Uncovering Nonlinear Predictors of Serum Biomarker Uric Acid Using Interpretable Machine Learning in Healthy Men
Abstract
1. Introduction
2. Materials and Methods
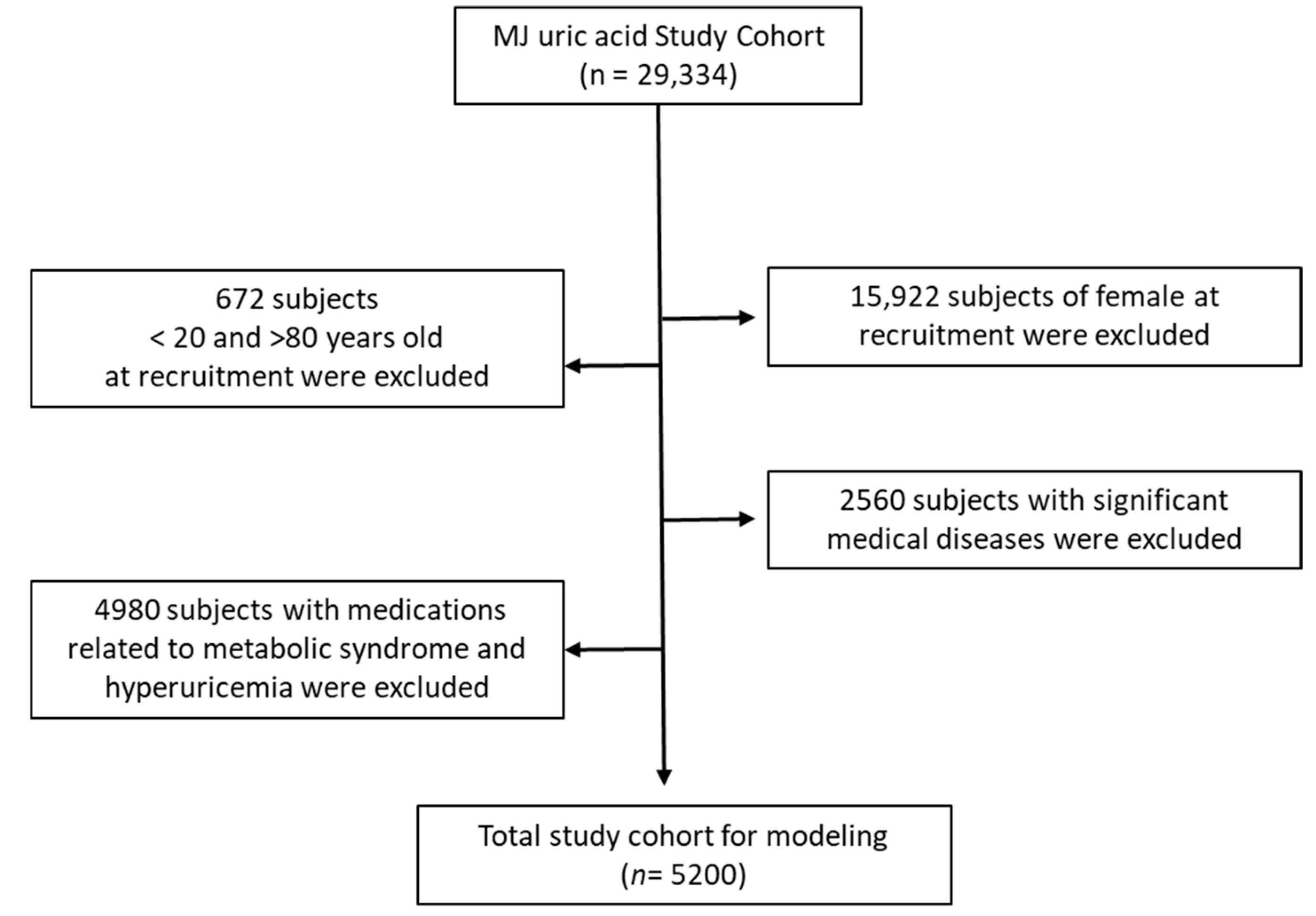
2.1. Participant and Study Design
2.2. Laboratory Tests
2.3. Traditional Statistics
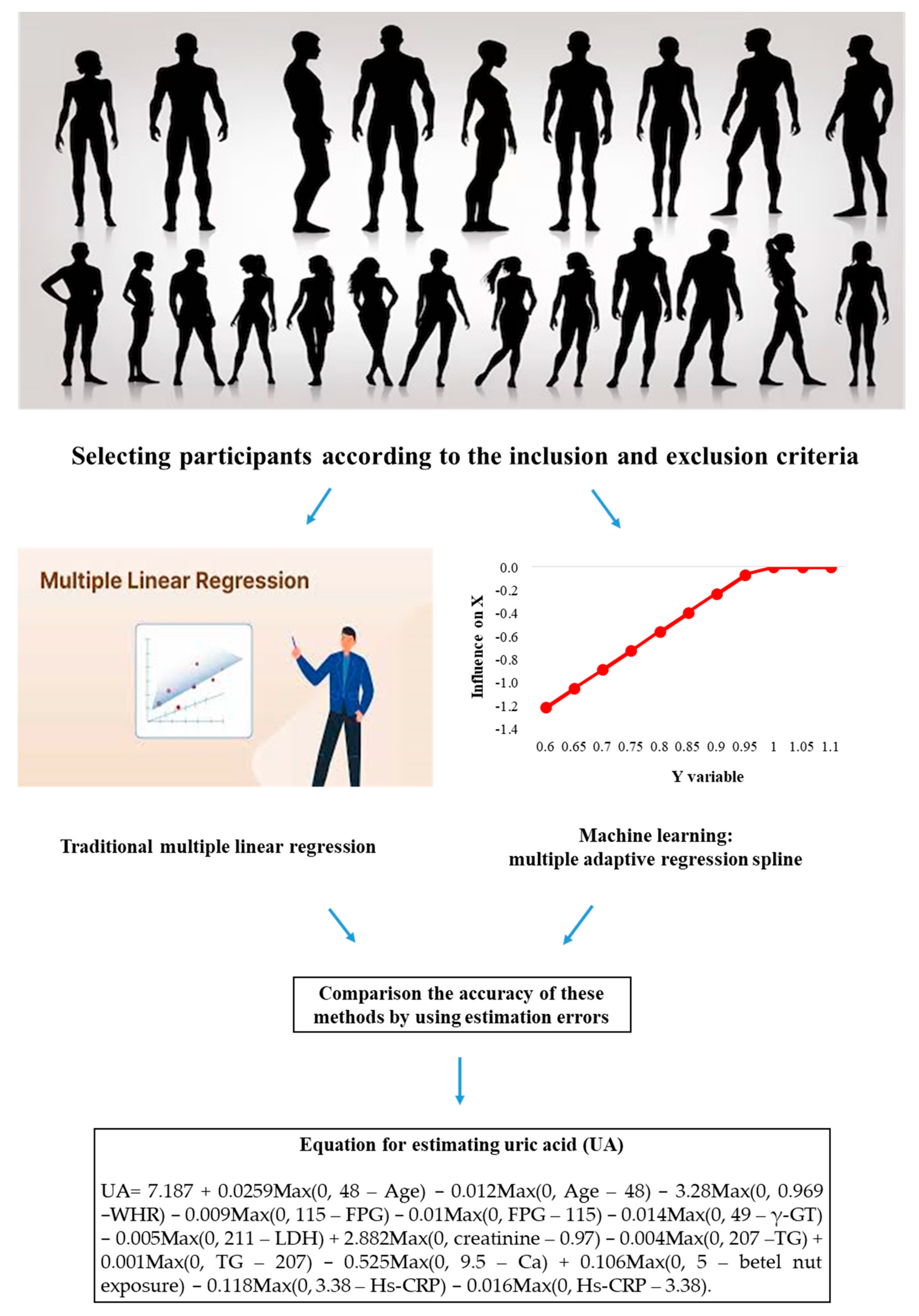
2.4. Machine Learning Method
3. Results
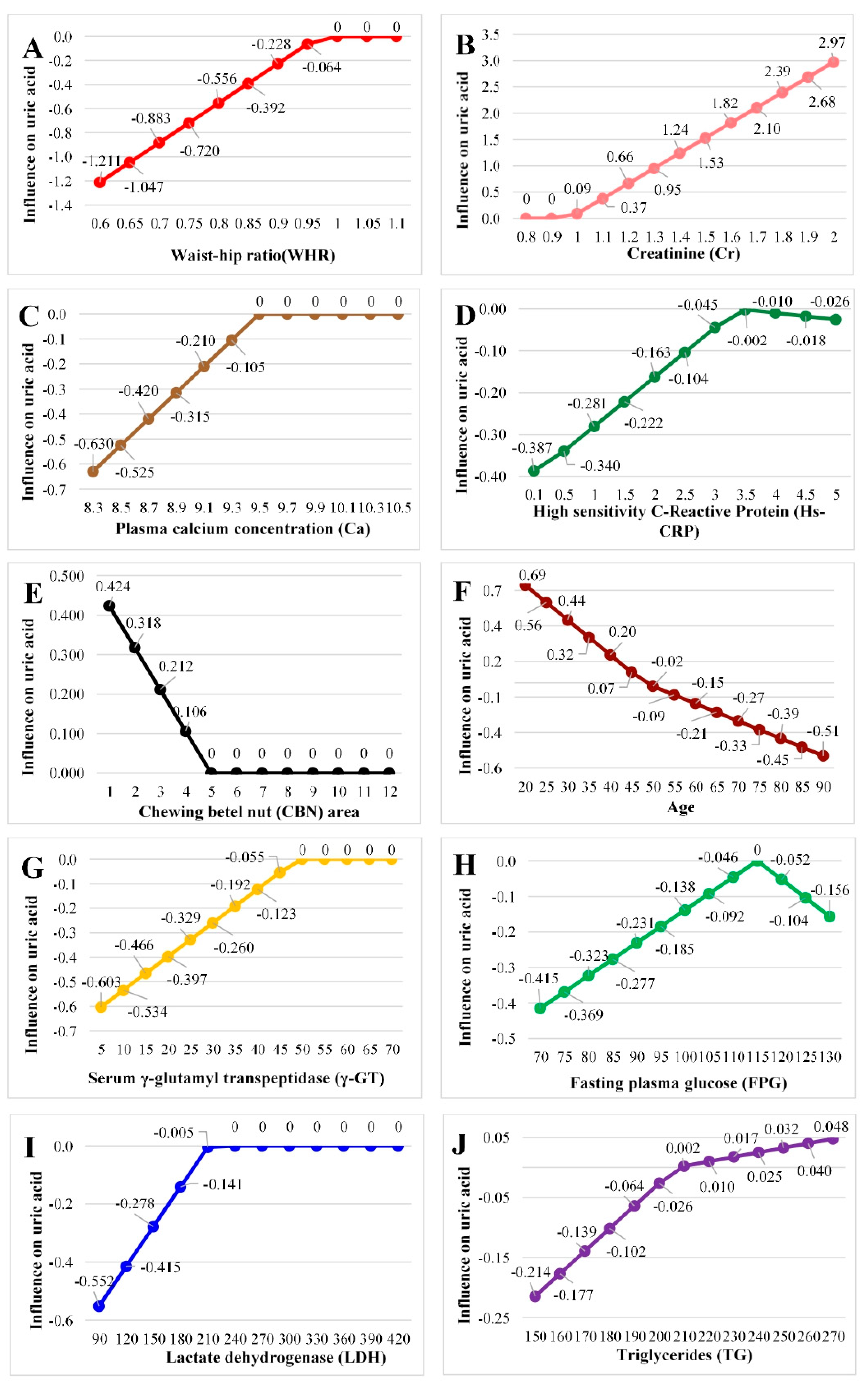
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El Ridi, R.; Tallima, H. Physiological functions and pathogenic potential of uric acid: A review. J. Adv. Res. 2017, 8, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Lin, S.-C.; Chang, H.-Y.; Lyu, L.-C.; Tsai, K.-S.; Pan, W.-H. High prevalence of hyperuricemia in elderly Taiwanese. Asia Pac. J. Clin. Nutr. 2005, 14, 285–292. [Google Scholar] [PubMed]
- Kuwabara, M.; Kodama, T.; Ae, R.; Kanbay, M.; Andres-Hernando, A.; Borghi, C.; Hisatome, I.; Lanaspa, M.A. Update in uric acid, hypertension, and cardiovascular diseases. Hypertens. Res. 2023, 46, 1714–1726. [Google Scholar] [CrossRef] [PubMed]
- Agabiti-Rosei, E.; Grassi, G. Beyond gout: Uric acid and cardiovascular diseases. Curr. Med Res. Opin. 2013, 29 (Suppl. 3), 33–39. [Google Scholar] [CrossRef]
- Sekizuka, H. Uric acid, xanthine oxidase, and vascular damage: Potential of xanthine oxidoreductase inhibitors to prevent cardiovascular diseases. Hypertens. Res. 2022, 45, 772–774. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Zeng, C. Update on the epidemiology, genetics, and therapeutic options of hyperuricemia. Am. J. Transl. Res. 2020, 12, 3167–3181. [Google Scholar]
- Ni, Q.; Lu, X.; Chen, C.; Du, H.; Zhang, R. Risk factors for the development of hyperuricemia: A STROBE-compliant cross-sectional and longitudinal study. Medicine 2019, 98, e17597. [Google Scholar] [CrossRef]
- Sampa, M.B.; Hossain, N.; Hoque, R.; Islam, R.; Yokota, F.; Nishikitani, M.; Ahmed, A. Blood Uric Acid Prediction with Machine Learning: Model Development and Performance Comparison. JMIR Public Health Surveill. 2020, 8, e18331. [Google Scholar] [CrossRef]
- Shivamurthy, G.; Smanjunath, N. Study of estimation of serum LDH and uric acid in preeclampsia and it’s clinical correlation. Int. J. Reprod. Contracept. Obstet. Gynecol. 2020, 9, 5012–5018. [Google Scholar] [CrossRef]
- Tzou, S.-J.; Peng, C.-H.; Huang, L.-Y.; Chen, F.-Y.; Kuo, C.-H.; Wu, C.-Z.; Chu, T.-W. Comparison between linear regression and four different machine learning methods in selecting risk factors for osteoporosis in a Chinese female aged cohort. J. Chin. Med Assoc. 2023, 86, 1028–1036. [Google Scholar] [CrossRef]
- Wu, X.; Tsai, S.P.; Tsao, C.K.; Chiu, M.L.; Tsai, M.K.; Lu, P.J.; Lee, J.H.; Chen, C.H.; Wen, C.; Chang, S.-S.; et al. Cohort Profile: The Taiwan MJ Cohort: Half a million Chinese with repeated health surveillance data. Int. J. Epidemiol. 2017, 46, 1744–1744g. [Google Scholar] [CrossRef] [PubMed]
- The Introduction of MJ Health Database. MJ Health Research Foundation Technical Report, MJHRF-TR-01. Available online: http://www.mjhrf.org/upload/files/MJHRF-TR-01%E7%BE%8E%E5%85%86%E5%81%A5%E5%BA%B7%E8%B3%87%E6%96%99%E5%BA%AB%E7%B0%A1%E4%BB%8B.pdf (accessed on 22 August 2016).
- ISO 9001:2015; Quality Management Systems—Requirements. ISO: Geneva, Switzerland, 2005.
- Friedman, J.H.; Roosen, C.B. An introduction to multivariate adaptive regression splines. Stat. Methods Med. Res. 1995, 4, 197–217. [Google Scholar] [CrossRef] [PubMed]
- Milborrow, S. Derived from Mda: MARS by T. Hastie and R. Tibshirani. Earth: Multivariate Adaptive Regression Splines. R Package Version 5.3.4. 2024. Available online: https://cran.r-project.org/package=earth (accessed on 5 October 2024).
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-94. 2024. Available online: http://CRAN.R-project.org/package=caret (accessed on 17 April 2019).
- The Lancet Diabetes Endocrinology. Obesity in China: Time to act. Lancet Diabetes Endocrinol. 2021, 9, 407. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, S.; Qiu, X.; Wu, J.; Tan, M.; Wang, M. Serum Uric Acid Levels and Metabolic Indices in an Obese Population: A Cross-Sectional Study. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 627–635. [Google Scholar] [CrossRef]
- Yoo, T.W.; Sung, K.C.; Shin, H.S.; Kim, B.J.; Kim, B.S.; Kang, J.H.; Lee, M.H.; Park, J.R.; Kim, H.; Rhee, E.J.; et al. Relationship Between Serum Uric Acid Concentration and Insulin Resistance and Metabolic Syndrome. Circ. J. 2005, 69, 928–933. [Google Scholar] [CrossRef]
- Joo, H.J.; Kim, G.R.; Choi, D.-W.; Joo, J.H.; Park, E.-C. Uric acid level and kidney function: A cross-sectional study of the Korean national health and nutrition examination survey (2016–2017). Sci. Rep. 2020, 10, 21672. [Google Scholar] [CrossRef]
- Obermayr, R.P.; Temml, C.; Gutjahr, G.; Knechtelsdorfer, M.; Oberbauer, R.; Klauser-Braun, R. Elevated Uric Acid Increases the Risk for Kidney Disease. J. Am. Soc. Nephrol. 2008, 19, 2407–2413. [Google Scholar] [CrossRef]
- Ryoo, J.-H.; Choi, J.-M.; Oh, C.-M.; Kim, M.-G. The Association between Uric Acid and Chronic Kidney Disease in Korean Men: A 4-Year Follow-up Study. J. Korean Med. Sci. 2013, 28, 855–860. [Google Scholar] [CrossRef]
- Satirapoj, B.; Supasyndh, O.; Chaiprasert, A.; Ruangkanchanasetr, P.; Kanjanakul, I.; Phulsuksombuti, D.; Utainam, D.; Choovichian, P. Relationship between serum uric acid levels with chronic kidney disease in a Southeast Asian population. Nephrology 2010, 15, 253–258. [Google Scholar] [CrossRef]
- Mohandas, R.; Johnson, R.J. Uric Acid Levels Increase Risk for New-Onset Kidney Disease. J. Am. Soc. Nephrol. 2008, 19, 2251–2253. [Google Scholar] [CrossRef]
- Chonchol, M.; Shlipak, M.G.; Katz, R.; Sarnak, M.J.; Newman, A.B.; Siscovick, D.S.; Kestenbaum, B.; Carney, J.K.; Fried, L.F. Relationship of Uric Acid With Progression of Kidney Disease. Am. J. Kidney Dis. 2007, 50, 239–247. [Google Scholar] [CrossRef]
- Khosla, U.M.; Zharikov, S.; Finch, J.L.; Nakagawa, T.; Roncal, C.; Mu, W.; Krotova, K.; Block, E.R.; Prabhakar, S.; Johnson, R.J. Hyperuricemia induces endothelial dysfunction. Kidney Int. 2005, 67, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Kanellis, J.; Kang, D.-H. Uric acid as a mediator of endothelial dysfunction, inflammation, and vascular disease. Semin. Nephrol. 2005, 25, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-H.; Nakagawa, T.; Feng, L.; Watanabe, S.; Han, L.; Mazzali, M.; Truong, L.; Harris, R.; Johnson, R.J. A Role for Uric Acid in the Progression of Renal Disease. J. Am. Soc. Nephrol. 2002, 13, 2888–2897. [Google Scholar] [CrossRef] [PubMed]
- Nordin, B.E. Calcium and osteoporosis. Nutrition 1997, 13, 664–686. [Google Scholar] [CrossRef]
- Peacock, M.; Robertson, W.G.; Schneider, H.J.; Vahlensieck, W. Urolithiasis: Etiology Diagnosis; Springer: Berlin/Heidelberg, Germany, 1985. [Google Scholar]
- Gouri, A.; Dekaken, A.; Bentorki, A.; Touaref, A.; Yakhlef, A.; Kouicem, N. Serum Uric Acid Level and Cardiovascular Risks in Hemodialysis Patients: An Algerian Cohort Study. Clin. Lab. 2014, 60, 751–758. [Google Scholar] [CrossRef]
- Coates, V.; Raiment, P.C. The Calcium Content of the Blood Serum in Cases of Gout. Biochem. J. 1924, 18, 921–924. [Google Scholar] [CrossRef]
- Saadat, P.; Ahangar, A.A.; Babaei, M.; Kalantar, M.; Bayani, M.A.; Barzegar, H.; Gholinia, H.; Tajrishi, F.Z.; Faraji, S.; Frajzadeh, F. Relationship of Serum Uric Acid Level with Demographic Features, Risk Factors, Severity, Prognosis, Serum Levels of Vitamin D, Calcium, and Magnesium in Stroke. Stroke Res. Treat. 2018, 2018, 6580178. [Google Scholar] [CrossRef]
- Kumar, A.U.A.; Browne, L.D.; Li, X.; Adeeb, F.; Perez-Ruiz, F.; Fraser, A.D.; Stack, A.G. Temporal trends in hyperuricaemia in the Irish health system from 2006–2014: A cohort study. PLoS ONE 2018, 13, e0198197. [Google Scholar] [CrossRef]
- Guessous, I.; Bonny, O.; Paccaud, F.; Mooser, V.; Waeber, G.; Vollenweider, P.; Bochud, M. Serum Calcium Levels Are Associated with Novel Cardiometabolic Risk Factors in the Population-Based CoLaus Study. PLoS ONE 2011, 6, e18865. [Google Scholar] [CrossRef]
- Bonora, E.; Targher, G.; Zenere, M.B.; Saggiani, F.; Cacciatori, V.; Tosi, F.; Travia, D.; Zenti, M.G.; Branzi, P.; Santi, L.; et al. Relationship of uric acid concentration to cardiovascular risk factors in young men. Role of obesity and central fat distribution. The Verona Young Men Atherosclerosis Risk Factors Study. Risk Factors Study Int. J. Obes. Relat. Metab. Disord. 1996, 20, 975–980. [Google Scholar]
- Lyngdoh, T.; Marques-Vidal, P.; Paccaud, F.; Preisig, M.; Waeber, G.; Bochud, M.; Vollenweider, P. Elevated Serum Uric Acid Is Associated with High Circulating Inflammatory Cytokines in the Population-Based Colaus Study. PLoS ONE 2011, 6, e19901. [Google Scholar] [CrossRef] [PubMed]
- Kirilmaz, B.; Asgun, F.; Alioglu, E.; Ercan, E.; Tengiz, I.; Turk, U.; Saygi, S.; Özerkan, F. High Inflammatory Activity Related to the Number of Metabolic Syndrome Components. J. Clin. Hypertens. 2010, 12, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Shrayyef, M.Z.; DePapp, Z.; Cave, W.T.; Wittlin, S.D. Hypercalcemia in Two Patients with Sarcoidosis and Mycobacterium avium intracellulare Not Mediated by Elevated Vitamin D Metabolites. Am. J. Med. Sci. 2011, 342, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Fuss, M.; Pepersack, T.; Gillet, C.; Karmali, R.; Corvilain, J. Calcium and vitamin D metabolism in granulomatous diseases. Clin. Rheumatol. 1992, 11, 28–36. [Google Scholar] [CrossRef]
- Kamath, D.Y.; Xavier, D.; Sigamani, A.; Pais, P. High sensitivity C-reactive protein (hsCRP) & cardiovascular disease: An Indian perspective. Indian J. Med. Res. 2015, 142, 261–268. [Google Scholar] [CrossRef]
- Spiga, R.; Marini, M.A.; Mancuso, E.; Di Fatta, C.; Fuoco, A.; Perticone, F.; Andreozzi, F.; Mannino, G.C.; Sesti, G. Uric Acid Is Associated with Inflammatory Biomarkers and Induces Inflammation via Activating the NF-κB Signaling Pathway in HepG2 Cells. Arter. Thromb. Vasc. Biol. 2017, 37, 1241–1249. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Geng, J.-H.; Wu, P.-Y.; Huang, J.-C.; Chen, S.-C.; Chang, J.-M.; Chen, H.-C. Betel Nut Chewing Increases the Risk of Metabolic Syndrome and Its Components in a Large Taiwanese Population Follow-Up Study Category: Original Investigation. Nutrients 2022, 14, 1018. [Google Scholar] [CrossRef]
- Chang, C.-K.; Lee, J.-I.; Chang, C.-F.; Lee, Y.-C.; Jhan, J.-H.; Wang, H.-S.; Shen, J.-T.; Tsao, Y.-H.; Huang, S.-P.; Geng, J.-H. Betel Nut Chewing Is Associated with the Risk of Kidney Stone Disease. J. Pers. Med. 2022, 12, 126. [Google Scholar] [CrossRef]
- Tai, T.-S.; Hsu, C.-C.; Pai, H.-C.; Liu, W.-H.; Hsu, Y.-H. The association between hyperuricemia and betel nut chewing in Taiwanese men: A cross-sectional study. BMC Public Health 2013, 13, 1136. [Google Scholar] [CrossRef]
- Kuzuya, M.; Ando, F.; Iguchi, A.; Shimokata, H. Effect of Aging on Serum Uric Acid Levels: Longitudinal Changes in a Large Japanese Population Group. J. Gerontol. A Biol. Sci. Med. Sci. 2002, 57, M660–M664. [Google Scholar] [CrossRef]
- Wu, J.; Qiu, L.; Yan, W.-H.; Cheng, X.-Q.; Wu, W.; Guo, X.-Z.; Ding, H.-T.; Han, H.-J.; Han, S.-M.; Zhu, G.-J. Serum γ-glutamyltransferase and uric acid levels are associated with impaired fasting glucose in adults from Inner Mongolia, China. BMC Public Health 2013, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Rubio, A.; Sanvisens, A.; Bolao, F.; Pérez-Mañá, C.; García-Marchena, N.; Fernández-Prendes, C.; Muñoz, A.; Muga, R. Association of hyperuricemia and gamma glutamyl transferase as a marker of metabolic risk in alcohol use disorder. Sci. Rep. 2020, 10, 20060. [Google Scholar] [CrossRef] [PubMed]
- Aygun, F.; Kirkoc, R.; Aygun, D.; Cam, H. Gamma Glutamyl Transferase and Uric Acid Levels Can Be Associated with the Prognosis of Patients in the Pediatric Intensive Care Unit. Children 2018, 5, 147. [Google Scholar] [CrossRef] [PubMed]
- Radovanovic, S.; Savic-Radojevic, A.; Pekmezovic, T.; Markovic, O.; Memon, L.; Jelic, S.; Simic, D.; Radic, T.; Pljesa-Ercegovac, M.; Simic, T. Uric Acid and Gamma-glutamyl Transferase Activity Are Associated with Left Ventricular Remodeling Indices in Patients with Chronic Heart Failure. Rev. Esp. Cardiol. 2014, 67, 632–642. [Google Scholar] [CrossRef]
- Chen, J.-H.; Chuang, S.-Y.; Chen, Y.-C.; Wang, W.-T.; Wu, Y.-C. Effect of allopurinol on blood pressure and glucose metabolism: A systematic review and meta-analysis of randomized controlled trials. Atherosclerosis 2015, 241, 674–681. [Google Scholar]
- Moharana, J.J.; Mishra, R.; Nayak, A.K. A Study on Serum Lactate Dehydrogenase and Uric Acid in Preeclampsia and Eclampsia: Can they Predict Adverse Fetomaternal Outcome? Int. J. Appl. Basic Med. Res. 2023, 13, 95–100. [Google Scholar] [CrossRef]
- Tariq, T.; Fatima, M.; Ali, A.; Maryam, H.; Ahmad, W. Correlation between Uric Acid and Lipid Profile in Untreated Dyslipidemic Patients. Saudi J. Pathol. Microbiol. 2022, 7, 338–344. [Google Scholar] [CrossRef]
- Hofmann, S.M.; Tschöp, M.H. Dietary sugars: A fat difference. J. Clin. Investig. 2009, 119, 1089–1092. [Google Scholar] [CrossRef]
- Ebrahimpour, P.; Fakhrzadeh, H.; Heshmat, R.; Bandarian, F.; Larijani, B. Serum uric acid levels and risk of metabolic syndrome in healthy adults. Endocr. Pract. 2008, 14, 298–304. [Google Scholar] [CrossRef]
| Metric | Description | Calculation |
|---|---|---|
| SMAPE | Symmetric Mean Absolute Percentage Error | |
| RAE | Relative absolute error | |
| RRSE | Root relative squared error | |
| RMSE | Root mean squared error |
| Variables | Unit | Mean ± SD |
|---|---|---|
| Age | year | 42.05 ± 11.48 |
| Waist–hip ratio | waist/hip circumference | 0.86 ± 0.06 |
| Leukocyte | ×103/μL | 6.16 ± 1.61 |
| Hemoglobin | ×106/μL | 15.29 ± 1.07 |
| Platelets | ×103/μL | 227.05 ± 47.99 |
| Fasting plasma glucose | mg/dL | 103.40 ± 18.70 |
| Systolic blood pressure | mmHg | 119.54 ± 15.24 |
| Diastolic blood pressure | mmHg | 76.86 ± 10.46 |
| Total bilirubin | mg/dL | 1.06 ± 0.41 |
| Albumin | mg/dL | 4.47 ± 0.20 |
| Globulin | g/dL | 3.02 ± 0.32 |
| Alkaline Phosphatase | IU/L | 62.41 ± 20.85 |
| Glutamic oxaloacetic transaminase | IU/L | 25.81 ± 12.23 |
| Glutamic pyruvic transaminase | IU/L | 34.87 ± 25.73 |
| γ-glutamyl transpeptidase | IU/L | 38.38 ± 50.17 |
| Lactate dehydrogenase | mg/dL | 162.52 ± 34.13 |
| Creatinine | mg/dL | 1.08 ± 0.18 |
| Triglycerides | mg/dL | 133.31 ± 131.20 |
| High density lipoprotein cholesterol | mg/dL | 52.63 ± 11.74 |
| Low density lipoprotein cholesterol | mg/dL | 123.38 ± 33.32 |
| Plasma calcium concentration | mg/dL | 9.49 ± 0.36 |
| Plasma phosphate concentration | mg/dL | 3.55 ± 0.48 |
| Alpha-fetoprotein | ng/mL | 2.93 ± 1.96 |
| Carcinoembryonic antigen | ng/mL | 3.47 ± 96.08 |
| Thyroid stimulating hormone | μIU/mL | 1.64 ± 1.38 |
| C reactive protein | mg/dL | 0.23 ± 0.45 |
| Bone mass density | T-score | 0.51 ± 1.22 |
| Alcohol consumption | - | 7.39 ± 18.96 |
| Smoking | - | 9.59 ± 20.65 |
| Betel nut exposure | - | 0.97 ± 7.41 |
| Sports | - | 7.62 ± 8.97 |
| Homocysteine | μmol/L | 10.46 ± 4.36 |
| High sensitivity C-Reactive Protein | mg/L | 2.08 ± 4.55 |
| Ferritin | ng/mL | 252.82 ± 166.95 |
| Fibrinogen | mg/dL | 274.86 ± 58.48 |
| Uric acid | mg/dL | 6.58 ± 1.32 |
| Marriage status | n (%) | |
| Single | 1600 (30.77%) | |
| With spouse | 3600 (69.23%) | |
| Education level | n (%) | |
| Illiterate | 7 (0.13%) | |
| Elementary school | 97 (1.87%) | |
| Junior high school (vocational) | 210 (4.04%) | |
| High school | 1049 (20.17%) | |
| Junior college | 993 (19.10%) | |
| University | 1900 (36.54%) | |
| Graduate school or above | 944 (18.15%) | |
| Sleep hours | n (%) | |
| <4 h/day | 82 (1.58%) | |
| 4–6 h/day | 1276 (24.54%) | |
| 6–7 h/day | 2528 (48.62%) | |
| 7–8 h/day | 1114 (21.42%) | |
| 8–9 h/day | 166 (3.19%) | |
| >9 h/day | 34 (0.65%) | |
| Variable | r | Variable | r | Variable | r | Variable | r |
|---|---|---|---|---|---|---|---|
| Age | −0.1057 *** | Leukocyte | 0.1311 *** | GOT | 0.1694 *** | Creatinine | 0.1708 *** |
| Alcohol | 0.0764 *** | Hemoglobin | 0.1095 *** | GPT | 0.2222 *** | CRP | 0.027 * |
| Smoking | 0.0151 | Platelets | 0.1064 *** | Albumin | 0.146 *** | Hs-CRP | 0.0445 ** |
| Betel nut | −0.0164 | TG | 0.1924 *** | Globulin | 0.1126 *** | CEA | 0.0061 |
| Sports | −0.0029 | HDL-C | −0.1461 *** | TBIL | −0.0497 *** | TSH | 0.0438 ** |
| WHR | 0.2256 *** | LDL-C | 0.1649 | γ-GT | 0.1781 *** | Hcys | 0.0766 |
| BMD | 0.1412 *** | FPG | −0.0309 * | LDH | 0.1366 *** | Ferritin | 0.1288 *** |
| SBP | 0.1458 *** | Ca | 0.1666 *** | ALP | 0.0309 * | Fibrinogen | 0.0593 |
| DBP | 0.1383 *** | P | 0.0695 | AFP | 0.0055 |
| Variables | Uric Acid (mg/dL) | p Value |
|---|---|---|
| Marriage status | ||
| Single | 6.62 ± 1.33 | 0.075 |
| With spouse | 6.56 ± 1.31 | |
| Education level | ||
| Illiterate | 7.53 ± 1.61 | 0.005 |
| Elementary school | 6.21 ± 1.11 | |
| Junior high school (vocational) | 6.60 ± 1.41 | |
| High school | 6.62 ± 1.40 | |
| Junior college | 6.59 ± 1.33 | |
| University | 6.58 ± 1.27 | |
| Graduate school or above | 6.53 ± 1.28 | |
| Sleep hours | ||
| <4 h/day | 6.62 ± 1.44 | 0.936 |
| 4–6 h/day | 6.59 ± 1.32 | |
| 6–7 h/day | 6.56 ± 1.31 | |
| 7–8 h/day | 6.59 ± 1.32 | |
| 8–9 h/day | 6.61 ± 1.35 | |
| Methods | SMAPE | RAE | RRSE | RMSE |
|---|---|---|---|---|
| MLR | 0.1797 | 1.1250 | 1.2542 | 1.6666 |
| MARS | 0.1789 | 1.1199 | 1.2563 | 1.6694 |
| Definition | a1 | |
|---|---|---|
| Intercept | 7.187 | |
| B1 | Max(0, 48–Age) | 0.025 |
| B2 | Max(0, Age–48) | −0.012 |
| B3 | Max(0, 0.969–WHR) | −3.280 |
| B4 | Max(0, 115–FPG) | −0.009 |
| B5 | Max(0, FPG–115) | −0.010 |
| B6 | Max(0, 49–γ-GT) | −0.014 |
| B7 | Max(0, 211–LDH) | −0.005 |
| B8 | Max(0, Cr–0.97) | 2.882 |
| B9 | Max(0, 207–TG) | −0.004 |
| B10 | Max(0, TG–207) | 0.001 |
| B11 | Max(0, 9.5–Ca) | −0.525 |
| B12 | Max(0, 5–BN) | 0.106 |
| B13 | Max(0, 3.38–Hs-CRP) | −0.118 |
| B14 | Max(0, Hs-CRP–3.38) | −0.016 |
| Variable | MARS Threshold (Present Study) | Clinical Threshold for Metabolic Syndrome/Obesity/Diabetes/Kidney Disease | Source/Guideline | Interpretation/Clinical Relevance |
|---|---|---|---|---|
| WHR | <0.969 (impact zone) | ≥0.90 (abdominal obesity, men) | IDF 2006; WHO 2008 | MARS identifies a protective” effect below 0.969—still above the obesity threshold (0.90) |
| Creatinine | >0.97 mg/dL | >1.1–1.3 mg/dL (mild renal impairment, men) | KDIGO 2012; Lab reference ranges | MARS detects UA elevation before creatinine reaches clinical “abnormal” range—supports early renal stress even within “normal” labs. |
| hs-CRP | >3.38 mg/L | >3.0 mg/L (high cardiovascular risk) | AHA/CDC 2003 | MARS threshold aligns closely with established CV risk stratification—reinforces inflammation as key UA driver. |
| Fasting Glucose | >115 mg/dL | ≥100 mg/dL (prediabetes); ≥126 mg/dL (diabetes) | ADA 2023 | MARS identifies nonlinear effect starting at 115 mg/dL —between prediabetes and diabetes thresholds, suggesting glucose dysregulation impacts UA before frank diabetes. |
| Calcium | <9.5 mg/dL | Normal range: 8.5–10.5 mg/dL; no direct MetS threshold | Lab reference ranges | MARS suggests low–normal calcium (still within “normal”) is associated with higher UA—possibly reflecting subclinical inflammation or bone turnover. |
| Triglycerides | >207 mg/dL | ≥150 mg/dL (MetS criterion) | NCEP ATP III 2001 | MARS effect activates well above MetS threshold— suggests UA is more sensitive to severe hypertriglyceridemia than mild elevations. |
| γ-GT | <49 IU/L | Normal: <55–65 IU/L (lab-dependent); no direct MetS threshold | Lab reference ranges | MARS identifies effect in low–normal range— may reflect hepatic oxidative stress before enzyme elevation becomes clinically apparent. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-C.; Shen, M.-C.; Lai, Z.-Y.; Ke, J.-C.; Chu, T.-W.; Chuang, Y.-J. Uncovering Nonlinear Predictors of Serum Biomarker Uric Acid Using Interpretable Machine Learning in Healthy Men. Biomedicines 2025, 13, 2469. https://doi.org/10.3390/biomedicines13102469
Yang C-C, Shen M-C, Lai Z-Y, Ke J-C, Chu T-W, Chuang Y-J. Uncovering Nonlinear Predictors of Serum Biomarker Uric Acid Using Interpretable Machine Learning in Healthy Men. Biomedicines. 2025; 13(10):2469. https://doi.org/10.3390/biomedicines13102469
Chicago/Turabian StyleYang, Chung-Chi, Min-Chung Shen, Zih-Yin Lai, Jyun-Cheng Ke, Ta-Wei Chu, and Yung-Jen Chuang. 2025. "Uncovering Nonlinear Predictors of Serum Biomarker Uric Acid Using Interpretable Machine Learning in Healthy Men" Biomedicines 13, no. 10: 2469. https://doi.org/10.3390/biomedicines13102469
APA StyleYang, C.-C., Shen, M.-C., Lai, Z.-Y., Ke, J.-C., Chu, T.-W., & Chuang, Y.-J. (2025). Uncovering Nonlinear Predictors of Serum Biomarker Uric Acid Using Interpretable Machine Learning in Healthy Men. Biomedicines, 13(10), 2469. https://doi.org/10.3390/biomedicines13102469







