Clinicopathological Significance of Transcription Factor p73 in Breast Cancers: Protein Expression and Transcriptomic Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tissue Microarray (TMA) and Immunohistochemistry (IHC)
2.3. Evaluation of Immunohistochemical Staining
2.4. Statistical Analysis
2.5. Transcriptomic Analysis
2.6. Bioinformatics Analysis
2.7. Western Blot Analyses
3. Results
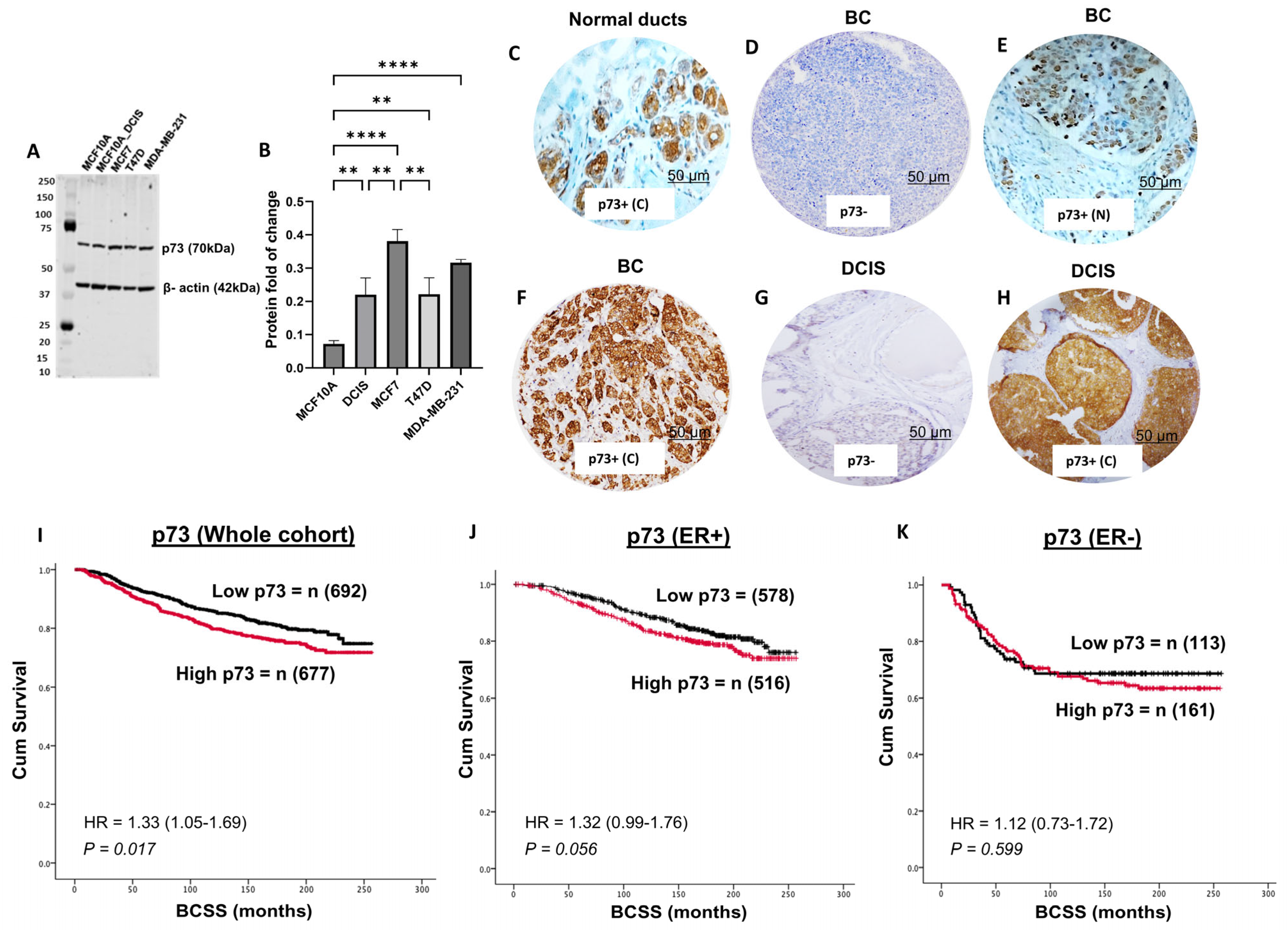
3.1. p73 Protein Expression and Invasive BC
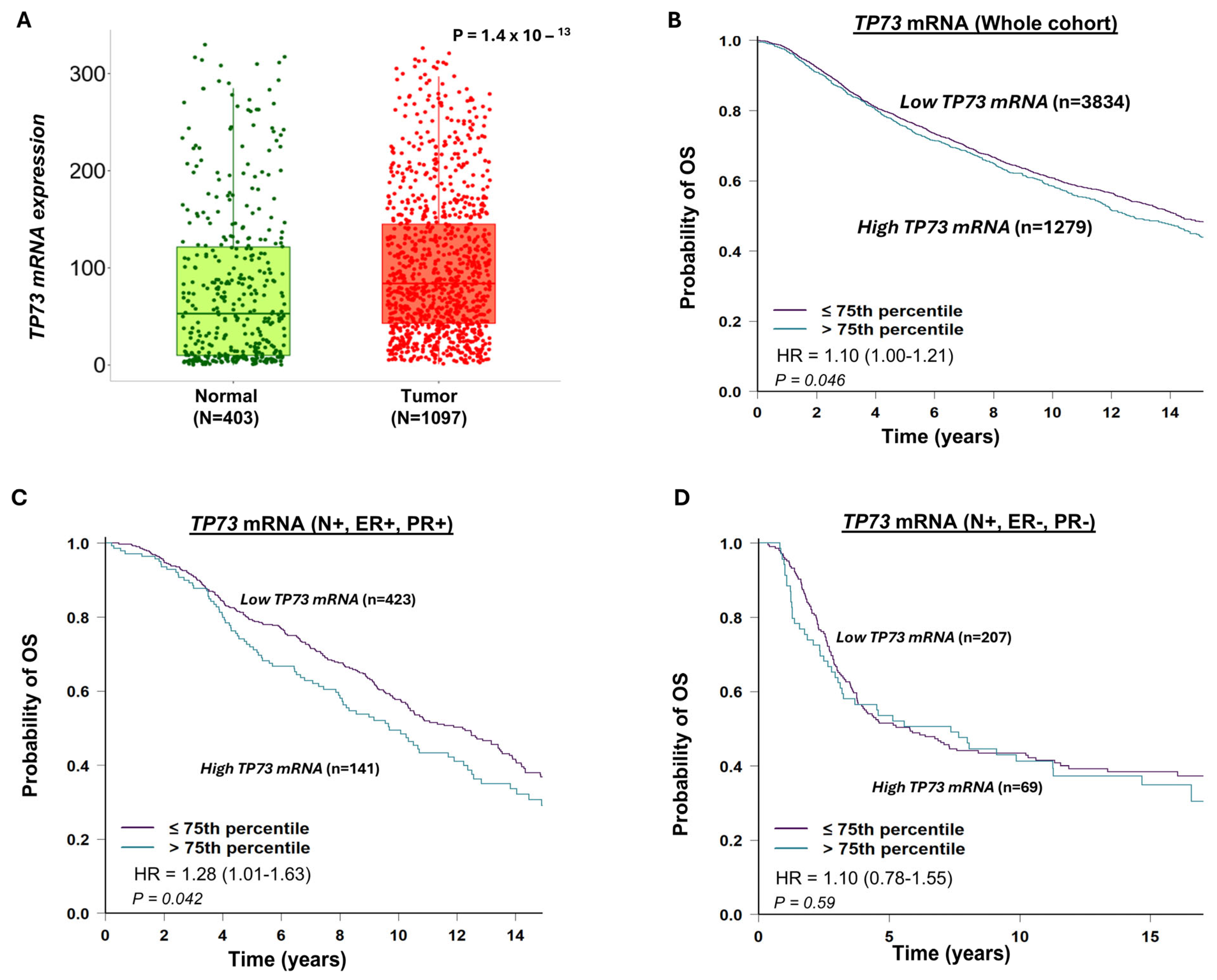
3.2. TP73 mRNA Expression and Invasive BC
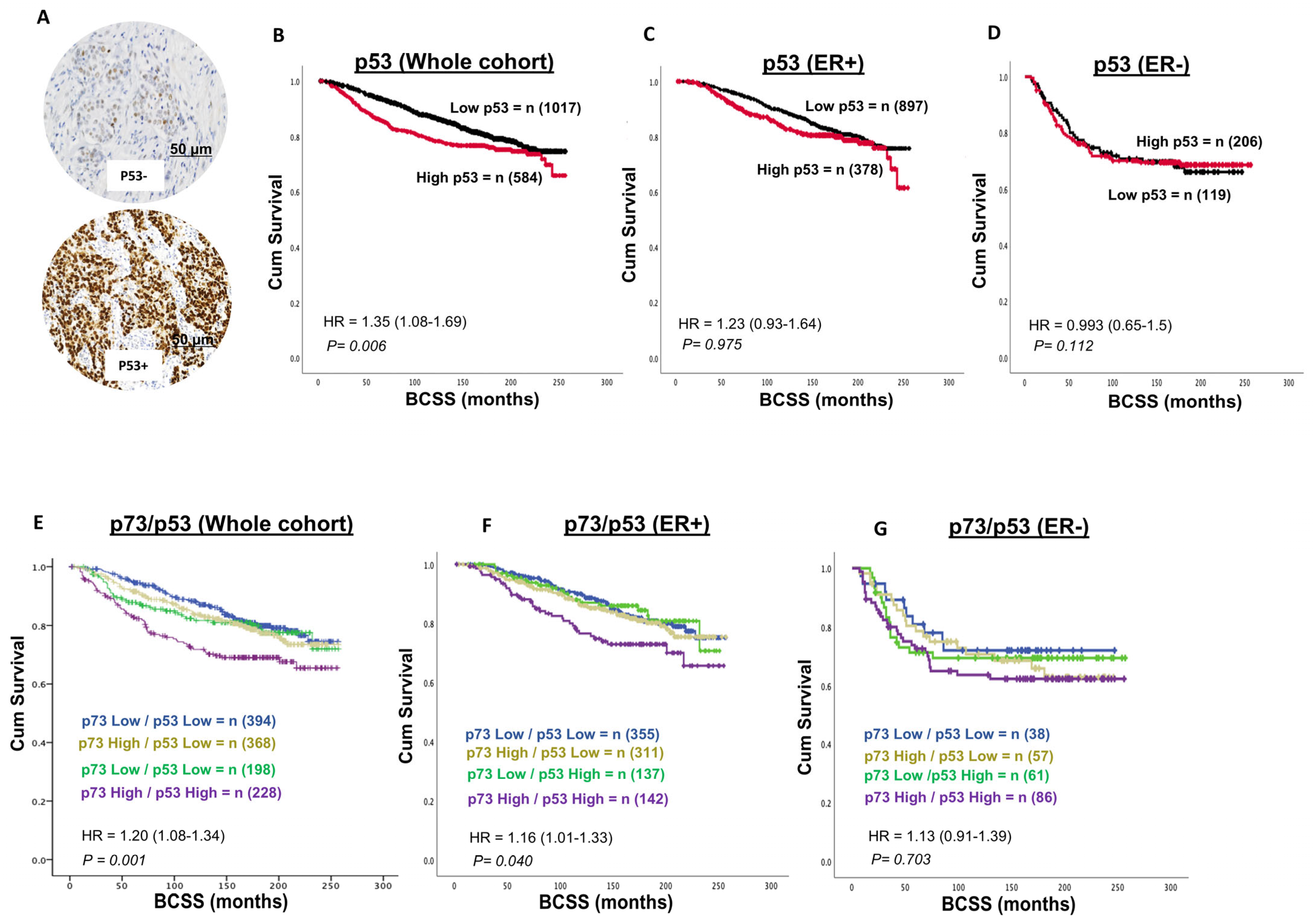
3.3. p53 and Invasive BC
3.4. p73-p53 Co-Expression in Invasive BC
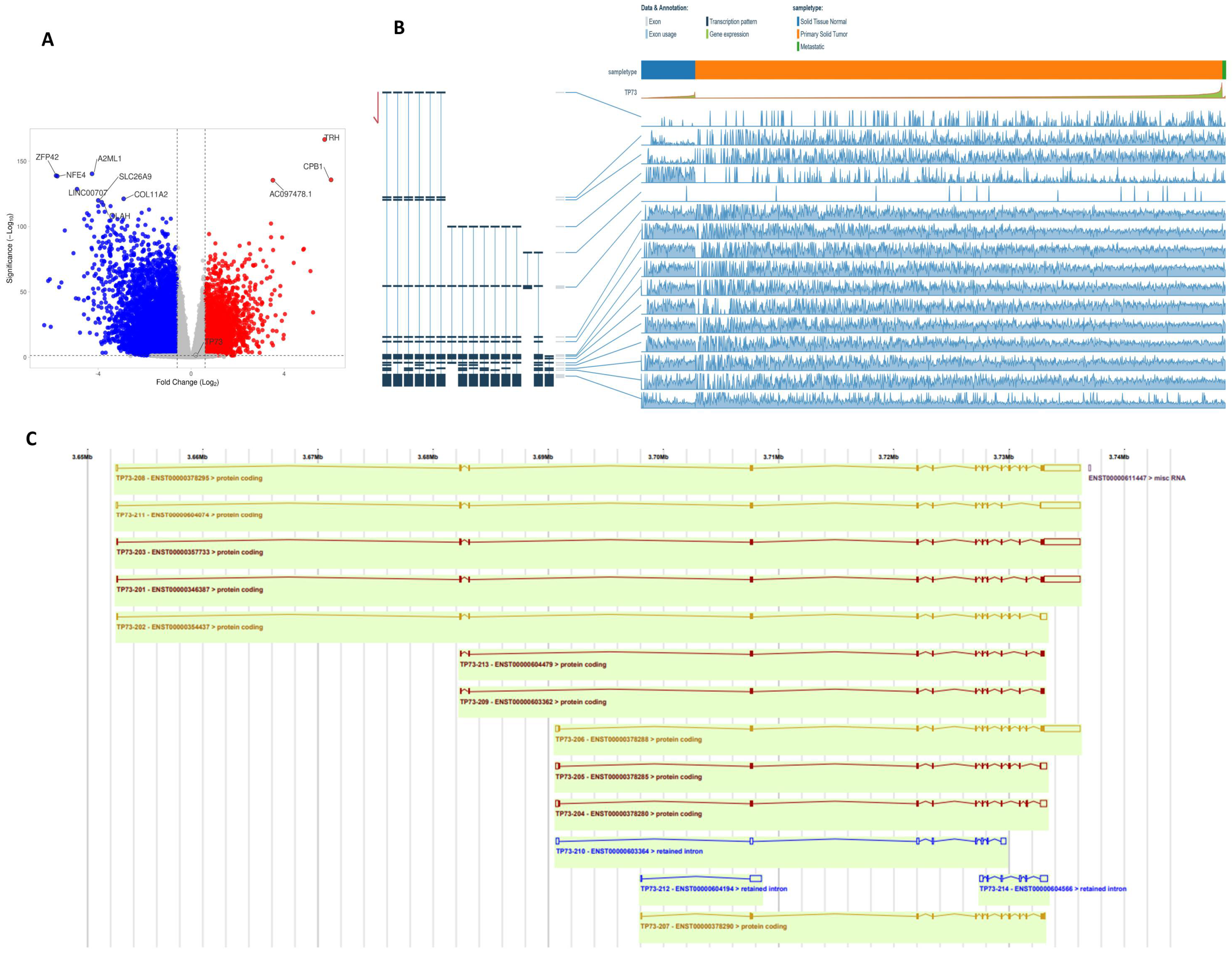
3.5. TP73 Transcripts in BC-TCGA
3.6. p73 and p53 Protein Expression in DCIS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCSS | Breast cancer-specific survival |
| TA | Transactivation domain |
| DCIS | Ductal carcinoma in situ |
| NPI | Nottingham Prognostic Index |
| IHC | Immunohistochemistry |
References
- Ghoncheh, M.; Mohammadian-Hafshejani, A.; Salehiniya, H. Incidence and Mortality of Breast Cancer and their Relationship to Development in Asia. Asian Pac. J. Cancer Prev. 2015, 16, 6081–6087. [Google Scholar] [CrossRef]
- Tao, Z.; Shi, A.; Lu, C.; Song, T.; Zhang, Z.; Zhao, J. Breast Cancer: Epidemiology and Etiology. Cell Biochem. Biophys. 2015, 72, 333–338. [Google Scholar] [CrossRef]
- Sharma, R. Global burden of breast cancer: Current statistics and projections. J. Glob. Oncol. 2021, 7, 567–579. [Google Scholar]
- Khalid, M.S.A.; Patel, D.; Sharma, S. Breast cancer subtypes: Incidence, distribution, and clinical significance. Front. Oncol. 2024, 14, 1123456. [Google Scholar]
- Bergholtz, H.; Lien, T.G.; Swanson, D.M.; Frigessi, A.; Oslo Breast Cancer Research Consortium (OSBREAC); Daidone, M.G.; Tost, J.; Wärnberg, F.; Sørlie, T. Contrasting DCIS and invasive breast cancer by subtype suggests basal-like DCIS as distinct lesions. NPJ Breast Cancer. 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Cowell, C.F.; Weigelt, B.; Sakr, R.A.; Ng, C.K.; Hicks, J.; King, T.A.; Reis-Filho, J.S. Progression from ductal carcinoma in situ to invasive breast cancer: Revisited. Mol. Oncol. 2013, 7, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P.T.; Reis-Filho, J.S.; Gale, T.; Lakhani, S.R. Molecular evolution of breast cancer. J. Pathol. 2005, 205, 248–254. [Google Scholar] [CrossRef]
- Kaplan, H.G.; Dowdell, A.K.; Berry, A.B.; Shimol, R.B.; Robinson, F.L.; Carney, C.A.; Piening, B.D. Multi-omic profiling of simultaneous ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res. Treat. 2024, 205, 451–464. [Google Scholar] [CrossRef]
- Marabese, M.; Vikhanskaya, F.; Broggini, M. p73: A chiaroscuro gene in cancer. Eur. J. Cancer 2007, 43, 1361–1372. [Google Scholar] [CrossRef]
- Oswald, C.; Stiewe, T. In good times and bad: p73 in cancer. Cell Cycle 2008, 7, 1726–1731. [Google Scholar] [CrossRef]
- Rufini, A.; Agostini, M.; Grespi, F.; Tomasini, R.; Sayan, B.S.; Niklison-Chirou, M.V.; Conforti, F.; Velletri, T.; Mastino, A.; Mak, T.W.; et al. p73 in Cancer. Genes. Cancer 2011, 2, 491–502. [Google Scholar] [CrossRef]
- Irwin, M.S.; Miller, F.D. p73: Regulator in cancer and neural development. Cell Death Differ. 2004, 11 Suppl. S1, S17–S22. [Google Scholar] [CrossRef]
- Logotheti, S.; Richter, C.; Murr, N.; Spitschak, A.; Marquardt, S.; Putzer, B.M. Mechanisms of Functional Pleiotropy of p73 in Cancer and Beyond. Front. Cell Dev. Biol. 2021, 9, 737735. [Google Scholar] [CrossRef] [PubMed]
- Maas, A.M.; Bretz, A.C.; Mack, E.; Stiewe, T. Targeting p73 in cancer. Cancer Lett. 2013, 332, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A. p73, a sophisticated p53 family member in the cancer world. Cancer Sci. 2005, 96, 729–737. [Google Scholar] [CrossRef]
- Rozenberg, J.M.; Zvereva, S.; Dalina, A.; Blatov, I.; Zubarev, I.; Luppov, D.; Bessmertnyi, A.; Romanishin, A.; Alsoulaiman, L.; Kumeiko, V.; et al. Dual Role of p73 in Cancer Microenvironment and DNA Damage Response. Cells 2021, 10, 3516. [Google Scholar] [CrossRef] [PubMed]
- Stiewe, T.; Pützer, B.M. Role of p73 in malignancy: Tumor suppressor or oncogene? Cell Death Differ. 2002, 9, 237–245. [Google Scholar] [CrossRef]
- Dominguez, G.; Silva, J.M.; Silva, J.; Garcia, J.M.; Sanchez, A.; Navarro, A.; Gallego, I.; Provencio, M.; España, P.; Bonilla, F. Wild type p73 overexpression and high-grade malignancy in breast cancer. Breast Cancer Res. Treat. 2001, 66, 183–190. [Google Scholar] [CrossRef]
- Schwartz, D.I.; Lindor, N.M.; Walsh-Vockley, C.; Roche, P.C.; Mai, M.; Smith, D.I.; Liu, W.; Couch, F.J. p73 mutations are not detected in sporadic and hereditary breast cancer. Breast Cancer Res. Treat. 1999, 58, 25–29. [Google Scholar] [CrossRef]
- Yao, J.; Xu, F.; Zhang, D.; Yi, W.; Chen, X.; Chen, G.; Zhou, E. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J. Cell Biochem. 2018, 119, 680–690. [Google Scholar] [CrossRef]
- Zou, Y.L.X.; Wang, Y.; Guo, J. TP73-AS1 functions as an oncogene in breast cancer via modulation of p73 isoforms. J. Cell Biochem. 2018, 119, 3797–3805. [Google Scholar]
- Tavakkol Afshari, Z.; Gholizadeh, Z.; Nikpoor, A.R.; Tavakkol, A.J.; Ganjali, R.; Homaei, S.F.; Jamialahmadi, K. A Case-Control Study on the p73 G4A Gene Polymorphism and Susceptibility to Breast Cancer in an Iranian Population. Iran J. Public Health. 2019, 48, 1855–1860. [Google Scholar] [CrossRef]
- Ronchi, C.; Haider, S.; Brisken, C. EMBER creates a unified space for independent breast cancer transcriptomic datasets enabling precision oncology. NPJ Breast Cancer. 2024, 10, 56. [Google Scholar] [CrossRef]
- Manjunath, S.S.R. A comprehensive study of mRNA and long noncoding RNAs in Indian breast cancer patients reveals subtype-specific alterations. BMC Cancer 2022, 22, 145. [Google Scholar]
- Llera, A.S.; Abdelhay, E.S.F.W.; Artagaveytia, N.; Daneri-Navarro, A.; Müller, B.; Velazquez, C.; Alcoba, E.B.; Alonso, I.; Alvesda, Q.D.B.; Binato, R.; et al. The Transcriptomic Portrait of Locally Advanced Breast Cancer and Its Prognostic Value in a Multi-Country Cohort of Latin American Patients. Front Oncol. 2022, 12, 835626. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Venet, D.; Lifrange, F.; Larsimont, D.; Rediti, M.; Stenbeck, L.; Dupont, F.; Rouas, G.; Garcia, A.J.; Craciun, L.; et al. Spatial transcriptomics reveals substantial heterogeneity in triple-negative breast cancer with potential clinical implications. Nat. Commun. 2024, 15, 10232. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Reporting recommendations for tumour MARKer prognostic studies (REMARK). J. Natl. Cancer Inst. 2005, 97, 1180–1184. [Google Scholar] [CrossRef] [PubMed]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Jézéquel, P.; Campone, M.; Gouraud, W.; Guérin-Charbonnel, C.; Leux, C.; Ricolleau, G.; Campion, L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2012, 131, 765–775. [Google Scholar] [CrossRef]
- Sun, W.; Duan, T.; Ye, P.; Chen, K.; Zhang, G.; Lai, M.; Zhang, H. TSVdb: A web-tool for TCGA splicing variants analysis. BMC Genom. 2018, 19, 405. [Google Scholar] [CrossRef]
- Aqeilan, R.I.; Pekarsky, Y.; Herrero, J.J.; Palamarchuk, A.; Letofsky, J.; Druck, T.; Trapasso, F.; Han, S.Y.; Melino, G.; Huebner, K.; et al. Functional association between Wwox tumour suppressor protein and p73, a p53 homolog. Proc. Natl. Acad. Sci. USA 2004, 101, 4401–4406. [Google Scholar] [CrossRef]
- Ozaki, T.; Kubo, N.; Nakagawara, A. p73-Binding Partners and Their Functional Significance. Int. J. Proteom. 2010, 2010, 283863. [Google Scholar] [CrossRef]
- Dominguez, G.; Garcia, J.M.; Pena, C.; Silva, J.; Garcia, V.; Martinez, L.; Maximiano, C.; Gomez, M.E.; Rivera, J.A.; Garcia-Andrade, C.; et al. DeltaTAp73 upregulation correlates with poor prognosis in human tumours: Putative in vivo network involving p73 isoforms, p53, and E2F-1. J. Clin. Oncol. 2006, 24, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Zaika, A.I.; Kovalev, S.; Marchenko, N.D.; Moll, U.M. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999, 59, 3257–3263. [Google Scholar]
- Loukopoulos, P.; Shibata, T.; Katoh, H.; Kokubu, A.; Sakamoto, M.; Yamazaki, K.; Kosuge, T.; Kanai, Y.; Hosoda, F.; Imoto, I.; et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: Identification of genetic indicators that predict patient outcome. Cancer Sci. 2007, 98, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, M.; Lee, A.; Choi, B.O.; Park, W.C.; Kim, S.H.; Lee, J.; Kang, J. Correlating p53 immunostaining patterns with somatic TP53 mutation and functional properties of mutant p53 in triple-negative breast cancer. Histopathology 2025, 87, 299–309. [Google Scholar] [CrossRef] [PubMed]
| Parameters | N % |
|---|---|
| Tumour size | |
| <2 cm | 852 (58%) |
| ≥2 cm | 628 (42%) |
| Grade | |
| 1 | 253 (17%) |
| 2 | 549 (37%) |
| 3 | 678 (46%) |
| Tubular formation | |
| 1 | 119 (8%) |
| 2 | 456 (31%) |
| 3 | 905 (61%) |
| Pleomorphism | |
| 1 | 44 (3%) |
| 2 | 500 (34%) |
| 3 | 936 (63%) |
| Mitosis | |
| 1 | 647 (44%) |
| 2 | 294 (20%) |
| 3 | 539 (36%) |
| Histological tumour type | |
| No special type (NST) | 923 (62%) |
| Lobular | 132 (9%) |
| Other special types | 73 (5%) |
| NST mixed | 352 (24%) |
| Lymphovascular invasion | |
| Absent | 1040 (70%) |
| Present | 440 (30%) |
| Lymph node status | |
| Negative | 937 (63%) |
| Positive | 543 (37%) |
| Nottingham Prognostic Index | |
| Good prognostic group | 491 (33%) |
| Moderate prognostic group | 769 (52%) |
| Poor prognostic group | 220 (15%) |
| ER status | |
| ER- | 347 (24%) |
| ER+ | 1128 (76%) |
| PgR status | |
| Negative | 601 (41%) |
| Positive | 857 (59%) |
| HER2 status | |
| Negative | 1264 (86%) |
| Positive | 199 (14%) |
| Ki67 index | |
| Low | 533 (48%) |
| High | 580 (52%) |
| Molecular classes | |
| Luminal A | 478 (38%) |
| Luminal B | 442 (36%) |
| HER2+ | 87 (7%) |
| Triple negative | 235 (19%) |
| Menopausal Status | |
| Pre-menopausal | 540 (36%) |
| Postmenopausal | 940 (64%) |
| Age: 50 years | |
| <50 | 481 (32%) |
| ≥50 | 999 (68%) |
| Parameters | p73 Cytoplasmic Expression | R2 p Value | |
|---|---|---|---|
| Low (N%) | High (N%) | ||
| Tumour size | |||
| <2 cm | 440 (52%) | 400 (48%) | 2.922 |
| ≥2 cm | 252 (48%) | 277 (52%) | 0.087 |
| Grade | |||
| 1 | 114 (53%) | 100 (47%) | 15.241 |
| 2 | 302 (56%) | 237 (44%) | <0.0001 |
| 3 | 276 (45%) | 340 (55%) | |
| Tubular formation | |||
| 1 | 52 (53%) | 46 (47%) | 0.508 |
| 2 | 207 (51%) | 196 (49%) | 0.776 |
| 3 | 433 (50%) | 435 (50%) | |
| Pleomorphism | |||
| 1 | 8 (44%) | 10 (56%) | 13.117 |
| 2 | 231 (58%) | 166 (42%) | 0.001 |
| 3 | 453 (47%) | 501 (53%) | |
| Mitosis | |||
| 1 | 362 (55%) | 298 (45%) | 11.729 |
| 2 | 135 (50%) | 134 (50%) | 0.003 |
| 3 | 195 (44%) | 245 (56%) | |
| Histological tumour type | |||
| No special type (NST) | 424 (48%) | 460 (52%) | 16.248 |
| Invasive lobular carcinoma (ILC) | 69 (62%) | 42 (38%) | 0.012 |
| Mixed NST and ILC | 45 (54%) | 38 (46%) | |
| Mixed NST and special type | 14 (37%) | 24 (63%) | |
| Pure special tumour type | 14 (70%) | 6 (30%) | |
| Metaplastic carcinoma | 2 (40%) | 3 (60%) | |
| Tubular and tubular mixed | 124 (54%) | 104 (46%) | |
| Lymphovascular invasion | |||
| Absent | 506 (52%) | 474 (48%) | 1.624 |
| Present | 186 (48%) | 203 (52%) | 0.203 |
| Lymph node status | |||
| Negative | 438 (52%) | 408 (48%) | 1.33 |
| Positive | 254 (49%) | 269 (51%) | 0.249 |
| Nottingham Prognostic Index | |||
| Good | 261 (56%) | 205 (44%) | |
| Moderate | 322 (47%) | 357 (53%) | 8.531 |
| Poor | 109 (49%) | 115 (51%) | 0.014 |
| Nodal stage | |||
| 1 | 438 (52%) | 408 (48%) | |
| 2 | 193 (51%) | 184 (49%) | 5.06 |
| 3 | 61 (42%) | 85 (58%) | 0.08 |
| ER status | |||
| ER- | 113 (41%) | 161 (59%) | 11.78 |
| ER+ | 578 (53%) | 516 (47%) | 0.001 |
| PgR status | |||
| Negative | 269 (48%) | 291 (52%) | 2.34 |
| Positive | 418 (52%) | 382 (48%) | 0.126 |
| HER2 status | |||
| Negative | 592 (50%) | 590 (50%) | 0.634 |
| Positive | 99 (53%) | 87 (47%) | 0.426 |
| Triple negative (TN) | |||
| Non-TN | 614 (53%) | 546 (47%) | 17.738 |
| TN | 71 (37%) | 123 (63%) | <0.0001 |
| Molecular classes | |||
| Luminal | 578 (53%) | 516 (47%) | |
| HER2 enriched | 36 (55%) | 30 (45%) | 17.811 |
| TNBC | 71 (37%) | 123 (63%) | <0.0001 |
| Menopausal status | |||
| Pre | 218 (47%) | 247 (53%) | 3.786 |
| Post | 474 (52%) | 430 (48%) | 0.052 |
| Age | |||
| <50 | 193 (45%) | 233 (55%) | 6.8 |
| ≥50 | 499 (53%) | 444 (47%) | 0.009 |
| Parameters | TP53 Expression | R2 p Value | |
|---|---|---|---|
| Low (N%) | High (N%) | ||
| Tumour size | |||
| <2 cm | 634 (65%) | 343 (35%) | 2.03 |
| ≥2 cm | 383 (61%) | 241 (39%) | 0.154 |
| Grade | |||
| 1 | 187 (78%) | 52 (22%) | |
| 2 | 474 (75%) | 157 (25%) | 128.278 |
| 3 | 356 (49%) | 375 (51%) | <0.0001 |
| Tubular formation | |||
| 1 | 84 (76%) | 26 (24%) | |
| 2 | 324 (68%) | 153 (32%) | 17.066 |
| 3 | 609 (60%) | 405 (40%) | <0.0001 |
| Pleomorphism | |||
| 1 | 17 (74%) | 6 (26%) | |
| 2 | 373 (81%) | 85 (19%) | 92.017 |
| 3 | 627 (56%) | 493 (44%) | <0.0001 |
| Mitosis | |||
| 1 | 581 (77%) | 173 (23%) | |
| 2 | 187 (60%) | 125 (40%) | 127.902 |
| 3 | 249 (46%) | 286 (54%) | <0.0001 |
| Histological tumour type | |||
| No special type (NST) | 598 (57%) | 459 (43%) | |
| Invasive lobular carcinoma (ILC) | 108 (86%) | 17 (14%) | |
| Mixed NST and ILC | 68 (73%) | 25 (27%) | |
| Mixed NST and special type | 27 (66%) | 14 (34%) | 73.222 |
| Pure special tumour type | 13 (72%) | 5 (28%) | <0.0001 |
| Metaplastic carcinoma | 3 (60%) | 2 (40%) | |
| Tubular and tubular mixed | 200 (76%) | 62 (24%) | |
| Lymphovascular invasion | |||
| Absent | 744 (65%) | 395 (35%) | 5.505 |
| Present | 273 (59%) | 189 (41%) | 0.019 |
| Lymph node status | |||
| Negative | 637 (65%) | 348 (35%) | 1.357 |
| Positive | 380 (62%) | 235 (38%) | 0.244 |
| Nottingham Prognostic Index | |||
| Good | 392 (73%) | 141 (27%) | |
| Moderate | 494 (61%) | 321 (39%) | 40.584 |
| Poor | 131 (52%) | 121 (48%) | <0.0001 |
| Nodal stage | |||
| 1 | 637 (65%) | 348 (35%) | |
| 2 | 291 (64%) | 164 (36%) | 4.905 |
| 3 | 89 (56%) | 71 (44%) | 0.086 |
| ER status | |||
| ER- | 119 (37%) | 206 (63%) | 127.185 |
| ER+ | 897 (70%) | 378 (30%) | <0.0001 |
| PgR status | |||
| Negative | 354 (54%) | 300 (46%) | 43.509 |
| Positive | 660 (70%) | 279 (30%) | <0.0001 |
| HER2 status | |||
| Negative | 914 (66%) | 469 (34%) | 29.329 |
| Positive | 101 (47%) | 114 (53%) | <0.0001 |
| Ki67 index | |||
| Low | 458 (76%) | 149 (24%) | 60.599 |
| High | 319 (54%) | 272 (46%) | <0.0001 |
| Molecular classes | |||
| Luminal | 897 (70%) | 378 (30%) | |
| HER2 enriched | 25 (32%) | 54 (68%) | 127.187 |
| TNBC | 89 (38%) | 146 (62%) | <0.0001 |
| Menopausal status | |||
| Pre | 337 (60%) | 225 (40%) | 4.732 |
| Post | 680 (65%) | 359 (35%) | 0.03 |
| Age | |||
| <50 | 296 (58%) | 215 (42%) | 10.148 |
| ≥50 | 721 (66%) | 369 (34%) | 0.001 |
| Categories | N (%) |
|---|---|
| Age at time of diagnosis | |
| <50 | 87 (27%) |
| ≥50 | 230 (73%) |
| Tumour size | |
| ≤2 cm | 133 (42%) |
| >2 cm | 181 (58%) |
| Tumour grade | |
| Low grade | 47 (15%) |
| Intermediate grade | 85 (27%) |
| High grade | 185 (58%) |
| Molecular subtype | |
| Luminal A | 128 (54%) |
| Luminal B | 44 (19%) |
| HER2 enriched | 37 (16%) |
| Triple negative | 27 (11%) |
| Comedo type necrosis | |
| Absent | 113 (36%) |
| Present | 204 (64%) |
| ER Status | |
| Negative | 68 (25%) |
| Positive | 207 (75%) |
| PR status | |
| Negative | 115 (42%) |
| Positive | 160 (58%) |
| HER2 status | |
| Negative | 226 (78%) |
| Positive | 64 (22%) |
| Recurrence | |
| No recurrence | 279 (88%) |
| Recurrence | 38 (12%) |
| Ki67 expression | |
| Low (≤14) | 192 (76%) |
| High (>14) | 60 (24%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shoqafi, A.; Ibrahim, A.; Lashen, A.; Toss, M.S.; Alqahtani, S.; Miligy, I.; Algethami, M.; Sheha, A.; Jeyapalan, J.N.; Mongan, N.P.; et al. Clinicopathological Significance of Transcription Factor p73 in Breast Cancers: Protein Expression and Transcriptomic Study. Biomedicines 2025, 13, 2484. https://doi.org/10.3390/biomedicines13102484
Shoqafi A, Ibrahim A, Lashen A, Toss MS, Alqahtani S, Miligy I, Algethami M, Sheha A, Jeyapalan JN, Mongan NP, et al. Clinicopathological Significance of Transcription Factor p73 in Breast Cancers: Protein Expression and Transcriptomic Study. Biomedicines. 2025; 13(10):2484. https://doi.org/10.3390/biomedicines13102484
Chicago/Turabian StyleShoqafi, Ahmed, Asmaa Ibrahim, Ayat Lashen, Michael S. Toss, Shatha Alqahtani, Islam Miligy, Mashael Algethami, Amera Sheha, Jennie N. Jeyapalan, Nigel P. Mongan, and et al. 2025. "Clinicopathological Significance of Transcription Factor p73 in Breast Cancers: Protein Expression and Transcriptomic Study" Biomedicines 13, no. 10: 2484. https://doi.org/10.3390/biomedicines13102484
APA StyleShoqafi, A., Ibrahim, A., Lashen, A., Toss, M. S., Alqahtani, S., Miligy, I., Algethami, M., Sheha, A., Jeyapalan, J. N., Mongan, N. P., Green, A. R., Rakha, E. A., & Madhusudan, S. (2025). Clinicopathological Significance of Transcription Factor p73 in Breast Cancers: Protein Expression and Transcriptomic Study. Biomedicines, 13(10), 2484. https://doi.org/10.3390/biomedicines13102484









