate chemotherapy protocols well, unlike adult patients, who have a slightly worse prognosis. Still, in recent years ALL has evolved in recent decades with standardized protocols and greater use of targeted therapies, such as blinatumomab and inotuzumab [3,119,120,121,122]. Even so, there are fewer studies on the pharmacogenetics of AML and the influence of genetic variants on the response to chemotherapy drugs than on ALL.
Regarding ALL, most studies address the pediatric population, but over the years there has been an increase in the absolute number of cases in adult patients, as reported by Yi et al. (2020) who state that between 1990 and 2017 there was an increase of around 31% in cases [123]. Given their poorer prognosis, further studies should explore how variants affect this patient group, with the aim of reducing severe toxicities and improving clinical outcomes.
In this study, it was possible to observe a predominance of non-synonymous variants (missense) in both types of leukemias, especially in ALL. These variants are generally associated with a greater functional potential, as they cause amino acid substitutions that can directly affect the structure and function of proteins involved in the metabolization, transport or action of chemotherapy drugs. Some studies have already observed that the presence of this type of variant influences the response to drugs used in the treatment of acute leukemia in different ways and may alter the effectiveness of the treatment or increase the risk of severe toxicities [122,124].
On the other hand, the high percentage of synonymous variants observed in AML should not be underestimated. Although these variants do not directly alter the protein sequence, there is growing evidence that they can influence drug response through altering mRNA stability, translation efficiency, and especially splicing. In cancer, synonymous variants in genes such as ABCB1 (MDR1) can modulate the expression of efflux proteins, influencing the response to chemotherapy drugs [125].
In this research, we identified ABCB1 as the gene with the highest frequency of single-nucleotide variants (SNVs) associated with chemotherapy response in acute leukemias, found in 12.5% of AML and 10.5% of ALL cases. ABCB1 (MDR1) encodes P-glycoprotein (P-gp), an ATP-dependent efflux pump that removes cytotoxic drugs from cells, such as anthracyclines, vincristine, and etoposide. Functional studies and pharmacogenetic analyses demonstrate that ABCB1 polymorphisms affect P-gp expression and activity, and are significantly correlated with treatment failure, refractory disease, relapse, reduced event-free and overall survival, as well as increased hematologic toxicity [126,127,128]. Furthermore, a meta-analyses indicated that variant alleles of ABCB1 influence drug accumulation in blast cells, improving remission rates but also elevating toxicity risk [129].
Although hematologic cancers typically exhibit relatively low P-gp levels, post-treatment genetic instability, and clonal selection in refractory or relapsed patients enhance ABCB1 activation and P-gp overexpression. Beyond drug efflux, P-gp appears to support leukemic blast survival through mechanisms independent of chemotherapy expulsion—such as regulation of apoptotic pathways [130,131,132]. These data collectively reinforce the critical role of ABCB1 SNVs in mediating chemoresistance and underscore the utility of integrating ABCB1 genotyping into precision therapy for acute leukemia.
The identification of high-risk ABCB1 variants raises the question of how to clinically overcome P-gp mediated resistance. One promising strategy involves co-administration of P-gp inhibitors to restore intracellular chemotherapeutic concentrations. While third-generation inhibitors like tariquidar have faced toxicity challenges in trials, repurposing established drugs such as verapamil and cyclosporine A has shown promise in preclinical models, and newer agents like elacridar represent a translational frontier for patients with resistance profiles [133,134].
A comprehensive understanding of pharmacogenomics in chemoresistance must also account for key genes with the highest levels of clinical evidence, such as TPMT and NUDT15, which are critical for thiopurine drug metabolism. Both genes carry a Level 1A evidence rating from the Clinical Pharmacogenetics Implementation Consortium (CPIC), underscoring their well-validated role in clinical practice. Thiopurine S-methyltransferase (TPMT) catalyzes the S-methylation of thiopurines, inactivating them and preventing excessive formation of cytotoxic thioguanine nucleotides (TGNs). Patients with loss-of-function alleles (e.g., TPMT2, 3A, 3C) accumulate TGNs, leading to high risk of severe myelosuppression [135].
Similarly, NUDT15 provides a critical detoxification pathway by hydrolyzing the active metabolite deoxythioguanosine triphosphate (dTGTP), preventing its misincorporation into DNA. Loss-of-function NUDT15 variants (e.g., rs116855232) result in dTGTP accumulation, causing DNA damage and leukopenia. For patients carrying these risk variants, pharmacological interventions are already clinically validated; for instance, allopurinol supplementation mitigates thiopurine-induced hematologic toxicity through “allopurinol-guided dosing,” allowing safe administration of therapeutic doses [136,137]. Pre-emptive genotyping for TPMT and NUDT15, enabling dose reductions or therapy switching, exemplifies the successful translation of pharmacogenetics into clinical practice.
It is noteworthy that none of the LOE 1A variants were synonymous, reinforcing that non-synonymous and splicing variants, which directly alter protein function, demonstrate stronger clinical associations [11,138]. However, the potential impact of synonymous and noncoding variants should not be disregarded. While de current pharmacogenomic landscape is dominated by functionally disruptive variants, emerging evidence highlights the role of synonymous and intronic variants in regulating gene expression through mechanisms such as altered mRNA stability, translation efficiency, splicing modulation, and epitranscriptomic modifications like N6-adenosine methylation (m6A) [139,140,141,142]. For example, intronic variants in ALL, though noncoding, can alter splicing or regulatory elements, influencing drug response [143]. Therefore, future studies should include functional annotation of synonymous and intronic polymorphisms to fully elucidate the genetic landscape of chemoresistance [125,144,145].
The adverse effects observed in the treatment of acute leukemia directly reflect the pharmacological nature of the chemotherapy agents, whose targets are tissues of high turnover and metabolic pathways that are fundamental for cell proliferation. The presence of genetic variants associated with the response to chemotherapy drugs can influence the frequency and intensity of these effects and can generate such toxicity that it can lead to the patient’s death even before the treatment protocol is completed [63,146,147].
Polymorphisms in genes such as TPMT (e.g., 3A, 3C) and NUDT15 (e.g., rs116855232) are strongly associated with severe myelotoxicity in patients with acute leukemia treated with thiopurines, such as mercaptopurine and thioguanine. The presence of these variants, even in heterozygosis, can increase the risk of severe myelosuppression by up to nine times, requiring significant dose reductions and may culminate in fatal outcomes in the absence of adequate therapeutic adjustment [36].
Similarly, variants in genes such as SLC28A3, ABCC1, RARG, UGT1A6, and ABCB4, involved in anthracycline transport and metabolism, have been implicated in the predisposition to cardiotoxicity in pediatric and adult patients, compromising treatment continuity and may negatively affect late survival [73,148,149,150]. These recent findings corroborate the importance of pre-therapy genetic evaluation to mitigate severe toxicities, individualizing the approach, and improving clinical outcomes in ALL and AML patients.
While transcriptomic analyses provide valuable insights into gene expression states, a comprehensive genomic profiling strategy must also incorporate the systematic SNV identification to fully predict treatment outcomes and toxicity risks. Gene expression levels are transient and influenced by various factors, whereas germline and somatic SNVs represent stable, patient-specific markers that directly alter protein function, drug metabolism, and transport. Relying solely on expression data may miss critical pharmacogenetic variants in genes such as DPYD, TPMT, NUDT15, or ABCB1, which may not correlate with mRNA abundance but profoundly impact drug efficacy and safety. Therefore, integrating SNV profiling with RNA-sequencing or whole-genome sequencing in clinical workflows enables more robust personalized risk assessment, combining correlative expression signatures with causative genetic factors to refine therapeutic decisions [11,151].
Overall, most reported variants have a LOE of 3, indicating limited supporting data from single studies, case reports, or in vitro assays [12]. This highlights the need for further research to validate these associations and elucidate their clinical impact. While the use of the ClinPGx/PharmGKB database was instrumental in curating clinically relevant pharmacogenetic associations, our study’s limitations reflect those of the source resource, including potential population bias due to underrepresentation of non-European cohorts, underrepresentation of rare variants, and the predominance of LOE 3 associations. Therefore, our results should be interpreted as a comprehensive yet evolving landscape, underscoring the imperative for further validation in diverse cohorts and functional studies to enable safer, more personalized chemotherapeutic interventions.
5. Conclusions
This study reinforces the critical role of genetic variants in modulating therapeutic response and toxicity to chemotherapeutic agents in acute leukemias. Our comprehensive survey of the ClinPGx/PharmGKB database revealed a diverse landscape of pharmacogenomic variants in AML and ALL, with the ABCB1 gene exhibiting the highest frequency in both subtypes, highlighting its central role in mediating resistance. More importantly, the numerous associations identified between specific variants and severe adverse effects highlight the tangible clinical value of pre-treatment genotyping. The implementation of point-of-care testing for high-evidence genes, such as TPMT and NUDT15, represents a readily actionable strategy to proactively guide initial dosing, prevent life-threatening toxicities like myelosuppression, and improve a patient’s quality of life during treatment.
To fully realize the potential of precision oncology in acute leukemias, future efforts must extend beyond current knowledge. It is essential to expand pharmacogenetic studies, particularly in underrepresented populations, to ensure the equity and global applicability of genotyping panels. Furthermore, the clinical utility of a broader set of variants, including those involved in resistance mechanisms like ABCB1 and other promising markers with preliminary evidence, must be rigorously validated in prospective clinical trials. In these trials, patients would be randomized to receive either standard therapy or a genotype-guided protocol, with endpoints focusing on the reduction in severe adverse events, improved dose intensity, and enhanced overall survival. Success of such initiatives would pave the way for the systematic integration of comprehensive pharmacogenomics into standard treatment protocols, ultimately shifting the paradigm from reactive toxicity management to proactive, personalized therapy that maximizes efficacy and safety for all patients.
Author Contributions
Invitation received, C.A.M.-N.; conceptualization, F.M.C.d.P.P., I.M.F., B.M.D.N., and C.A.M.-N.; provision of data and sub-sequent analysis and interpretation, F.M.C.d.P.P., I.M.F., B.M.D.N., C.B.M., I.V.B., A.K.d.C.M., G.P.d.M., L.S.d.C., D.d.S.O., R.M.R., A.P.T., P.M.P.T., M.O.d.M.F., and M.E.A.d.M.; writing—original draft preparation, F.M.C.d.P.P., I.M.F., B.M.D.N., and C.A.M.-N.; writing—review and editing, F.M.C.d.P.P., I.M.F., B.M.D.N., and C.A.M.-N.; funding acquisition, C.A.M.-N. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Brazilian funding agencies: Coordination for the Improvement of Higher Education Personnel (CAPES; to F.M.C.d.P.P., C.B.M., L.S.d.C., and B.M.D.N.), National Council of Technological and Scientific Development (CNPq grant number 404213/2021-9 to C.A.M.-N.; and Productivity in Research PQ scholarships to M.O.d.M.F., M.E.A.d.M. and C.A.M.-N.), and Cearense Foundation of Scientific and Technological Support (FUNCAP; to I.M.F., A.K.d.C.M., and G.P.d.M.).
Data Availability Statement
The data presented in this study are publicly available. Information on pharmacogenetic variants was obtained from the PharmGKB database (https://www.clinpgx.org/), and variant frequency data were obtained from the 1000 Genomes Project (https://www.coriell.org/). All data sources are properly cited in the manuscript.
Conflicts of Interest
A.P.T. was employed by the company Clementino Fraga Group. The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or data interpretation; in the writing of the manuscript, or in the decision to publish the results.
References
- Pagliaro, L.; Chen, S.J.; Herranz, D.; Mecucci, C.; Harrison, C.J.; Mullighan, C.G.; Zhang, M.; Chen, Z.; Boissel, N.; Winter, S.S.; et al. Acute Lymphoblastic Leukaemia. Nat. Rev. Dis. Prim. 2024, 10, 41. [Google Scholar] [CrossRef]
- Tebbi, C.K. Etiology of Acute Leukemia: A Review. Cancers 2021, 13, 2256. [Google Scholar] [CrossRef]
- Hunger, S.P.; Mullighan, C.G. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 2015, 373, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Vakiti, A.; Reynolds, S.B.; Mewawalla, P. Acute Myeloid Leukemia; StatPearls Publishing: Orlando, FL, USA, 2024. [Google Scholar]
- Tallman, M.S.; Gilliland, D.G.; Rowe, J.M. Drug Therapy for Acute Myeloid Leukemia. Blood 2005, 106, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Palomero, T.; Ferrando, A. Therapeutic Targeting of NOTCH1 Signaling in T-ALL Teresa. Clin. Lymphoma Myeloma 2009, 9, S205–S210. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. When a Synonymous Variant Is Nonsynonymous. Genes 2022, 13, 1485. [Google Scholar] [CrossRef]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A Meta-Analysis of Nonsense Mutations Causing Human Genetic Disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef]
- Sadee, W.; Wang, D.; Hartmann, K.; Toland, A.E. Pharmacogenomics: Driving Personalized Medicine. Pharmacol. Rev. 2023, 75, 789–814. [Google Scholar] [CrossRef]
- Haga, S.B.; Burke, W. Using Pharmacogenetics to Improve Drug Safety and Efficacy. Jama 2004, 291, 2869–2871. [Google Scholar] [CrossRef]
- Relling, M.V.; Evans, W.E. Pharmacogenomics in the Clinic. Nature 2015, 526, 343–350. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Gréen, H.; Falk, I.J.; Lotfi, K.; Paul, E.; Hermansson, M.; Rosenquist, R.; Paul, C.; Nahi, H. Association of ABCB1 Polymorphisms with Survival and in Vitro Cytotoxicty in de Novo Acute Myeloid Leukemia with Normal Karyotype. Pharmacogenom. J. 2012, 12, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, J.Y.; Sohn, S.K.; Lee, N.Y.; Baek, J.H.; Jeon, S.B.; Kim, J.G.; Suh, J.S.; Do, Y.R.; Lee, K.B. Multidrug Resistance-1 Gene Polymorphisms Associated with Treatment Outcomes in de Novo Acute Myeloid Leukemia. Int. J. Cancer 2006, 118, 2195–2201. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Yin, J.Y.; Xu, Y.J.; Li, X.; Zhang, Y.; Liu, Z.G.; Zhou, F.; Zhai, M.; Li, Y.; Li, X.P.; et al. Association of ABCB1 Polymorphisms with the Efficacy of Ondansetron in Chemotherapy-Induced Nausea and Vomiting. Clin. Ther. 2014, 36, 1242–1252. [Google Scholar] [CrossRef]
- Megías-Vericat, J.E.; Rojas, L.; Herrero, M.J.; Bosó, V.; Montesinos, P.; Moscardó, F.; Poveda, J.L.; Sanz, M.Á.; Aliño, S.F. Influence of ABCB1 Polymorphisms upon the Effectiveness of Standard Treatment for Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis of Observational Studies. Pharmacogenom. J. 2015, 15, 109–118. [Google Scholar] [CrossRef]
- Mortland, L.; Alonzo, T.A.; Walter, R.B.; Gerbing, R.B.; Mitra, A.K.; Pollard, J.A.; Loken, M.R.; Hirsch, B.; Raimondi, S.; Franklin, J.; et al. Clinical Significance of CD33 Nonsynonymous Single-Nucleotide Polymorphisms in Pediatric Patients with Acute Myeloid Leukemia Treated with Gemtuzumab-Ozogamicin-Containing Chemotherapy. Clin. Cancer Res. 2013, 19, 1620–1627. [Google Scholar] [CrossRef]
- He, H.; Xu, Y.-J.; Yin, J.-Y.; Li, X.; Qu, J.; Xu, X.-J.; Liu, Z.-G.; Zhou, F.; Zhai, M.; Li, Y.; et al. Association of Nitric Oxide Synthase 3 (NOS3) 894 G>T Polymorphism with Prognostic Outcomes of Anthracycline in Chinese Patients with Acute Myeloid Leukaemia. Clin. Exp. Pharmacol. Physiol. 2014, 41, 400–407. [Google Scholar] [CrossRef]
- Cao, X.; Mitra, A.K.; Pounds, S.; Crews, K.R.; Gandhi, V.; Plunkett, W.; Dolan, M.E.; Hartford, C.; Raimondi, S.; Campana, D.; et al. RRM1 and RRM2 Pharmacogenetics: Association with Phenotypes in HapMap Cell Lines and Acute Myeloid Leukemia Patients. Pharmacogenomics 2013, 14, 1449–1466. [Google Scholar] [CrossRef]
- Iacobucci, I.; Lonetti, A.; Candoni, A.; Sazzini, M.; Papayannidis, C.; Formica, S.; Ottaviani, E.; Ferrari, A.; Michelutti, A.; Simeone, E.; et al. Profiling of Drug-Metabolizing Enzymes/Transporters in CD33+ Acute Myeloid Leukemia Patients Treated with Gemtuzumab-Ozogamicin and Fludarabine, Cytarabine and Idarubicin. Pharmacogenom. J. 2013, 13, 335–341. [Google Scholar] [CrossRef]
- Yunis, L.K.; Linares-Ballesteros, A.; Aponte, N.; Barros, G.; García, J.; Niño, L.; Uribe, G.; Quintero, E.; Yunis, J.J. Pharmacogenetics of ABCB1, CDA, DCK, GSTT1, GSTM1 and Outcomes in a Cohort of Pediatric Acute Myeloid Leukemia Patients from Colombia. Cancer Rep. 2023, 6, e1744. [Google Scholar] [CrossRef]
- Cheong, H.S.; Koh, Y.; Ahn, K.S.; Lee, C.; Shin, H.D.; Yoon, S.S. NT5C3 Polymorphisms and Outcome of First Induction Chemotherapy in Acute Myeloid Leukemia. Pharmacogenet. Genom. 2014, 24, 436–441. [Google Scholar] [CrossRef]
- Drenberg, C.D.; Paugh, S.W.; Pounds, S.B.; Shi, L.; Orwick, S.J.; Li, L.; Hu, S.; Gibson, A.A.; Ribeiro, R.C.; Rubnitz, J.E.; et al. Inherited Variation in OATP1B1 Is Associated with Treatment Outcome in Acute Myeloid Leukemia. Clin. Pharmacol. Ther. 2016, 99, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.; Varatharajan, S.; Abbas, S.; Zhang, W.; Shaji, R.V.; Ahmed, R.; Abraham, A.; George, B.; Srivastava, A.; Chandy, M.; et al. Cytidine Deaminase Genetic Variants Influence RNA Expression and Cytarabine Cytotoxicity in Acute Myeloid Leukemia. Pharmacogenomics 2012, 13, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, U.; Dransfeld, C.L.; Bulut, N.; Kramer, M.; Thiede, C.; Ehninger, G.; Schaich, M. SNP Analyses in Cytarabine Metabolizing Enzymes in AML Patients and Their Impact on Treatment Response and Patient Survival: Identification of CDA SNP C-451T as an Independent Prognostic Parameter for Survival. Leukemia 2009, 23, 1929–1932. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shi, J.Y.; Shi, Z.Z.; Zhang, S.J.; Zhu, Y.M.; Gu, B.W.; Li, G.; Bai, X.T.; Gao, X.D.; Hu, J.; Jin, W.; et al. Association between Single Nucleotide Polymorphisms in Deoxycytidine Kinase and Treatment Response among Acute Myeloid Leukaemia Patients. Pharmacogenetics 2004, 14, 759–768. [Google Scholar] [CrossRef]
- Yee, S.W.; Mefford, J.A.; Singh, N.; Percival, M.-E.; Stecula, A.; Yang, K.; Witte, J.S.; Takahashi, A.; Kubo, M.; Matsuda, K.; et al. Impact of Polymorphisms in Drug Pathway Genes on Disease—Free Survival in Adults with Acute Myeloid Leukemia. J. Hum. Genet. 2013, 58, 353–361. [Google Scholar] [CrossRef]
- Larkin, T.; Kashif, R.; Elsayed, A.H.; Greer, B.; Mangrola, K.; Rafiee, R.; Nguyen, N.; Shastri, V.; Horn, B.; Lamba, J.K. Polygenic Pharmacogenomic Markers as Predictors of Toxicity Phenotypes in the Treatment of Acute Lymphoblastic Leukemia: A Single-Center Study. JCO Precis. Oncol. 2023, 7, e2200580. [Google Scholar] [CrossRef]
- Megías-Vericat, J.E.; Montesinos, P.; Herrero, M.J.; Moscardó, F.; Bosó, V.; Rojas, L.; Martínez-Cuadrón, D.; Rodríguez-Veiga, R.; Sendra, L.; Cervera, J.; et al. Impact of NADPH Oxidase Functional Polymorphisms in Acute Myeloid Leukemia Induction Chemotherapy. Pharmacogenom. J. 2018, 18, 301–307. [Google Scholar] [CrossRef]
- Cargnin, S.; Genazzani, A.A.; Canonico, P.L.; Terrazzino, S. Diagnostic Accuracy of NUDT15 Gene Variants for Thiopurine-Induced Leukopenia: A Systematic Review and Meta-Analysis. Pharmalog. Res. 2018, 135, 102–111. [Google Scholar] [CrossRef]
- Pai, A.A.; Mohan, A.; Benjamin, E.S.B.; Illangeswaran, R.S.S.; Raj, I.X.; Janet, N.B.; Arunachalam, A.K.; Kavitha, M.L.; Kulkarni, U.; Devasia, A.J.; et al. Nudt15 c.415c>t Polymorphism Predicts 6-Mp Induced Early Myelotoxicity in Patients with Acute Lymphoblastic Leukemia Undergoing Maintenance Therapy. Pharmgenomics. Pers. Med. 2021, 14, 1303–1313. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kato, M.; Hasegawa, D.; Urayama, K.Y.; Nakadate, H.; Kondoh, K.; Nakamura, K.; Koh, K.; Komiyama, T.; Manabe, A. Susceptibility to 6-MP Toxicity Conferred by a NUDT15 Variant in Japanese Children with Acute Lymphoblastic Leukaemia. Br. J. Haematol. 2015, 171, 109–115. [Google Scholar] [CrossRef]
- Zhou, H.; Li, L.; Yang, P.; Yang, L.; Zheng, J.E.; Zhou, Y.; Han, Y. Optimal Predictor for 6-Mercaptopurine Intolerance in Chinese Children with Acute Lymphoblastic Leukemia: NUDT15, TPMT, or ITPA Genetic Variants? BMC Cancer 2018, 18, 516. [Google Scholar] [CrossRef]
- Ramalingam, R.; Kaur, H.; Scott, J.X.; Sneha, L.M.; Arunkumar, G.; Srinivasan, A.; Paul, S.F.D. Evaluation of Cytogenetic and Molecular Markers with MTX-Mediated Toxicity in Pediatric Acute Lymphoblastic Leukemia Patients. Cancer Chemother. Pharmacol. 2022, 89, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Huang, X.; He, X.; Mao, M.; Chen, M.; Zhang, R.; Shao, H.; Lv, Z.; Liu, X.; Chuan, J. Association of NUDT15 Gene Polymorphism with Adverse Reaction, Treatment Efficacy, and Dose of 6-Mercaptopurine in Patients with Acute Lymphoblastic Leukemia: A Systematic Review and Meta-Analysis. Haematologica 2024, 109, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Hawwa, A.F.; Millership, J.S.; Collier, P.S.; Vandenbroeck, K.; McCarthy, A.; Dempsey, S.; Cairns, C.; Collins, J.; Rodgers, C.; McElnay, J.C. Pharmacogenomic Studies of the Anticancer and Immunosuppressive Thiopurines Mercaptopurine and Azathioprine. Br. J. Clin. Pharmacol. 2008, 66, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Szumlanski, C.; Weinshilboum, R.; Schmiegelow, K. Pharmacokinetics, Dose Adjustments, and 6-Mercaptopurine/ Methotrexate Drug Interactions in Two Patients with Thiopurine Methyltransferase Deficiency. Acta Paediatr. 1998, 87, 108–111. [Google Scholar] [CrossRef]
- Karas-Kuzelicki, N.; Jazbec, J.; Milek, M.; Mlinaric-Rascan, I. Heterozygosity at the TPMT Gene Locus, Augmented by Mutated MTHFR Gene, Predisposes to 6-MP Related Toxicities in Childhood ALL Patients. Leukemia 2009, 23, 971–974. [Google Scholar] [CrossRef]
- Lennard, L.; Cartwright, C.S.; Wade, R.; Vora, A. Thiopurine Dose Intensity and Treatment Outcome in Childhood Lymphoblastic Leukaemia: The Influence of Thiopurine Methyltransferase Pharmacogenetics. Br. J. Haematol. 2015, 169, 228–240. [Google Scholar] [CrossRef]
- Voelz, K.; Miller, G.; Lee-Miller, C. 6-Mercaptopurine-Associated Sinusoidal Obstructive Syndrome During Interim Maintenance I: A Case Report. J. Pediatr. Hematol. Oncol. 2024, 46, 317–321. [Google Scholar] [CrossRef]
- Krynetski, E.Y.; Schuetz, J.D.; Galpin, A.J.; Pui, C.H.; Relling, M.V.; Evans, W.E. A Single Point Mutation Leading to Loss of Catalytic Activity in Human Thiopurine S-Methyltransferase. Proc. Natl. Acad. Sci. USA 1995, 92, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Correa-Jimenez, O.; Yunis, J.J.; Linares-Ballesteros, A.; Sarmiento-Urbina, I. Susceptibility to Thiopurine Toxicity by TPMT and NUDT15 Variants in Colombian Children with Acute Lymphoblastic Leukemia. Colomb. Med. 2021, 52, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Soler, A.M.; Olano, N.; Méndez, Y.; Lopes, A.; Silveira, A.; Dabezies, A.; Castillo, L.; da Luz, J.A. TPMT and NUDT15 Genes Are Both Related to Mercaptopurine Intolerance in Acute Lymphoblastic Leukaemia Patients from Uruguay. Br. J. Haematol. 2018, 181, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Esmaili, M.A.; Kazemi, A.; Faranoush, M.; Mellstedt, H.; Zaker, F.; Safa, M.; Mehrvar, N.; Rezvany, M.R. Polymorphisms within Methotrexate Pathwaygenes: Relationship between Plasma Methotrexate Levels, Toxicity Experienced and Outcome in Pediatric Acute Lymphoblastic Leukemia. Iran. J. Basic Med. Sci. 2020, 23, 800–809. [Google Scholar] [CrossRef]
- Shimasaki, N.; Mori, T.; Torii, C.; Sato, R.; Shimada, H.; Tanigawara, Y.; Kosaki, K.; Takahashi, T. Influence of MTHFR and RFC1 Polymorphisms on Toxicities during Maintenance Chemotherapy for Childhood Acute Lymphoblastic Leukemia or Lymphoma. J. Pediatr. Hematol. Oncol. 2008, 30, 347–352. [Google Scholar] [CrossRef]
- Zgheib, N.K.; Akra-Ismail, M.; Aridi, C.; Mahfouz, R.; Abboud, M.R.; Solh, H.; Muwakkit, S.A. Genetic Polymorphisms in Candidate Genes Predict Increased Toxicity with Methotrexate Therapy in Lebanese Children with Acute Lymphoblastic Leukemia. Pharmacogenet. Genom. 2014, 24, 387–396. [Google Scholar] [CrossRef]
- Tan, Y.; Kong, Q.; Li, X.; Tang, Y.; Mai, H.; Zhen, Z.; Zhou, D.; Chen, H. Relationship between Methylenetetrahydrofolate Reductase Gene Polymorphisms and Methotrexate Drug Metabolism and Toxicity. Transl. Pediatr. 2023, 12, 31–45. [Google Scholar] [CrossRef]
- Aráoz, H.V.; D’Aloi, K.; Foncuberta, M.E.; Sanchez La Rosa, C.G.; Alonso, C.N.; Chertkoff, L.; Felice, M. Pharmacogenetic Studies in Children with Acute Lymphoblastic Leukemia in Argentina. Leuk. Lymphoma 2015, 56, 1370–1378. [Google Scholar] [CrossRef]
- EL-Khodary, N.M.; EL-Haggar, S.M.; Eid, M.A.; Ebeid, E.N. Study of the Pharmacokinetic and Pharmacogenetic Contribution to the Toxicity of High-Dose Methotrexate in Children with Acute Lymphoblastic Leukemia. Med. Oncol. 2012, 29, 2053–2062. [Google Scholar] [CrossRef]
- Salazar, J.; Altés, A.; del Río, E.; Estella, J.; Rives, S.; Tasso, M.; Navajas, A.; Molina, J.; Villa, M.; Vivanco, J.L.; et al. Methotrexate Consolidation Treatment According to Pharmacogenetics of MTHFR Ameliorates Event-Free Survival in Childhood Acute Lymphoblastic Leukaemia. Pharmacogenom. J. 2012, 12, 379–385. [Google Scholar] [CrossRef]
- Liu, S.-G.; Li, Z.-G.; Cui, L.; Gao, C.; Li, W.-J.; Zhao, X.-X. Effects of Methylenetetrahydrofolate Reductase Gene Polymorphisms on Toxicities during Consolidation Therapy in Pediatric Acute Lymphoblastic Leukemia in a Chinese Population. Leuk. Lymphoma 2011, 52, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Sun, J.-L.; Li, R.; Li, X. Involvement of the ABCB1 C3435T Variant but Not the MTHFR C677T or MTHFR A1298C Variant in High-Dose Methotrexate-Induced Toxicity in Pediatric Acute Lymphoblastic Leukemia Patients in China. Int. J. Gen. Med. 2024, 17, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.D.; Taylor, O.A.; Gramatges, M.M.; Hughes, A.E.; Zobeck, M.; Pruitt, S.; Bernhardt, M.B.; Chavana, A.; Huynh, V.; Ludwig, K.; et al. Evaluation of Methotrexate Pharmacogenomic Variation to Predict Acute Neurotoxicity in Children with Acute Lymphoblastic Leukemia. Pharmacotherapy 2025, 45, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Suthandiram, S.; Gan, G.G.; Zain, S.M.; Bee, P.C.; Lian, L.H.; Chang, K.M.; Ong, T.C.; Mohamed, Z. Effect of Polymorphisms within Methotrexate Pathway Genes on Methotrexate Toxicity and Plasma Levels in Adults with Hematological Malignancies. Pharmacogenomics 2014, 15, 1479–1494. [Google Scholar] [CrossRef]
- Gregers, J.; Christensen, I.J.; Dalhoff, K.; Lausen, B.; Schroeder, H.; Rosthoej, S.; Carlsen, N.; Schmiegelow, K.; Peterson, C. The Association of Reduced Folate Carrier 80G>A Polymorphism to Outcome in Childhood Acute Lymphoblastic Leukemia Interacts with Chromosome 21 Copy Number. Blood 2010, 115, 4671–4677. [Google Scholar] [CrossRef][Green Version]
- Lin, C.; Ma, R.; Zeng, X.; Zhang, B.; Cao, T.; Jiao, S.; Chen, H.; He, Y.; Liu, M.; Cai, H. Integration of Genomics, Clinical Characteristics and Baseline Biological Profiles to Predict the Risk of Liver Injury Induced by High-Dose Methotrexate. Front. Pharmacol. 2024, 15, 1423214. [Google Scholar] [CrossRef]
- Gong, Y.; Luo, L.; Wang, L.; Chen, J.; Chen, F.; Ma, Y.; Xu, Z.; Sun, Y.; Luo, L.; Shi, C.; et al. Association of MTHFR and ABCB1 Polymorphisms with MTX-Induced Mucositis in Chinese Paediatric Patients with Acute Lymphoblastic Leukaemia, Lymphoma or Osteosarcoma—A Retrospective Cohort Study. J. Clin. Pharm. Ther. 2021, 46, 1557–1563. [Google Scholar] [CrossRef]
- Gurieva, O.D.; Savelyeva, M.I.; Valiev, T.T.; Sozaeva, Z.A.; Kondratenko, S.N.; Ilyin, M.V. Pharmacogenetic Aspects of Efficacy and Safety of Methotrexate Treatment in Pediatric Acute Lymphoblastic Leukemia. Drug Metab. Pers. Ther. 2023, 38, 349–357. [Google Scholar] [CrossRef]
- Dorababu, P.; Naushad, S.M.; Linga, V.G.; Gundeti, S.; Nagesh, N.; Kutala, V.K.; Reddanna, P.; Digumarti, R. Genetic Variants of Thiopurine and Folate Metabolic Pathways Determine 6-MP-Mediated Hematological Toxicity in Childhood ALL. Pharmacogenomics 2012, 13, 1001–1008. [Google Scholar] [CrossRef]
- Garcia-Bournissen, F.; Moghrabi, A.; Krajinovic, M. Therapeutic Responses in Childhood Acute Lymphoblas—Tic Leukemia (ALL) and Haplotypes of Gamma Glutamyl Hydrolase (GGH) Gene. Leuk. Res. 2007, 31, 1023–1025. [Google Scholar] [CrossRef]
- Li, M.; Wang, S.M.; Wu, W.S.; Yan, D.; Zhang, L.P.; Zheng, H.Y. Frequency Distribution of Five SNPs in Human GGH Gene and Their Effects on Clinical Outcomes of Chinese Pediatric Patients with Acute Lymphoblastic Leukemia. Pharmazie 2020, 75, 142–146. [Google Scholar] [CrossRef]
- Kishi, S.; Cheng, C.; French, D.; Pei, D.; Das, S.; Cook, E.H.; Hijiya, N.; Rizzari, C.; Rosner, G.L.; Frudakis, T.; et al. Ancestry and Pharmacogenetics of Antileukemic Drug Toxicity. Blood 2007, 109, 4151–4157. [Google Scholar] [CrossRef] [PubMed]
- Franca, R.; Rebora, P.; Bertorello, N.; Fagioli, F.; Conter, V.; Biondi, A.; Colombini, A.; Micalizzi, C.; Zecca, M.; Parasole, R.; et al. Pharmacogenetics and Induction/Consolidation Therapy Toxicities in Acute Lymphoblastic Leukemia Patients Treated with AIEOP-BFM ALL 2000 Protocol. Pharmacogenom. J. 2017, 17, 4–10. [Google Scholar] [CrossRef]
- Kim, H.; Kang, H.J.; Kim, H.J.; Jang, M.K.; Kim, N.H.; Oh, Y.; Han, B.D.; Choi, J.Y.; Kim, C.W.; Lee, J.W.; et al. Pharmacogenetic Analysis of Pediatric Patients with Acute Lymphoblastic Leukemia: A Possible Association between Survival Rate and ITPA Polymorphism. PLoS ONE 2012, 7, e45558. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.; Adam, H.; Hailu, D.; Engidawork, E.; Howe, R.; Abula, T.; Coenen, M.J.H. Genetic Variants of Genes Involved in Thiopurine Metabolism Pathway Are Associated with 6-Mercaptopurine Toxicity in Pediatric Acute Lymphoblastic Leukemia Patients from Ethiopia. Front. Pharmacol. 2023, 14, 1159307. [Google Scholar] [CrossRef] [PubMed]
- Stocco, G.; Cheok, M.; Crews, K.; Dervieux, T.; French, D.; Pei, D.; Yang, W.; Cheng, C.; Pui, C.-H.; Relling, M.; et al. Genetic Polymorphism of Inosine Triphosphate Pyrophosphatase Is a Determinant of Mercaptopurine Metabolism and Toxicity During Treatment for Acute Lymphoblastic Leukemia. Clin. Pharmacol. Ther. 2009, 2385, 164–172. [Google Scholar] [CrossRef]
- Hareedy, M.S.; El Desoky, E.S.; Woillard, J.B.; Thabet, R.H.; Ali, A.M.; Marquet, P.; Picard, N. Genetic Variants in 6-Mercaptopurine Pathway as Potential Factors of Hematological Toxicity in Acute Lymphoblastic Leukemia Patients. Pharmacogenomics 2015, 16, 1119–1134. [Google Scholar] [CrossRef]
- Erčulj, N.; Kotnik, B.F.; Debeljak, M.; Jazbec, J.; Dolžan, V. Influence of Folate Pathway Polymorphisms on High-Dose Methotrexate-Related Toxicity and Survival in Childhood Acute Lymphoblastic Leukemia. Leuk. Lymphoma 2012, 53, 1096–1104. [Google Scholar] [CrossRef]
- Krajinovic, M.; Lemieux-Blanchard, É.; Chiasson, S.; Primeau, M.; Costea, I.; Moghrabi, A. Role of Polymorphism in MTHFR and MTHFD1 Genes in the Outcome of Childhood Acute Lymphoblastic Leukemia. Pharmacogenom. J. 2004, 4, 66–72. [Google Scholar] [CrossRef]
- Hao, Q.; Song, Y.; Fang, Q.; Lin, Y.; Chen, L.; Wang, X.; Zhang, P.; Wang, Z.; Gong, X.; Liu, K.; et al. Effects of Genetic Polymorphisms on Methotrexate Levels and Toxicity in Chinese Patients with Acute Lymphoblastic Leukemia. Blood Sci. 2023, 5, 32–38. [Google Scholar] [CrossRef]
- Huang, L.; Tissing, W.J.E.; de Jonge, R.; van Zelst, B.D.; Pieters, R. Polymorphisms in Folate-Related Genes: Association with Side Effects of High-Dose Methotrexate in Childhood Acute Lymphoblastic Leukemia. Leukemia 2008, 22, 1798–1800. [Google Scholar] [CrossRef]
- Yang, W.; Karol, S.E.; Hoshitsuki, K.; Lee, S.; Larsen, E.C.; Winick, N.; Carroll, W.L.; Loh, M.L.; Raetz, E.A.; Hunger, S.P.; et al. Association of Inherited Genetic Factors with Drug-Induced Hepatic Damage among Children with Acute Lymphoblastic Leukemia. JAMA Netw. Open 2022, 5, E2248803. [Google Scholar] [CrossRef]
- Liu, S.G.; Gao, C.; Zhang, R.D.; Zhao, X.X.; Cui, L.; Li, W.J.; Chen, Z.P.; Yue, Z.X.; Zhang, Y.Y.; Wu, M.Y.; et al. Polymorphisms in Methotrexate Transporters and Their Relationship to Plasma Methotrexate Levels, Toxicity of High-Dose Methotrexate, and Outcome of Pediatric Acute Lymphoblastic Leukemia. Oncotarget 2017, 8, 37761–37772. [Google Scholar] [CrossRef]
- Gutierrez-Camino, A.; Martin-Guerrero, I.; Garcia-Orad, A. PNPLA3 Rs738409 and Hepatotoxicity in Children with B-Cell Acute Lymphoblastic Leukemia: A Validation Study in a Spanish Cohort. Clin. Pharmacol. Ther. 2017, 102, 906. [Google Scholar] [CrossRef] [PubMed]
- Radtke, S.; Zolk, O.; Renner, B.; Paulides, M.; Zimmermann, M.; Möricke, A.; Stanulla, M.; Schrappe, M.; Langer, T. Germline Genetic Variations in Methotrexate Candidate Genes Are Associated with Pharmacokinetics, Toxicity, and Outcome in Childhood Acute Lymphoblastic Leukemia. Blood 2013, 121, 5145–5153. [Google Scholar] [CrossRef] [PubMed]
- Braidotti, S.; Zudeh, G.; Franca, R.; Kiren, V.; Colombini, A.; Bettini, L.R.; Brivio, E.; Locatelli, F.; Vinti, L.; Bertorello, N.; et al. The Role of Candidate Polymorphisms in Drug Transporter Genes on High-Dose Methotrexate in the Consolidation Phase of the AIEOP-BFM ALL 2009 Protocol. Clin. Transl. Sci. 2025, 18, e70136. [Google Scholar] [CrossRef]
- Wu, S.; Wang, M.; Alqahtani, A.; Lou, M.; Stock, W.; Bhojwani, D.; Alachkar, H. Hispanic Ethnicity and the Rs4880 Variant in SOD2 Are Associated with Elevated Liver Enzymes and Bilirubin Levels in Children Receiving Asparaginase-Containing Chemotherapy for Acute Lymphoblastic Leukemia. Biomed. Pharmacother. 2022, 150, 113000. [Google Scholar] [CrossRef]
- Alachkar, H.; Fulton, N.; Sanford, B.; Malnassy, G.; Mutonga, M.; Larson, R.A.; Bloomfield, C.D.; Marcucci, G.; Nakamura, Y.; Stock, W. Expression and Polymorphism (Rs4880) of Mitochondrial Superoxide Dismutase (SOD2) and Asparaginase Induced Hepatotoxicity in Adult Patients with Acute Lymphoblastic Leukemia. Pharmacogenom. J. 2017, 17, 274–279. [Google Scholar] [CrossRef]
- Costea, I.; Moghrabi, A.; Laverdiere, C.; Graziani, A.; Krajinovic, M. Folate Cycle Gene Variants and Chemotherapy Toxicity in Pediatric Patients with Acute Lymphoblastic Leukemia. Haematologica 2006, 91, 1113–1116. [Google Scholar]
- Costea, I.; Moghrabi, A.; Krajinovic, M. The Influence of Cyclin D1 (CCND1) 870A>G Polymorphism and CCND1-Thymidylate Synthase (TS) Gene-Gene Interaction on the Outcome of Childhood Acute Lymphoblastic Leukaemia. Pharmacogenetics 2003, 13, 577–580. [Google Scholar] [CrossRef]
- Liu, C.; Yang, W.; Devidas, M.; Cheng, C.; Pei, D.; Smith, C.; Carroll, W.L.; Raetz, E.A.; Bowman, W.P.; Larsen, E.C.; et al. Clinical and Genetic Risk Factors for Acute Pancreatitis in Patients with Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2016, 34, 2133–2140. [Google Scholar] [CrossRef]
- Karol, S.E.; Mattano, L.A.; Yang, W.; Maloney, K.W.; Smith, C.; Liu, C.C.; Ramsey, L.B.; Fernandez, C.A.; Chang, T.Y.; Neale, G.; et al. Genetic Risk Factors for the Development of Osteonecrosis in Children under Age 10 Treated for Acute Lymphoblastic Leukemia. Blood 2016, 127, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Tong, H.F.; Li, Y.; Qian, J.C.; Wang, J.X.; Wang, Z.; Ruan, J.C. Effect of the Polymorphism of Folylpolyglutamate Synthetase on Treatment of High-Dose Methotrexate in Pediatric Patients with Acute Lymphocytic Leukemia. Med. Sci. Monit. 2016, 22, 4967–4973. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-H.; Pei, D.; Yang, W.; Cheng, C.; Jeha, S.; Cox, N.J.; Evans, W.E.; Pui, C.-H.; Relling, M.V. Genetic Variations in GRIA1 on Chromosome 5q33 Related to Asparaginase Hypersensitivity. Clin. Pharmacol. Ther. 2010, 88, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cheng, C.; Yang, W.; Pei, D.; Cao, X.; Fan, Y.; Pounds, S.; Treviño, L.R.; French, D.; Campana, D.; et al. Genome-Wide Interrogation of Germline Genetic Variation Associated with Treatment Response in Childhood Acute Lymphoblastic Leukemia. Jama 2009, 301, 393–403. [Google Scholar] [CrossRef][Green Version]
- Ceppi, F.; Langlois-Pelletier, C.; Gagné, V.; Rousseau, J.; Ciolino, C.; Lorenzo, S.D.; Kevin, K.M.; Cijov, D.; Sallan, S.E.; Silverman, L.B.; et al. Polymorphisms of the Vincristine Pathway and Response to Treatment in Children with Childhood Acute Lymphoblastic Leukemia. Pharmacogenomics 2014, 15, 1105–1116. [Google Scholar] [CrossRef]
- Lopez-Lopez, E.; Ballesteros, J.; Piñan, M.A.; Sanchez De Toledo, J.; Garcia De Andoin, N.; Garcia-Miguel, P.; Navajas, A.; Garcia-Orad, A. Polymorphisms in the Methotrexate Transport Pathway: A New Tool for MTX Plasma Level Prediction in Pediatric Acute Lymphoblastic Leukemia. Pharmacogenet. Genom. 2013, 23, 53–61. [Google Scholar] [CrossRef]
- Den Hoed, M.A.H.; Lopez-Lopez, E.; Te Winkel, M.L.; Tissing, W.; De Rooij, J.D.E.; Gutierrez-Camino, A.; Garcia-Orad, A.; Den Boer, E.; Pieters, R.; Pluijm, S.M.F.; et al. Genetic and Metabolic Determinants of Methotrexate-Induced Mucositis in Pediatric Acute Lymphoblastic Leukemia. Pharmacogenom. J. 2015, 15, 248–254. [Google Scholar] [CrossRef]
- Zobeck, M.C.; Bernhardt, M.B.; Kamdar, K.Y.; Rabin, K.R.; Lupo, P.J.; Scheurer, M.E. Novel Risk Factors for Glucarpidase Use in Pediatric Acute Lymphoblastic Leukemia: Hispanic Ethnicity, Age, and the ABCC4 Gene. Pediatr. Blood Cancer 2021, 68, e29036. [Google Scholar] [CrossRef]
- Razali, R.H.; Noorizhab, M.N.F.; Jamari, H.; James, R.J.; Teh, K.H.; Ibrahim, H.M.; Teh, L.K.; Salleh, M.Z. Association of ABCC2 with Levels and Toxicity of Methotrexate in Malaysian Childhood Acute Lymphoblastic Leukemia (ALL). Pediatr. Hematol. Oncol. 2020, 37, 185–197. [Google Scholar] [CrossRef]
- Rajić, V.; Aplenc, R.; Debeljak, M.; Prestor, V.V.; Karas-Kuželicki, N.; Mlinarič-Raščan, I.; Jazbec, J. Influence of the Polymorphism in Candidate Genes on Late Cardiac Damage in Patients Treated Due to Acute Leukemia in Childhood. Leuk. Lymphoma 2009, 50, 1693–1698. [Google Scholar] [CrossRef] [PubMed]
- Ceppi, F.; Gagné, V.; Douyon, L.; Quintin, C.J.; Colombini, A.; Parasole, R.; Buldini, B.; Basso, G.; Conter, V.; Cazzaniga, G.; et al. DNA Variants in DHFR Gene and Response to Treatment in Children with Childhood B ALL: Revisited in AIEOP-BFM Protocol. Pharmacogenomics 2018, 19, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Lee, C.H.; Park, H.S.; Lee, J.H.; Kang, Y.A.; Kim, S.K.; Chang, J.; Kim, D.J.; Rha, S.Y.; Kim, J.H.; et al. Pharmacogenomic Assessment of Outcomes of Pemetrexed-Treated Patients with Adenocarcinoma of the Lung. Yonsei Med. J. 2013, 54, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Kodidela, S.; Pradhan, S.C.; Dubashi, B.; Basu, D. Influence of Dihydrofolate Reductase Gene Polymorphisms Rs408626 (-317A>G) and Rs442767 (-680C>A) on the Outcome of Methotrexate-Based Maintenance Therapy in South Indian Patients with Acute Lymphoblastic Leukemia. Eur. J. Clin. Pharmacol. 2015, 71, 1349–1358. [Google Scholar] [CrossRef]
- Ongaro, A.; De Mattei, M.; Della Porta, M.G.; Rigolin, G.; Ambrosio, C.; Di Raimondo, F.; Pellati, A.; Masieri, F.F.; Caruso, A.; Catozzi, L.; et al. Gene Polymorphisms in Folate Metabolizing Enzymes in Adult Acute Lymphoblastic Leukemia: Effects on Methotrexate-Related Toxicity and Survival. Haematologica 2009, 94, 1391–1398. [Google Scholar] [CrossRef]
- Yousef, A.M.; Farhad, R.; Alshamaseen, D.; Alsheikh, A.; Zawiah, M.; Kadi, T. Folate Pathway Genetic Polymorphisms Modulate Methotrexate-Induced Toxicity in Childhood Acute Lymphoblastic Leukemia. Cancer Chemother. Pharmacol. 2019, 83, 755–762. [Google Scholar] [CrossRef]
- López-López, E.; Gutiérrez-Camino, Á.; Piñán, M.Á.; Sánchez-Toledo, J.; Uriz, J.J.; Ballesteros, J.; García-Miguel, P.; Navajas, A.; García-Orad, Á. Pharmacogenetics of MicroRNAs and MicroRNAs Biogenesis Machinery in Pediatric Acute Lymphoblastic Leukemia. PLoS ONE 2014, 9, e91261. [Google Scholar] [CrossRef]
- Fernandez, C.A.; Smith, C.; Yang, W.; Mullighan, C.G.; Qu, C.; Larsen, E.; Bowman, W.P.; Liu, C.; Ramsey, L.B.; Chang, T.; et al. Genome-Wide Analysis Links NFATC2 with Asparaginase Hypersensitivity. Blood 2015, 126, 69–75. [Google Scholar] [CrossRef]
- Franca, R.; Stocco, G.; Favretto, D.; Giurici, N.; del Rizzo, I.; Locatelli, F.; Vinti, L.; Biondi, A.; Colombini, A.; Fagioli, F.; et al. PACSIN2 Rs2413739 Influence on Thiopurine Pharmacokinetics: Validation Studies in Pediatric Patients. Pharmacogenom. J. 2020, 20, 415–425. [Google Scholar] [CrossRef]
- Stocco, G.; Yang, W.; Crews, K.R.; Thierfelder, W.E.; Decorti, G.; Londero, M.; Franca, R.; Rabusin, M.; Valsecchi, M.G.; Pei, D.; et al. PACSIN2 Polymorphism Influences TPMT Activity and Mercaptopurine-Related Gastrointestinal Toxicity. Hum. Mol. Genet. 2012, 21, 4793–4804. [Google Scholar] [CrossRef]
- Smid, A.; Karas-Kuzelicki, N.; Jazbec, J.; Mlinaric-Rascan, I. PACSIN2 Polymorphism Is Associated with Thiopurine-Induced Hematological Toxicity in Children with Acute Lymphoblastic Leukaemia Undergoing Maintenance Therapy. Sci. Rep. 2016, 6, 30244. [Google Scholar] [CrossRef]
- Franca, R.; Stocco, G.; Kiren, V.; Tessitore, A.; Fagioli, F.; Quarello, P.; Bertorello, N.; Rizzari, C.; Colombini, A.; Bettini, L.R.; et al. Impact of Mercaptopurine Metabolites on Disease Outcome in the AIEOP-BFM ALL 2009 Protocol for Acute Lymphoblastic Leukemia. Clin. Pharmacol. Ther. 2023, 114, 1082–1092. [Google Scholar] [CrossRef]
- Yang, J.J.; Cheng, C.; Devidas, M. Genome-Wide Association Study Identifies Germline Polymorphisms Associated with Relapse of Childhood Acute Lymphoblastic Leukemia. Transfuze A Hematol. Dnes. 2013, 19, 54. [Google Scholar] [CrossRef]
- Treviño, L.R.; Shimasaki, N.; Yang, W.; Panetta, J.C.; Cheng, C.; Pei, D.; Chan, D.; Sparreboom, A.; Giacomini, K.M.; Pui, C.H.; et al. Germline Genetic Variation in an Organic Anion Transporter Polypeptide Associated with Methotrexate Pharmacokinetics and Clinical Effects. J. Clin. Oncol. 2009, 27, 5972–5978. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Panetta, J.C.; Smith, C.; Yang, W.; Fan, Y.; Winick, N.J.; Martin, P.L.; Cheng, C.; Devidas, M.; Pui, C.H.; et al. Genome-Wide Study of Methotrexate Clearance Replicates SLCO1B1. Blood 2013, 121, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.; Sauty, G.; Labuda, M.; Gagné, V.; Rousseau, J.; Moghrabi, A.; Laverdière, C.; Sinnett, D.; Krajinovic, M. Polymorphism in Multidrug Resistance-Associated Protein Gene 3 Is Associated with Outcomes in Childhood Acute Lymphoblastic Leukemia. Pharmacogenom. J. 2012, 12, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Shendy, K.; Abdelkawy, K.; Ali, A.A.; Belal, F.; Abdelhakiem, M.; Magdy, G.; Anber, N.; Elbarbry, F. The Effects of Genetic Polymorphism on Toxicity and Pharmacokinetics of Methotrexate in Egyptian Adult Patients with Leukaemia or Lymphoma. Xenobiotica 2024, 54, 95–105. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, Y.; Sheng, Q.; Lu, X.; Wang, F.; Lin, Z.; Tian, H.; Xu, A.; Zhang, J. Association of ABCC2 -24C>T Polymorphism with High-Dose Methotrexate Plasma Concentrations and Toxicities in Childhood Acute Lymphoblastic Leukemia. PLoS ONE 2014, 9, e82681. [Google Scholar] [CrossRef]
- Al-Shakfa, F.; Dulucq, S.; Brukner, I.; Milacic, I.; Ansari, M.; Beaulieu, P.; Moghrabi, A.; Laverdière, C.; Sallan, S.E.; Silverman, L.B.; et al. DNA Variants in Region for Noncoding Interfering Transcript of Dihydrofolate Reductase Gene and Outcome in Childhood Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2009, 15, 6931–6938. [Google Scholar] [CrossRef]
- Kotur, N.; Lazic, J.; Ristivojevic, B.; Stankovic, B.; Gasic, V.; Dokmanovic, L.; Krstovski, N.; Milosevic, G.; Janic, D.; Zukic, B.; et al. Pharmacogenomic Markers of Methotrexate Response in the Consolidation Phase of Pediatric Acute Lymphoblastic Leukemia Treatment. Genes 2020, 11, 468. [Google Scholar] [CrossRef]
- Nguyen, M.L.; Hoang, A.V.; Duong, B.T.; Phung, N.T.N. The Influence of NUDT15 Variants on 6-Mercaptopurine-Induced Neutropenia in Vietnamese Pediatric Acute Lymphoblastic Leukemia. Hum. Genet. Genom. Adv. 2023, 4, 100183. [Google Scholar] [CrossRef]
- French, D.; Hamilton, L.H.; Mattano, L.A.; Sather, H.N.; Devidas, M.; Nachman, J.B.; Relling, M.V. A PAI-1 (SERPINEI) Polymorphism Predicts Osteonecrosis in Children with Acute Lymphoblastic Leukemia: A Report from the Children’s Oncology Group. Blood 2008, 111, 4496–4499. [Google Scholar] [CrossRef]
- Gregers, J.; Green, H.; Christensen, I.J.; Dalhoff, K.; Schroeder, H.; Carlsen, N.; Rosthoej, S.; Lausen, B.; Schmiegelow, K.; Peterson, C. Polymorphisms in the ABCB1 Gene and Effect on Outcome and Toxicity in Childhood Acute Lymphoblastic Leukemia. Pharmacogenom. J. 2015, 15, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Roy Moulik, N.; Kumar, A.; Agrawal, S.; Awasthi, S.; Mahdi, A.A.; Kumar, A. Role of Folate Status and Methylenetetrahydrofolate Reductase Genotype on the Toxicity and Outcome of Induction Chemotherapy in Children with Acute Lymphoblastic Leukemia. Leuk. Lymphoma 2015, 56, 1379–1384. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.; Fukushima, T.; Sakai, A.; Suzuki, R.; Nakajima-Yamaguchi, R.; Kobayashi, C.; Iwabuchi, A.; Saito, M.; Yoshimi, A.; Nakao, T.; et al. Polymorphisms of MTHFR Associated with Higher Relapse/Death Ratio and Delayed Weekly MTX Administration in Pediatric Lymphoid Malignancies. Leuk. Res. Treat. 2013, 2013, 238528. [Google Scholar] [CrossRef]
- Laverdière, C.; Chiasson, S.; Costea, I.; Moghrabi, A.; Krajinovic, M. Polymorphism G80A in the Reduced Folate Carrier Gene and Its Relationship to Methotrexate Plasma Levels and Outcome of Childhood Acute Lymphoblastic Leukemia. Blood 2002, 100, 3832–3834. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, H.; Okamura, N.; Yagi, M.; Noro, Y.; Moriya, Y.; Nakamura, T.; Hayakawa, A.; Takeshima, Y.; Sakaeda, T.; Matsuo, M.; et al. Genetic Polymorphisms Associated with Adverse Events and Elimination of Methotrexate in Childhood Acute Lymphoblastic Leukemia and Malignant Lymphoma. J. Hum. Genet. 2007, 52, 166–171. [Google Scholar] [CrossRef]
- Health, N.I. of National Cancer Institute: Survaillance, Epidemiology, and End Results Program. Available online: https://seer.cancer.gov/statistics-network/explorer/ (accessed on 3 September 2025).
- Albuquerque, K.M.C.; de Joventino, C.B.; Moreira, L.C.; Rocha, H.A.L.; Gurgel, L.A.; Oliveira, D.d.S.; Rodrigues, C.E.M. Clinical Outcome and Prognosis of Patients with Acute Myeloid Leukemia Submitted to Chemotherapy with 5 Years of Follow-Up. Hematol. Transfus. Cell Ther. 2024, 46, 8–13. [Google Scholar] [CrossRef]
- Jaime-Pérez, J.C.; Hernández-Coronado, M.; Hernández-De Los Santos, J.A.; Marfil-Rivera, L.J.; Gómez-Almaguer, D. Monthly Variation in Diagnosis of Acute Lymphoblastic Leukemia and Survival Outcome in Children and Adults: 15-Year Trends at a Single Center. Hematol. Transfus. Cell Ther. 2022, 44, 314–320. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 Recommendations from an International Expert Panel on Behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Yi, M.; Zhou, L.; Li, A.; Luo, S.; Wu, K. Global Burden and Trend of Acute Lymphoblastic Leukemia from 1990 to 2017. Aging 2020, 12, 22869–22891. [Google Scholar] [CrossRef] [PubMed]
- Paugh, S.W.; Bonten, E.J.; Savic, D.; Ramsey, L.B.; Thierfelder, W.E.; Gurung, P.; Malireddi, R.K.S.; Actis, M.; Mayasundari, A.; Min, J.; et al. NALP3 Inflammasome Upregulation and CASP1 Cleavage of the Glucocorticoid Receptor Cause Glucocorticoid Resistance in Leukemia Cells. Nat. Genet. 2015, 47, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Miñana, B.; Valcárcel, J.; Gabaldón, T.; Lehner, B. Synonymous Mutations Frequently Act as Driver Mutations in Human Cancers. Cell 2014, 156, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Ernst, S.; Verhalen, B.; Zarrabi, N.; Wilkens, S.; Börsch, M. Drug Transport Mechanism of P-Glycoprotein Monitored by Single Molecule Fluorescence Resonance Energy Transfer. Multiphot. Microsc. Biomed. Sci. XI 2011, 7903, 790328. [Google Scholar] [CrossRef][Green Version]
- Ankathil, R. ABCB1 Genetic Variants in Leukemias: Current Insights into Treatment Outcomes. Pharmgenom. Pers. Med. 2017, 10, 169–181. [Google Scholar] [CrossRef]
- Han, J.; Liu, L.; Meng, L.; Guo, H.; Zhang, J.; Han, Z.Q.; Hong, Z.Y. Effect of Polymorphisms of ABCB1 and MTHFR on Methotrexate-Related Toxicities in Adults with Hematological Malignancies. Front. Oncol. 2021, 11, 759805. [Google Scholar] [CrossRef]
- Megías-Vericat, J.E.; Martínez-Cuadrón, D.; Solana-Altabella, A.; Poveda, J.L.; Montesinos, P. Systematic Review of Pharmacogenetics of ABC and SLC Transporter Genes in Acute Myeloid Leukemia. Pharmaceutics 2022, 14, 878. [Google Scholar] [CrossRef]
- Ferguson, P.J.; Cheng, Y.C. Phenotypic Instability of Drug Sensitivity in a Human Colon Carcinoma Cell Line. Cancer Res. 1989, 49, 1148–1153. [Google Scholar]
- Jamroziak, K.; Robak, T. Pharmacogenomics of MDR1/ABCB1 Gene: The Influence on Risk and Clinical Outcome of Haemotological Malignancies. Hematology 2004, 9, 91–105. [Google Scholar] [CrossRef][Green Version]
- Pallis, M.; Russell, N. P-Glycoprotein Plays a Drug-Efflux-Independent Role in Augmenting Cell Survival in Acute Myeloblastic Leukemia and Is Associated with Modulation of a Sphingomyelin-Ceramide Apoptotic Pathway. Blood 2000, 95, 2897–2904. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the Role of Efflux Pumps in Multidrug-Resistant Cancer. Nat. Rev. Cancer 2019, 18, 452–464. [Google Scholar] [CrossRef]
- Sharom, F.J. The P-Glycoprotein Multidrug Transporter. Essays Biochem. 2011, 50, 161–178. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Moriyama, T.; Nishii, R.; Perez-Andreu, V.; Yang, W.; Klussmann, F.A.; Zhao, X.; Lin, T.N.; Nersting, J. NUDT15 Polymorphisms Alter Thiopurine Metabolism and Hematopoietic Toxicity HHS Public Access Author Manuscript. Nat. Genet. 2016, 48, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Dean, L.; Kane, M.; Pratt, V.M.; Scott, S.A.; Pirmohamed, M.; Esquivel, B.; Kattman, B.L.; Malheiro, A.J. Mercaptopurine Therapy and TPMT and NUDT15 Genotype. In Medical Genetics Summaries; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2012. Available online: https://www.ncbi.nlm.nih.gov/books/NBK100660/ (accessed on 13 October 2025).
- Daly, A.K. Pharmacogenetics: A General Review on Progress to Date. Br. Med. Bull. 2017, 124, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Driest, S.L. Van Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, S.; Deng, Y.; Yi, P.; Yu, J. Targeting the RNA M6A Modification for Cancer Immunotherapy. Mol. Cancer 2022, 21, 76. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing MRNA Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA Modification Landscape in Human Disease. Rna 2017, 23, 1754–1769. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent Pathway Mutations of Splicing Machinery in Myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.; Lee, J.; Park, D.; Kim, Y.J.; Park, W.Y.; Hong, D.; Park, P.J.; Lee, E. Intron Retention Is a Widespread Mechanism of Tumor-Suppressor Inactivation. Nat. Genet. 2015, 47, 1242–1248. [Google Scholar] [CrossRef]
- Gotea, V.; Gartner, J.J.; Qutob, N.; Elnitski, L.; Samuels, Y. The Functional Relevance of Somatic Synonymous Mutations in Melanoma and Other Cancers. Pigment Cell Melanoma Res. 2015, 28, 673–684. [Google Scholar] [CrossRef]
- Cheok, M.H.; Lugthart, S.; Evans, W.E. Pharmacogenomics of Acute Leukemia. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 317–353. [Google Scholar] [CrossRef] [PubMed]
- Maamari, D.; El-Khoury, H.; Saifi, O.; Muwakkit, S.A.; Zgheib, N.K. Implementation of Pharmacogenetics to Individualize Treatment Regimens for Children with Acute Lymphoblastic Leukemia. Pharmgenom. Pers. Med. 2020, 13, 295–317. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, S.; Kotur, N.; Stankovic, B.; Zukic, B.; Gasic, V.; Dokmanovic, L. Pharmacogenomic and Pharmacotranscriptomic Profiling of Childhood Acute Lymphoblastic Leukemia: Paving the Way to Personalized Treatment. Genes 2019, 10, 191. [Google Scholar] [CrossRef] [PubMed]
- Chow, E.J.; Doody, D.R.; Wilkes, J.J.; Becker, L.K.; Chennupati, S.; Morin, P.E.; Winestone, L.E.; Henk, H.J.; Lyman, G.H. Adverse Events among Chronic Myelogenous Leukemia Patients Treated with Tyrosine Kinase Inhibitors: A Real-World Analysis of Health Plan Enrollees. Leuk. Lymphoma 2021, 62, 1203–1210. [Google Scholar] [CrossRef]
- Escherich, C.; Chen, W.; Miyamoto, S.; Namikawa, Y.; Yang, W.; Teachey, D.T.; Li, Z.; Raetz, E.A.; Larsen, E.; Devidas, M.; et al. Identification of TCF3 Germline Variants in Pediatric B-Cell Acute Lymphoblastic Leukemia. Blood Adv. 2023, 7, 2177–2180. [Google Scholar] [CrossRef]
- Lauschke, V.M.; Ingelman-Sundberg, M. Prediction of Drug Response and Adverse Drug Reactions: From Twin Studies to Next Generation Sequencing. Eur. J. Pharm. Sci. 2019, 130, 65–77. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).