Proteomic Characterisation of Heart Failure Reveals a Unique Molecular Phenotype for Hypertrophic Cardiomyopathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Cohort
2.2. Sample Preparation
2.3. Mass Spectrometry Analysis
2.4. Data Analysis
3. Results
3.1. Proteomic Characterisation of Heart Failure
3.2. Differential Protein Expression between Heart Failure and Non-Failing Heart Tissue
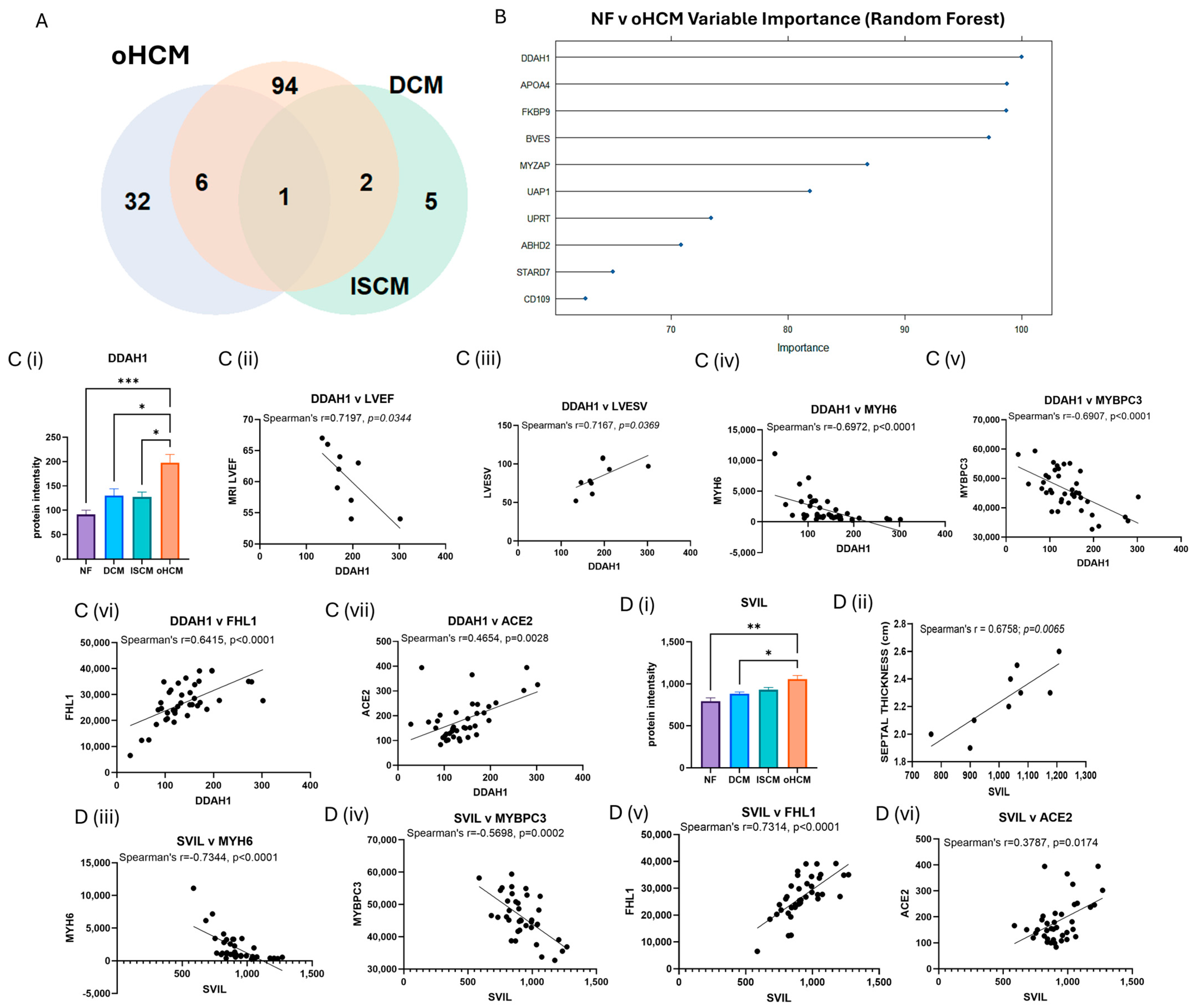
3.3. Differential Protein Expression in Obstructive HCM Reveals Unique Pathology of Disease
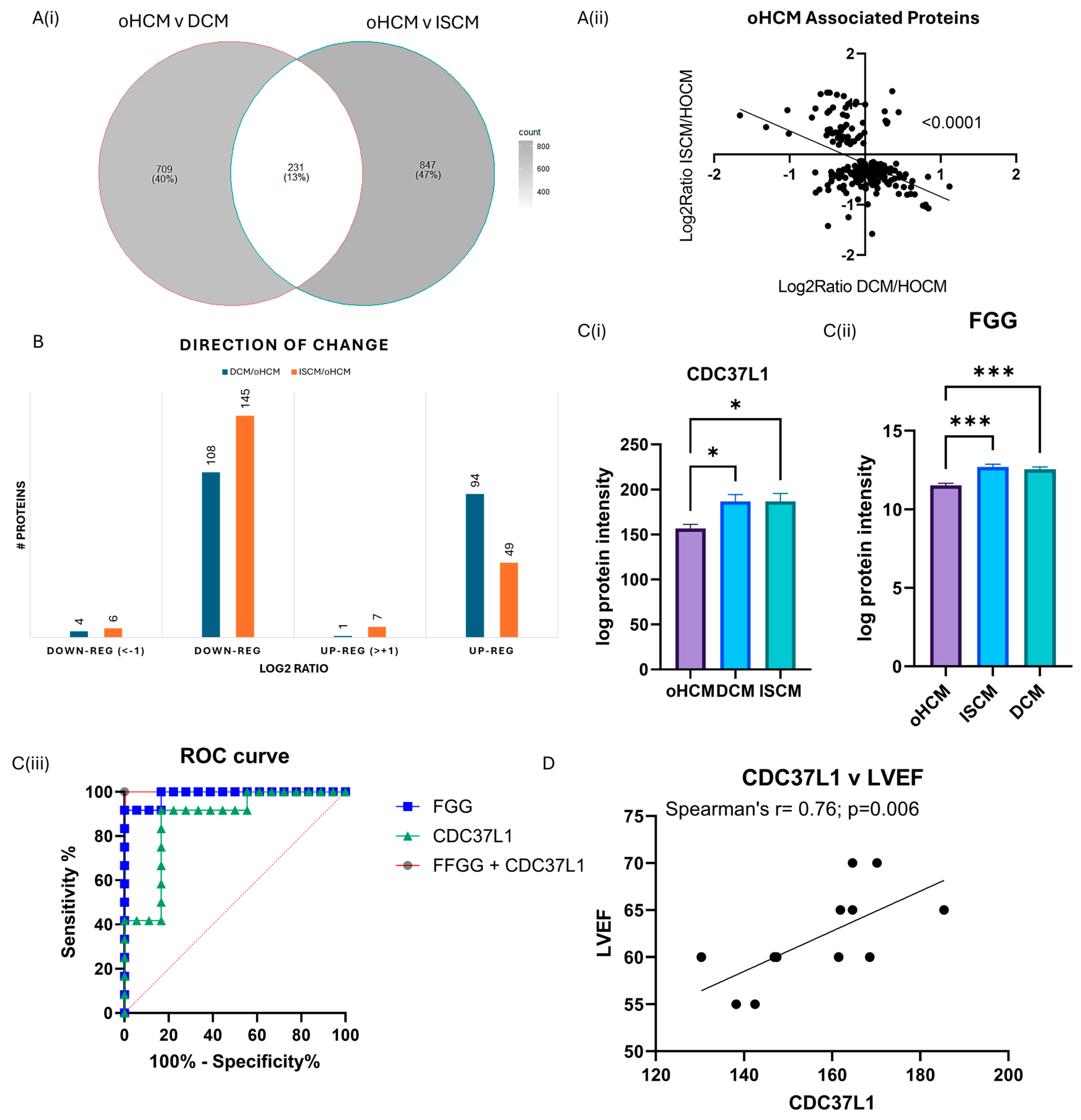
3.4. Differential Protein Expression between Obstructive HCM and Other Heart Failure Subtypes
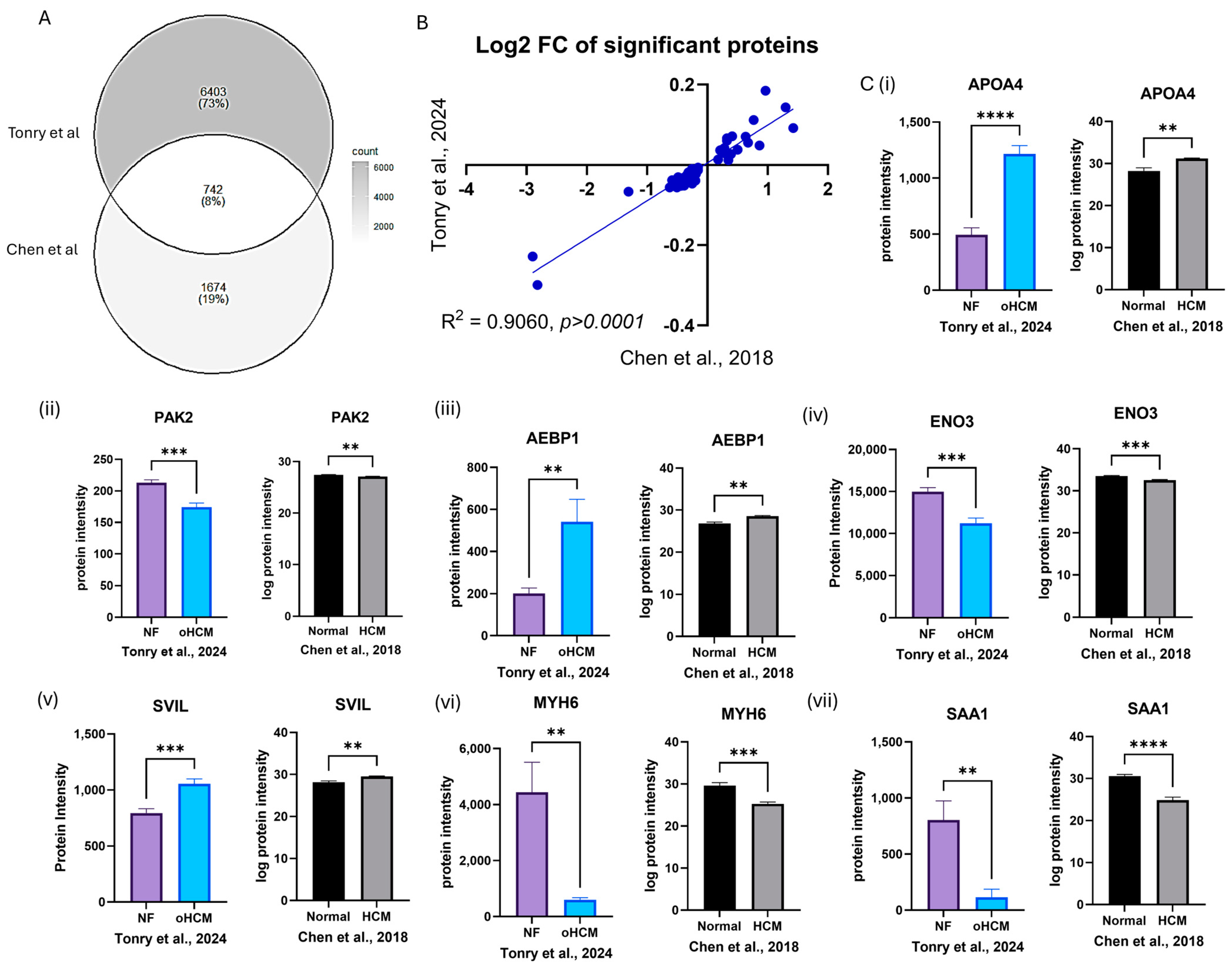
3.5. In Silico Validation of HCM-Associated Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef] [PubMed]
- Metra, M.; Teerlink, J.R. Heart failure. Lancet 2017, 390, 1981–1995. [Google Scholar] [CrossRef]
- Heidenreich, P.A.; Albert, N.M.; Allen, L.A.; Bluemke, D.A.; Butler, J.; Fonarow, G.C.; Ikonomidis, J.S.; Khavjou, O.; Konstam, M.A.; Maddox, T.M.; et al. Forecasting the impact of heart failure in the United States: A policy statement from the American Heart Association. Circ. Heart Fail. 2013, 6, 606–619. [Google Scholar] [CrossRef]
- Ziaeian, B.; Fonarow, G.C. Epidemiology and aetiology of heart failure. Nat. Rev. Cardiol. 2016, 13, 368–378. [Google Scholar] [CrossRef]
- Del Buono, M.G.; Moroni, F.; Montone, R.A.; Azzalini, L.; Sanna, T.; Abbate, A. Ischemic cardiomyopathy and heart failure after acute myocardial infarction. Curr. Cardiol. Rep. 2022, 24, 1505–1515. [Google Scholar] [CrossRef]
- Japp, A.G.; Gulati, A.; Cook, S.A.; Cowie, M.R.; Prasad, S.K. The Diagnosis and Evaluation of Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2016, 67, 2996–3010. [Google Scholar] [CrossRef]
- Veselka, J.; Anavekar, N.S.; Charron, P. Hypertrophic obstructive cardiomyopathy. Lancet 2017, 389, 1253–1267. [Google Scholar] [CrossRef] [PubMed]
- Teekakirikul, P.; Zhu, W.; Huang, H.C.; Fung, E. Hypertrophic Cardiomyopathy: An Overview of Genetics and Management. Biomolecules 2019, 9, 878. [Google Scholar] [CrossRef] [PubMed]
- Landstrom, A.P.; Ackerman, M.J. Mutation type is not clinically useful in predicting prognosis in hypertrophic cardiomyopathy. Circulation 2010, 122, 2441–2449; discussion 50. [Google Scholar] [CrossRef]
- Castiglione, V.; Aimo, A.; Vergaro, G.; Saccaro, L.; Passino, C.; Emdin, M. Biomarkers for the diagnosis and management of heart failure. Heart Fail. Rev. 2022, 27, 625–643. [Google Scholar] [CrossRef]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Brunner, A.-D.; Frank, M.; Ha, A.; Bludau, I.; Voytik, E.; Kaspar-Schoenefeld, S.; Lubeck, M.; Raether, O.; Bache, N.; et al. diaPASEF: Parallel accumulation–serial fragmentation combined with data-independent acquisition. Nat. Methods 2020, 17, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Brunner, A.-D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M.A.; et al. Online Parallel Accumulation–Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteom. 2018, 17, 2534–2545. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Caporizzo, M.A.; Bedi, K.; Vite, A.; Bogush, A.I.; Robison, P.; Heffler, J.G.; Salomon, A.K.; Kelly, N.A.; Babu, A.; et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat. Med. 2018, 24, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Tucholski, T.; Cai, W.; Gregorich, Z.R.; Bayne, E.F.; Mitchell, S.D.; McIlwain, S.J.; de Lange, W.J.; Wrobbel, M.; Karp, H.; Hite, Z.; et al. Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics. Proc. Natl. Acad. Sci. USA 2020, 117, 24691–24700. [Google Scholar] [CrossRef] [PubMed]
- Garmany, R.; Bos, J.M.; Dasari, S.; Johnson, K.L.; Tester, D.J.; Giudicessi, J.R.; Dos Remedios, C.; Maleszewski, J.J.; Ommen, S.R.; Dearani, J.A.; et al. Proteomic and phosphoproteomic analyses of myectomy tissue reveals difference between sarcomeric and genotype-negative hypertrophic cardiomyopathy. Sci. Rep. 2023, 13, 14341. [Google Scholar] [CrossRef]
- Captur, G.; Heywood, W.E.; Coats, C.; Rosmini, S.; Patel, V.; Lopes, L.R.; Collis, R.; Patel, N.; Syrris, P.; Bassett, P.; et al. Identification of a Multiplex Biomarker Panel for Hypertrophic Cardiomyopathy Using Quantitative Proteomics and Machine Learning. Mol. Cell. Proteom. 2020, 19, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Ewoldt, J.; Venturini, G.; Pereira, A.C.; Padilha, K.; Lawton, M.; Lin, W.; Goel, R.; Luptak, I.; Perissi, V.; et al. Multi-Omics Profiling of Hypertrophic Cardiomyopathy Reveals Altered Mechanisms in Mitochondrial Dynamics and Excitation-Contraction Coupling. Int. J. Mol. Sci. 2023, 24, 4724. [Google Scholar] [CrossRef]
- Coats, C.J.; Heywood, W.E.; Virasami, A.; Ashrafi, N.; Syrris, P.; Dos Remedios, C.; Treibel, T.A.; Moon, J.C.; Lopes, L.R.; McGregor, C.G.; et al. Proteomic Analysis of the Myocardium in Hypertrophic Obstructive Cardiomyopathy. Circ. Genom. Precis. Med. 2018, 11, e001974. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, D.; Kong, L.; Shi, H.; Tian, X.; Gao, L.; Liu, Y.; Wu, L.; Du, B.; Huang, Z.; et al. Aldolase promotes the development of cardiac hypertrophy by targeting AMPK signaling. Exp. Cell Res. 2018, 370, 78–86. [Google Scholar] [CrossRef]
- Zhuo, J.; Geng, H.; Wu, X.; Fan, M.; Sheng, H.; Yao, J. AKT-mTOR signaling-mediated rescue of PRKAG2 R302Q mutant-induced familial hypertrophic cardiomyopathy by treatment with β-adrenergic receptor (β-AR) blocker metoprolol. Cardiovasc. Diagn. Ther. 2022, 12, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, P.; Kwak, D.; Fassett, J.; Yue, W.; Atzler, D.; Hu, X.; Liu, X.; Wang, H.; Lu, Z.; et al. Cardiomyocyte dimethylarginine dimethylaminohydrolase-1 (DDAH1) plays an important role in attenuating ventricular hypertrophy and dysfunction. Basic. Res. Cardiol. 2017, 112, 55. [Google Scholar] [CrossRef] [PubMed]
- Linscheid, N.; Poulsen, P.C.; Pedersen, I.D.; Gregers, E.; Svendsen, J.H.; Olesen, M.S.; Olsen, J.V.; Delmar, M.; Lundby, A. Quantitative Proteomics of Human Heart Samples Collected In Vivo Reveal the Remodeled Protein Landscape of Dilated Left Atrium Without Atrial Fibrillation. Mol. Cell. Proteom. 2020, 19, 1132–1144. [Google Scholar] [CrossRef]
- Kopaliani, I.; Jarzebska, N.; Billoff, S.; Kolouschek, A.; Martens-Lobenhoffer, J.; Bornstein, S.R.; Bode-Böger, S.M.; Ragavan, V.N.; Weiss, N.; Mangoni, A.A.; et al. Overexpression of dimethylarginine dimethylaminohydrolase 1 protects from angiotensin II-induced cardiac hypertrophy and vascular remodeling. Am. J. Physiol. Heart Circ. Physiol. 2021, 321, H825–H838. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Guo, J.; Xu, F.; Weng, X.; Yue, W.; Ge, J. Cardiomyocyte dimethylarginine dimethylaminohydrolase1 attenuates left-ventricular remodeling after acute myocardial infarction: Involvement in oxidative stress and apoptosis. Basic Res. Cardiol. 2018, 113, 28. [Google Scholar] [CrossRef] [PubMed]
- Tadros, R.; Zheng, S.L.; Grace, C.; Jorda, P.; Francis, C.; Jurgens, S.J.; Thomson, K.L.; Harper, A.R.; Ormondroyd, E.; West, D.M.; et al. Large scale genome-wide association analyses identify novel genetic loci and mechanisms in hypertrophic cardiomyopathy. medRxiv 2023. [Google Scholar] [CrossRef]
- He, G.-W.; Hou, H.-T.; Xuan, C.; Wang, J.; Liu, L.-X.; Zhang, J.-F.; Liu, X.-C.; Yang, Q. Corrective surgery alters plasma protein profiling in congenital heart diseases and clinical perspectives. Am. J. Transl. Res. 2020, 12, 1319–1337. [Google Scholar]
- Zhang, H.; Li, C.; Song, X.; Cheng, L.; Liu, Q.; Zhang, N.; Wei, L.; Chung, K.; Adcock, I.M.; Ling, C.; et al. Integrated analysis reveals lung fibrinogen gamma chain as a biomarker for chronic obstructive pulmonary disease. Ann. Transl. Med. 2021, 9, 1765. [Google Scholar] [CrossRef]
- Peak, S.L.; Gracia, L.; Lora, G.; Jinwal, U.K. Hsp90-interacting Co-chaperones and their Family Proteins in Tau Regulation: Intro-ducing a Novel Role for Cdc37L1. Neuroscience 2021, 453, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Chen, J.; Wu, J.; Li, T.; Yi, C.; Wang, K.; Wang, P.; Sun, C.; Ye, H. Combination of an Autoantibody Panel and Alpha-Fetoprotein for Early Detection of Hepatitis B Virus-Associated Hepatocellular Carcinoma. Cancer Prev. Res. 2024, 17, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Dreßen, M.; Geyer, P.E.; Itzhak, D.N.; Braun, C.; Doppler, S.A.; Meier, F.; Deutsch, M.-A.; Lahm, H.; Lange, R.; et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Hewapathirana, S.; García-Seisdedos, D.; Kamatchinathan, S.; Kundu, D.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]

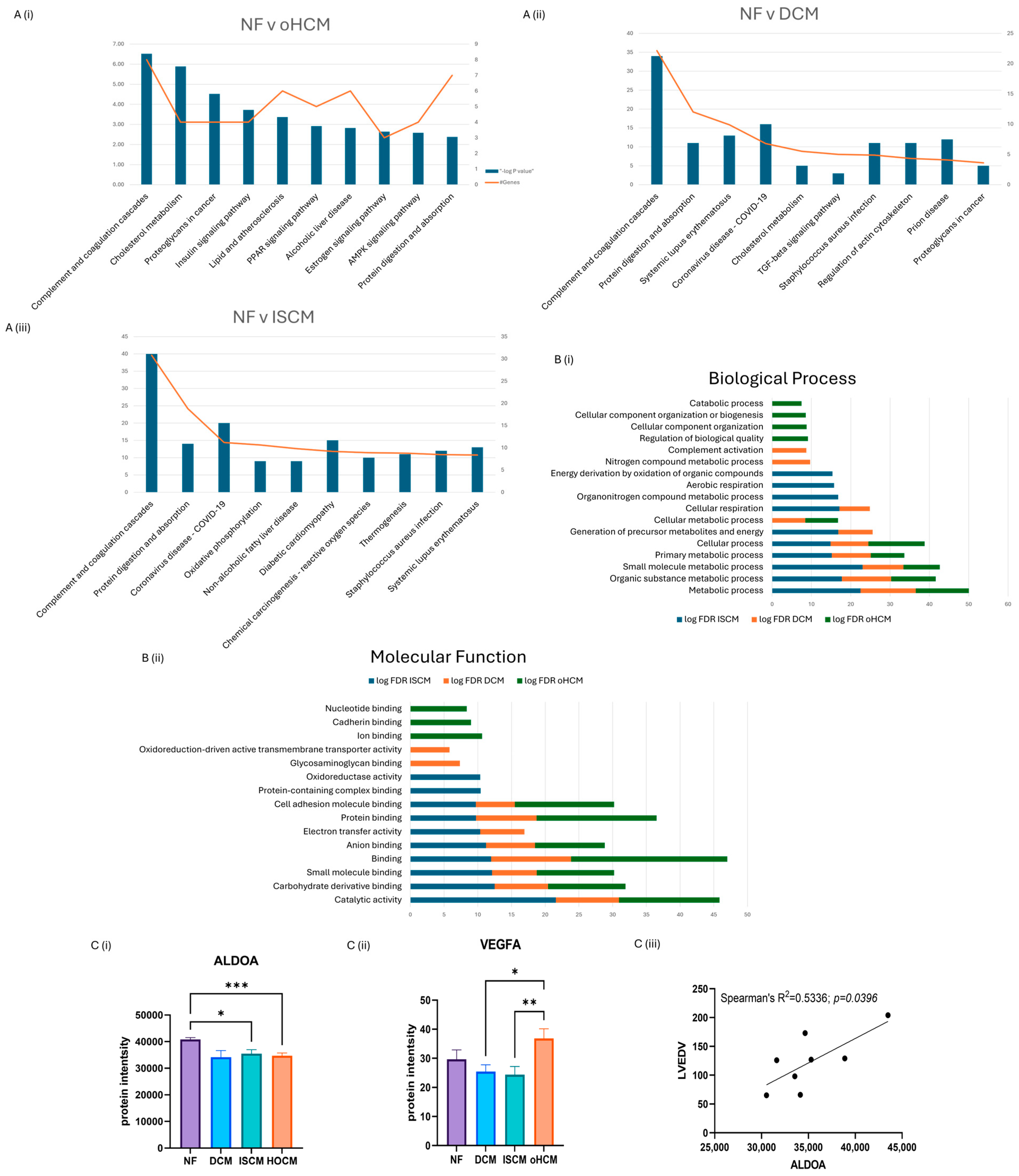
| Protein Name (Gene ID) | Median (NF, DCM, ISCM, oHCM) | p | p.adj | log2ratio | Clinical Correlations (r) | |
|---|---|---|---|---|---|---|
| Unique oHCM + validated | Apolipoprotein A-IV (APOA4) * | 1067.9 | 6.00 × 10−7 | 0.004 | 1.299 | |
| N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 (DDAH1) * | 132.4 | 4.00 × 10−5 | 0.027 | 1.116 | LVEDV: r = −0.9048, p = 0.0046 | |
| LVEF: r = −0.7197, p = 0.0344 | ||||||
| LVESV: r = 0.7167, p = 0.0369 | ||||||
| Fructose-bisphosphate aldolase A (ALDOA) | 36,406.44 | 1.00 × 10−4 | 0.045 | −0.235 | LVEDV: r = 0.8333, p = 0.0154 | |
| Ubiquitin domain-containing protein UBFD1 (UBFD1) | 182.3 | 1.00 × 10−4 | 0.045 | −0.487 | ||
| Serine/threonine-protein kinase PAK 2 (PAK2) | 197.35 | 2.00 × 10−4 | 0.047 | −0.289 | ||
| Beta-enolase (ENO3) | 12,210.88 | 2.00 × 10−4 | 0.047 | −0.419 | ||
| Large ribosomal subunit protein uL15 (RPL27A) | 739.05 | 2.00 × 10−4 | 0.047 | 0.56 | ||
| Myocardial zonula adherens protein (MYZAP) | 1227.59 | 2.00 × 10−4 | 0.047 | 0.514 | ||
| Palladin (PALLD) | 1687.3 | 3.00 × 10−4 | 0.048 | −0.326 | ||
| Supervillin (SVIL) | 903.31 | 3.00 × 10−4 | 0.049 | 0.411 | Septal thickness: r = 0.8452, p = 0.0062 |
| Protein Name (Gene) | %CV | p adj DCM v oHCM | log2ratio DCM/oHCM | p adj ISCM v oHCM | log2ratio ISCM/oHCM | Clinical Correlations |
|---|---|---|---|---|---|---|
| Cytochrome c oxidase subunit 7A-related protein, mitochondrial (COX7A2L) | 5.6 | 0.001 | 0.091 | 0.004 | −0.476 | RVSP: r = −0.7381; p = 0.0458 |
| Complement C1q subcomponent subunit A (C1QA) | 6.1 | 0.044 | −0.243 | 0.010 | 0.766 | LV Mass: r = 0.8333; p = 0.0154 |
| Complement C2 (C2) | 3.1 | 0.023 | 0.444 | 0.007 | 0.838 | LV Mass: r = 0.7857, p = 0.0279 |
| Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial (PDAH1) | 2.4 | 0.033 | 0.047 | 0.017 | −0.280 | LVEDV: r = 0.7667, p = 0.0214 |
| Complement component C7 (C7) | 6.1 | 0.004 | −0.374 | 0.007 | 1.184 | LVEDD: r = 0.7888, p = 0.0036 |
| Collagen alpha-2(VI) chain (COL6A2) | 2.6 | 0.046 | −0.118 | 0.001 | 1.045 | LVEF: r = 0.7224, p = 0.0098 |
| Enoyl-CoA hydratase domain-containing protein 2, mitochondrial (ECHDC2) | 6.1 | 0.037 | −0.310 | 0.010 | −0.415 | BNP: r = 0–1.000, p = 0.0167 |
| Zinc finger protein 397 (ZNF397) | 20.7 | 0.041 | 0.256 | 0.013 | −0.444 | LVEDV: r = 0.8833, p = 0.0031 |
| Complement factor D (CFD) | 9.9 | 0.034 | 0.358 | 0.008 | 1.255 | LV Mass: r = 0.7619; p = 0.0368 Septal thickness: r = 0.7197, p = 0.0338 |
| Complement C1q subcomponent subunit C (C1QC) | 6.0 | 0.026 | −0.260 | 0.008 | 0.928 | LVEDD: r = 0.6991, p = 0.0143 Post-amyl LVOT gradient: r = 0.8214, p = 0.0341 |
| Complement component C9 (C9) | 5.0 | 0.009 | −0.129 | 0.008 | 0.998 | LVEDV: r = −0.7381, p = 0.0458 RVSP: r = 0.8571, p = 0.0107 |
| C4b-binding protein alpha chain (C4BPA) | 4.5 | 0.031 | 0.000 | 0.005 | 0.991 | LVEDD: r = 0.5988, p = 0.0431 |
| Attractin (ATRN) | 7.4 | 0.016 | −0.009 | 0.015 | 0.433 | LV Mass: r = 0.9048, p = 0.0046 |
| Ceruloplasmin (CP) | 6.1 | 0.005 | 0.259 | 0.020 | 0.854 | |
| Alpha-1-antitrypsin (SERPINA1) | 2.9 | 0.004 | −0.426 | 0.005 | 0.932 | |
| Alpha-1-antichymotrypsin (SERPINA3) | 4.5 | 0.019 | −1.031 | 0.046 | 0.877 | |
| Complement C3 (C3) | 2.8 | 0.038 | 0.287 | 0.036 | 0.615 | |
| Plasma protease C1 inhibitor (SERPING1) | 5.1 | 0.007 | −0.295 | 0.008 | 0.791 | |
| Inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) | 1.4 | 0.012 | 0.295 | 0.021 | 0.641 | |
| Homeodomain-interacting protein kinase 2 (HIPK2) | 21.7 | 0.001 | −0.009 | 0.000 | −0.648 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tonry, C.; Linden, K.; Collier, P.; Ledwidge, M.; McDonald, K.; Collins, B.C.; Watson, C.J. Proteomic Characterisation of Heart Failure Reveals a Unique Molecular Phenotype for Hypertrophic Cardiomyopathy. Biomedicines 2024, 12, 1712. https://doi.org/10.3390/biomedicines12081712
Tonry C, Linden K, Collier P, Ledwidge M, McDonald K, Collins BC, Watson CJ. Proteomic Characterisation of Heart Failure Reveals a Unique Molecular Phenotype for Hypertrophic Cardiomyopathy. Biomedicines. 2024; 12(8):1712. https://doi.org/10.3390/biomedicines12081712
Chicago/Turabian StyleTonry, Claire, Katie Linden, Patrick Collier, Mark Ledwidge, Ken McDonald, Ben C. Collins, and Chris J. Watson. 2024. "Proteomic Characterisation of Heart Failure Reveals a Unique Molecular Phenotype for Hypertrophic Cardiomyopathy" Biomedicines 12, no. 8: 1712. https://doi.org/10.3390/biomedicines12081712
APA StyleTonry, C., Linden, K., Collier, P., Ledwidge, M., McDonald, K., Collins, B. C., & Watson, C. J. (2024). Proteomic Characterisation of Heart Failure Reveals a Unique Molecular Phenotype for Hypertrophic Cardiomyopathy. Biomedicines, 12(8), 1712. https://doi.org/10.3390/biomedicines12081712






