The Abundance of FOXP3, FOXP3/CD4 and CD8 Cells in the Microenvironment of Nodular Sclerosis and Mixed Cellularity Subtypes Is Associated with the Epstein–Barr Virus Status of Classic Hodgkin Lymphoma
Abstract
1. Introduction
2. Materials and Method
2.1. Patients and Data Collection
2.2. Methods
2.3. Statistical Analysis
2.4. Ethical Approval
3. Results
3.1. Descriptive Data
3.2. Clinical and Pathological Parameters According to EBV Status of CHL
3.3. Clinical and Pathological Parameters According to EBV Status of Common Subtypes
3.4. Clinical and Pathological Parameters According to Relapse
3.5. Clinical and Pathological Parameters According to B Symptoms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aldinucci, D.; Gloghini, A.; Pinto, A.; De Filippi, R.; Carbone, A. The classical Hodgkin’s lymphoma microenvironment and its role in promoting tumour growth and immune escape. J. Pathol. 2010, 221, 248–263. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.S.; Stein, H.; Swerlow, S.H. Classic Hodgkin lymphoma. In WHO Classification of Tumours of Haematopoetic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; IARC Lion: Lyon, France, 2017; pp. 35–442. [Google Scholar]
- Murray, P.G.; Young, L.S. An etiological role for the Epstein-Barr virus in the pathogenesis of classical Hodgkin lymphoma. Blood 2019, 134, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Poppema, S.; Visser, L. Absence of HLA class I expression by Reed-Sternberg cells. Am. J. Pathol. 1994, 145, 37–41. [Google Scholar]
- Sausen, D.G.; Basith, A.; Muqeemuddin, S. EBV and Lymphomagenesis. Cancers 2023, 15, 2133. [Google Scholar] [CrossRef] [PubMed]
- Cossman, J.; Messineo, C.; Bagg, A. Reed-Sternberg cell: Survival in a hostile sea. Lab. Investig. 1998, 78, 229–235. [Google Scholar] [PubMed]
- Marshall, N.A.; Culligan, D.J.; Tighe, J.; Johnston, P.W.; Barker, R.N.; Vickers, M.A. Relationships between Epstein-Barr virus latent membrane protein 1 and regulatory T cells in Hodgkin’s lymphoma. Exp. Hematol. 2007, 35, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Vignali, D.A.A.; Collison, L.W.; Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008, 8, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Yamaguchi, T.; Nomura, T.; Ono, M. Regulatory T cells and immune tolerance. Cell 2008, 133, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Rudensky, A.Y. Foxp3 in control of the regulatory T cell lineage. Nat. Immunol. 2007, 8, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Tai, X.; Hodes, R.J. CD28-CD80/86 and CD40-CD40L Interactions Promote Thymic Tolerance by Regulating Medullary Epithelial Cell and Thymocyte Development. Crit. Rev. Immunol. 2015, 35, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, N.; Sakaguchi, S. Transcriptional and epigenetic basis of Treg cell development and function: Its genetic anomalies or variations in autoimmune diseases. Cell Res. 2020, 30, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Povoleri, G.A.; Scottà, C.; Nova-Lamperti, E.A.; John, S.; Lombardi, G.; Afzali, B. Thymic versus induced regulatory T cells-who regulates the regulators? Front. Immunol. 2013, 4, 169. [Google Scholar] [CrossRef] [PubMed]
- Bluestone, J.A.; Abbas, A.K. Natural versus adaptive regulatory T cells. Nat. Rev. Immunol. 2003, 3, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Harris, F.; Berdugo, Y.A.; Tree, T. IL-2-based approaches to Treg enhancement. Clin. Exp. Immunol. 2023, 211, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Human regulatory T cells (Treg) and their response to cancer. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Tanchot, C.; Terme, M.; Pere, H.; Tran, T.; Benhamouda, N.; Strioga, M.; Banissi, C.; Galluzzi, L.; Kroemer, G.; Tartour, E. Tumor-infiltrating regulatory T cells: Phenotype, role, mechanism of expansion in situ and clinical significance. Cancer Microenviron. 2013, 6, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Furgiuele, S.; Descamps, G.; Lechien, J.R.; Dequanter, D.; Journe, F.; Saussez, S. Immunoscore Combining CD8, FoxP3, and CD68-Positive Cells Density and Distribution Predicts the Prognosis of Head and Neck Cancer Patients. Cells 2022, 11, 2050. [Google Scholar] [CrossRef] [PubMed]
- Tzankov, A.; Meier, C.; Hirschmann, P.; Went, P.; Pileri, S.A.; Dirnhofer, S. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin’s lymphoma. Haematologica 2008, 93, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ishii, T.; Inagaki, A.; Yano, H.; Komatsu, H.; Iida, S.; Inagaki, H.; Ueda, R. Specific recruitment of CC chemokine receptor 4-positive regulatory T cells in Hodgkin lymphoma fosters immune privilege. Cancer Res. 2006, 66, 5716–5722. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, B.; Kreher, S.; Anagnostopoulos, I.; Jöhrens, K.; Monteleone, G.; Jundt, F.; Stein, H.; Janz, M.; Dörken, B.; Mathas, S. Aberrant expression of the Th2 cytokine IL-21 in Hodgkin lymphoma cells regulates STAT3 signaling and attracts Treg cells via regulation of MIP-3alpha. Blood 2008, 112, 3339–3347. [Google Scholar] [CrossRef] [PubMed]
- Morales, O.; Mrizak, D.; François, V.; Mustapha, R.; Miroux, C.; Depil, S.; Decouvelaere, A.V.; Lionne-Huyghe, P.; Auriault, C.; de Launoit, Y.; et al. Epstein-Barr virus infection induces an increase of T regulatory type 1 cells in Hodgkin lymphoma patients. Br. J. Haematol. 2014, 166, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, A.; Glavina-Durdov, M.; Capkun, V.; Jakelic Pitesa, J.; Bozic Sakic, M. Classical Hodgkin Lymphoma with Positive Epstein-Barr Virus Status is Associated with More FOXP3 Regulatory T Cells. Med. Sci. Monit. 2016, 22, 2340–2346. [Google Scholar] [CrossRef] [PubMed]
- Wein, F.; Küppers, R. The role of T cells in the microenvironment of Hodgkin lymphoma. J. Leukoc. Biol. 2016, 99, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Glavina-Durdov, M.; Jakic-Razumovic, J.; Capkun, V.; Murray, P. Assessment of the prognostic impact of the Epstein-Barr virus-encoded latent membrane protein-1 expression in Hodgkin’s disease. Br. J. Cancer 2001, 84, 1227–1234. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Massini, G.; Siemer, D.; Hohaus, S. EBV in Hodgkin Lymphoma. Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009013. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.A.; Christie, L.E.; Munro, L.R.; Culligan, D.J.; Johnston, P.W.; Barker, R.N.; Vickers, M.A. Immunosuppressive regulatory T cells are abundant in the reactive lymphocytes of Hodgkin lymphoma. Blood 2004, 103, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Assis, M.C.; Campos, A.H.; Oliveira, J.S.; Soares, F.A.; Silva, J.M.; Silva, P.B.; Penna, A.D.; Souza, E.M.; Baiocchi, O.C. Increased expression of CD4+CD25+FOXP3+ regulatory T cells correlates with Epstein-Barr virus and has no impact on survival in patients with classical Hodgkin lymphoma in Brazil. Med. Oncol. 2012, 29, 3614–3619. [Google Scholar] [CrossRef]
- Alvaro, T.; Lejeune, M.; Salvadó, M.T.; Bosch, R.; García, J.F.; Jaén, J.; Banhamm, A.H.; Roncador, G.; Montalbán, C.; Piris, M.A. Outcome in Hodgkin’s lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin. Cancer Res. 2005, 11, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Baumforth, K.R.; Birgersdotter, A.; Reynolds, G.M.; Wei, W.; Kapatai, G.; Flavell, J.R.; Kalk, E.; Piper, K.; Lee, S.; Machado, L.; et al. Expression of the Epstein-Barr virus-encoded Epstein-Barr virus nuclear antigen 1 in Hodgkin’s lymphoma cells mediates Up-regulation of CCL20 and the migration of regulatory T cells. Am. J. Pathol. 2008, 173, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Meij, P.; Leen, A.; Rickinson, A.B.; Verkoeijen, S.; Vervoort, M.B.; Bloemena, E.; Middeldorp, J.M. Identification and prevalence of CD8+ T-cell responses directed against Epstein-Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. Int. J. Cancer 2002, 99, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ohga, S.; Nomura, A.; Takada, H.; Hara, T. Immunological aspects of Epstein-Barr virus infection. Crit. Rev. Oncol. Hematol. 2002, 44, 203–215. [Google Scholar] [CrossRef]
- Landais, E.; Saulquin, X.; Houssaint, E. The human T cell immune response to Epstein-Barr virus. Int. J. Dev. Biol. 2005, 49, 285–292. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz-Merino, L.; Lejeune, M.; Nogales Fernández, E.; Henao Carrasco, F.; Grueso López, A.; Illescas Vacas, A.; Pulla, M.P.; Callau, C.; Álvaro, T. Role of immune escape mechanisms in Hodgkin’s lymphoma development and progression: A whole new world with therapeutic implications. Clin. Dev. Immunol. 2012, 2012, 756353. [Google Scholar] [CrossRef] [PubMed]
- Kiyasu, J.; Aoki, R.; Tanaka, P.Y.; Pracchia, L.F.; Calore, E.E.; Perez, N.M.; Kimura, Y.; Niino, D.; Sugita, Y.; Takayanagi, R.; et al. FOXP3+ regulatory and TIA-1+ cytotoxic T lymphocytes in HIV-associated Hodgkin lymphoma. Pathol. Int. 2012, 62, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.R.; Shipp, M.A. Signaling pathways and immune evasion mechanisms in classical Hodgkin lymphoma. Blood 2017, 130, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Kosydar, S.; Ansell, S.M. The biology of classical Hodgkin lymphoma. Semin Hematol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Scarmozzino, F.; Angotzi, F.; Ruggeri, E.; Dei Tos, A.P.; Trentin, L.; Pizzi, M.; Visentin, A. Tackling the dysregulated immune-checkpoints in classical Hodgkin lymphoma: Bidirectional regulations between the microenvironment and Hodgkin/Reed-Sternberg cells. Front. Oncol. 2023, 13, 1203470. [Google Scholar] [CrossRef] [PubMed]
- Moy, R.H.; Younes, A. Immune Checkpoint Inhibition in Hodgkin Lymphoma. Hemasphere 2018, 2, e20. [Google Scholar] [CrossRef] [PubMed]

| Variable | Mean (SD) | Median (Q1–Q3) Min–Max |
|---|---|---|
| FOXP3 | 468 ± 428 | 327 (Q1–Q3) 153–707 min–max 17–2368 |
| CD8 | 555 ± 434 | 435 (Q1–Q3) 266–707 min–max 33–2899 |
| FOXP3/CD8 | 23.9 ± 51 | 8 (Q1–Q3) 2–24 min–max 0–431 |
| CD4 | 2037 ± 1141 | 1898 (Q1–Q3) 995–2962 min–max 4–5318 |
| FOXP3/CD4 | 404 ± 402 | 263 (Q1–Q3) 110–599 min–max 16–2355 |
| Variable | N (%) | |
|---|---|---|
| Gender | female | 86 (47) |
| male | 96 (53) | |
| Subtype | nodular sclerosis | 124 (68.1) |
| mixed cellularity | 44 (24.2) | |
| lymphocyte rich | 8 (4.4) | |
| lymphocyte depleted | 6 (3.3) | |
| Epstein–Barr virus | positive | 51 (28) |
| negative | 131 (72) | |
| Stage | I | 21 (13.8) |
| II | 70 (46.1) | |
| III | 37 (24.3) | |
| IV | 24 (15.8) | |
| B symptoms | no | 72 (46.7) |
| yes | 82 (53.2) | |
| Relapse | no | 96 (52.7) |
| yes | 28 (15.4) | |
| unknown | 58 (31.9) | |
| Survival status | alive | 116 (63.7) |
| dead | 36 (19.8) | |
| unknown | 30 (16.6) | |
| Classic Hodgkin Lymphoma | ||||
|---|---|---|---|---|
| Variable | EBV-Positive | EBV-Negative | p * | |
| N(%) or Median (Q1–Q3) Min–Max | ||||
| Gender | female | 22 (43) | 64 (49) | 0.631 * |
| male | 29 (57) | 67 (51) | ||
| Age | years | 54.5 (37–68; 6–87) | 31 (21–46; 5–86) | <0.001 ** |
| Subtype | NS | 20 (39.3) | 104 (80) | <0.001 * |
| MC | 27 (52.9) | 16 (12.3) | ||
| LR + LD | 4 (7.8) | 10 (10.7) | ||
| B symptoms | no | 21 (55) | 50 (43) | 0.282 * |
| yes | 17 (44) | 65 (57) | ||
| Stage | I | 9 (24.3) | 12 (10.4) | 0.089 * |
| II | 13 (35.1) | 57 (49.6) | ||
| III | 11(29.7) | 26 (22.6) | ||
| IV | 4 (10.8) | 20 (17.4) | ||
| Survival status | alive | 26 (51) | 89 (68.5) | 0.038 * |
| dead | 16 (31.4) | 20 (15.4) | ||
| unknown | 9 (17.6) | 21 (16.2) | ||
| FOXP3 | 560(183–902; 22–1413) | 292(150–536; 17–2368) | 0.049 ** | |
| CD8 | 620(386–893; 33–2899) | 363(246–603; 55–1509) | 0.001 ** | |
| FOXP3/CD8 | 12(2.7–37; 0–280) | 7(2–20; 0–431) | 0.193 ** | |
| CD4 | 2009(968–2919; 587–4221) | 1802(995–3053; 4–5318) | 0.875 ** | |
| FOXP3/CD4 | 348.5(115–772; 16–1347) | 223(109–480; 16–2355) | 0.1 ** | |
| Classic Hodgkin Lymphoma | ||||||
|---|---|---|---|---|---|---|
| EBV-Positive | EBV-Negative | |||||
| Phenotype | NS | MC | NS | MC | χ2 | p * |
| Median (Range) | ||||||
| CD8 | 700 (160–2899) | 500 (33–2600) | 336 (55–1509) | 558 (142–1433) | 19.1 | 0.001 |
| CD4 | 1893 (587–4221) | 1994 (859–4100) | 1727 (4–5318) | 1402 (779–4946) | 0.551 | 0.908 |
| FOXP3 | 329 (22–412) | 573 (39–1324) | 316 (19–368) | 163 (17–982) | 8.6 | 0.035 |
| FOXP3/CD8 | 13 (0–218) | 10 (0–117) | 7 (0–431) | 13 (0–57) | 1.8 | 0.607 |
| FOXP3/CD4 | 270 (16–1347) | 558 (35–1144) | 273 (18–2355) | 143 (16–878) | 8.2 | 0.041 |
| Relapse | ||||
|---|---|---|---|---|
| Variable | No (N = 96) | Yes (N = 28) | p | OR (95% CI); p *** |
| N(%) or Median (Q1–Q3) Min–Max | ||||
| Gender | 0.846 * | |||
| female | 46 (48) | 14 (50) | ||
| male | 50 (52) | 14 (50) | ||
| Age | 32 (22–50; 10–83) | 33 (20–49; 13–86) | 0.894 ** | |
| Subtype | 0.416 * | |||
| NS | 67 (69.8) | 23 (82.1) | ||
| MC | 25 (26) | 4 (14.3) | ||
| LR + LD | 4 (4.2) | 1 (3.6) | ||
| EBV status- | 0.978 * | |||
| positive | 24 (25) | 7 (25) | ||
| negative | 71 (75) | 21 (75) | ||
| Stage | 0.134 * | |||
| I | 15 (16) | 0 | ||
| II | 44 (46.8) | 15 (53.6) | ||
| III | 18 (19.1)1 | 8 (28.6) | ||
| IV | 7 (18.1) | 5 (17.9) | ||
| B symptoms | 0.016 * | 3.4 (95%CI: 1.3–8.8); p = 0.011 *** | ||
| no | 50 (53.2) | 7 (25) | ||
| yes | 44 (46.8) | 21 (75) | ||
| Survival status | <0.001 * | 10.3 (95% CI: 3.2–32); p < 0.001 *** | ||
| alive | 84 (87.5) | 15 (53.6) | ||
| dead | 6 (6.2) | 11 (39.3) | ||
| unknown | 6 | 2 | ||
| FOXP3 | 299 (124–627; 17–1960) | 263 (82–379; 40–1560) | 0.268 ** | |
| CD8 | 362 (246–542; 33–2899) | 342 (245–531; 167–1509) | 0.790 ** | |
| FOXP3/CD8 | 12 (5–27; 0–431) | 10 (14–23; 1–92) | 0.742 ** | |
| CD4 | 1385 (946–2381; 4–4946) | 1006 (871–1399; 377–3691) | 0.94 ** | |
| FOXP3/CD4 | 202 (84.494; 16–1514) | 188 (72–331; 19–1564) | 0.53 ** | |
| Variable | HR(95% CI); p * |
|---|---|
| CD4 | 4.8 (1.7–13); 0.002 |
| FOXP3 | 1.5 (0.7–3.5); 0.312 |
| Variable | B Symptoms | ||
|---|---|---|---|
| No (N = 72) | Yes (N = 82) | p | |
| N(%) or Median (Q1–Q3) Min–Max | |||
| Gender | 0.258 * | ||
| female | 40 (56) | 37 (45) | |
| male | 32 (44) | 45 (55) | |
| Age (years) | 35 (24–51; 5–78) | 35 (22–50; 13–86) | 0.963 ** |
| Subtype | 0.037 * | ||
| NS | 45 (62.5) | 65 (79,3) | |
| MC | 23 (31.9) | 12 (14.6) | |
| Epstein–Barr virus | 0.282 * | ||
| positive | 21 (30) | 17 (21) | |
| negative | 50 (70) | 65 (79) | |
| Stage | <0.001 * | ||
| I | 18 (25.4) | 3 (3.7) | |
| II | 36 (25.7) | 35 (42.7) | |
| III | 10 (14.1) | 27 (32.9) | |
| IV | 7 (9.9) | 17 (20.7) | |
| Survival status | 0.043 * | ||
| alive | 57 (88) | 49 (72) | |
| dead | 8 (12) | 19 (28) | |
| FOXP3 | 310 (138–795; 17–1960) | 327 (137–548; 22–1560) | 0.747 ** |
| CD8 | 459 (277–687; 55–2899) | 346 (243–593; 33–1722) | 0.066 ** |
| FOXP3/CD8 | 12.5 (3–28; 0–431) | 8 (2–22; 0–126) | 0.246 ** |
| CD4 | 1899 (1099–3048;5 81–5318) | 1346 (867–2510; 4–4241) | 0.017 ** |
| FOXP3/CD4 | 209 (101–599; 16–1514) | 274 (108–528; 16–1564) | 0.984 ** |
| Progression Free Survival (Months) | |||
|---|---|---|---|
| Variable | Mean (SE; 95%CI) | LR; p | HR (95% CI); p * |
| Gender | 0; 0.998 | ||
| female | 237 (11; 1; 215–260) | ||
| male | 232 (12; 209–255) | ||
| Age | 0.115; 0.734 | ||
| ≤35 years | 236 (11; 213–258) | ||
| >35 years | 230 (11; 208–252) | ||
| Subtype | 1.8; 0.182 | ||
| Nodular sclerosis | 223 (10; 202–243) | ||
| Mixed cellularity | 244(13; 218–269) | ||
| Epstein–Barr virus- | 0; 0.994 | ||
| positive | 227 (15; 197–257) | ||
| negative | 236 (9; 217–256) | ||
| Stage | 0.267; 0.875 | ||
| I ** | / | ||
| II | 217 (14; 190–245) | ||
| III | 198 (20; 158–238) | ||
| IV | 175 (17; 141–209) | ||
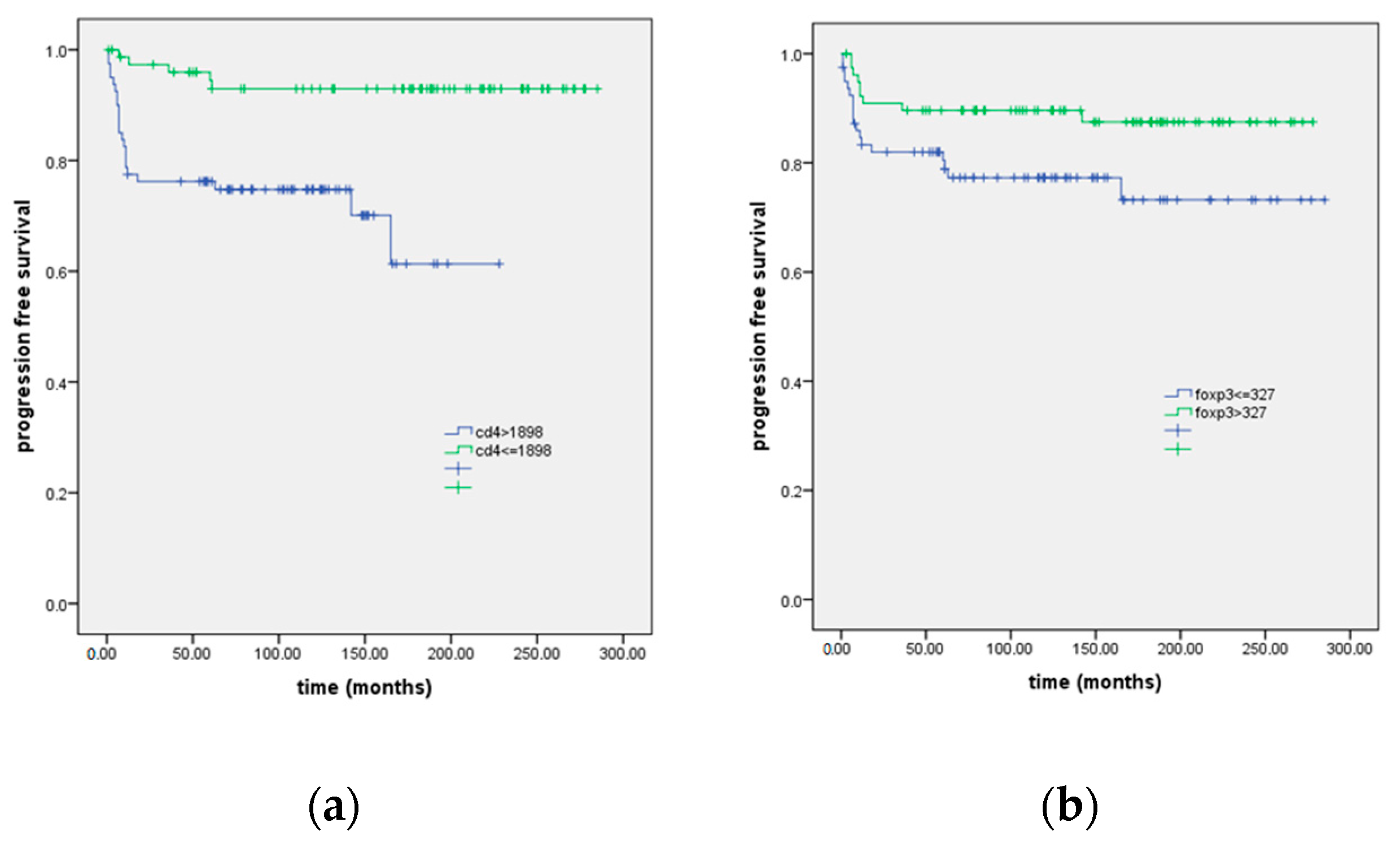
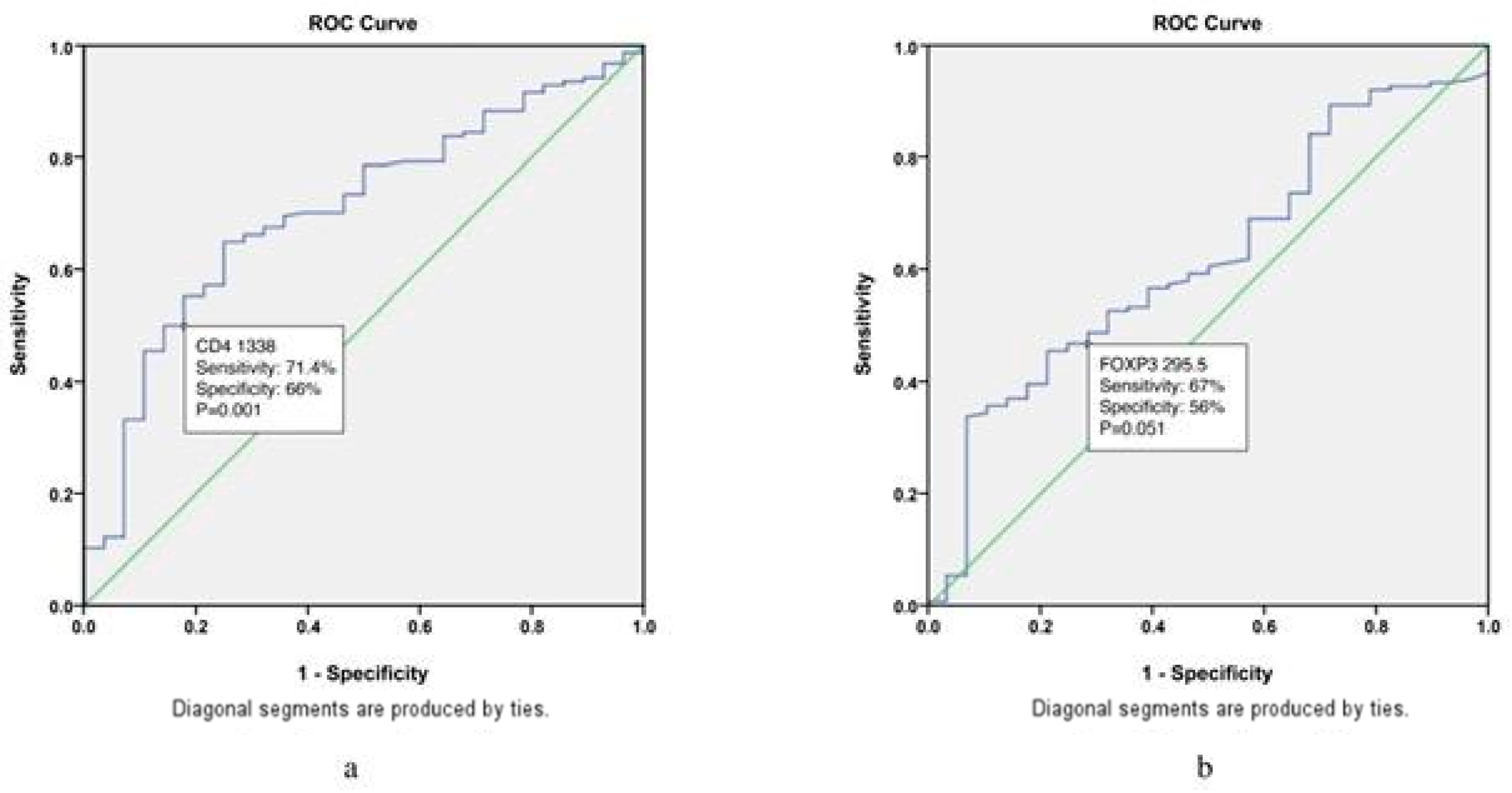
| FOXP3 | 4.3; 0.039 | 2.3 (1–5); 0.046 | |
| ≤327 | 219 (13; 193–246) | ||
| >327 † | 247 (9; 229–246) | ||
| CD8 | 1.5; 0.214 | ||
| ≤435 | 217 (12; 194–240) | ||
| >435 | 248 (11; 227–269) | ||
| FOXP3/CD8 | 0.101; 0.750 | ||
| ≤8 | 235 (12; 211–258) | ||
| >8 | 228 (11; 27–250) | ||
| CD4 | 14.1; p < 0.001 | 5.5 (2–14.7); 0.001 | |
| ≤1898 | 163 (12; 140–187) | ||
| >1898 † | 267 (7; 253–272) | ||
| FOXP3/CD4 | 2.6; 0.104 | ||
| ≤263 | 223 (13; 196–249) | ||
| >263 | 244 (10; 224–264) |
| Overall Survival (Months) | |||||
|---|---|---|---|---|---|
| Relapse | N | Mean (SE) | 95% CI | LR; p * | DIED |
| no | 96 | 241 (6.4) | 22.8–253 | 26.6; <0.001 | 17 |
| yes | 28 | 119 (16) | 87–151 | 11 | |
| unknown | 35 | 198 (16.5) | 165–230 | 18 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlović, A.; Miljak, A.; Brzica, K.; Glavina Durdov, M. The Abundance of FOXP3, FOXP3/CD4 and CD8 Cells in the Microenvironment of Nodular Sclerosis and Mixed Cellularity Subtypes Is Associated with the Epstein–Barr Virus Status of Classic Hodgkin Lymphoma. Biomedicines 2024, 12, 1680. https://doi.org/10.3390/biomedicines12081680
Pavlović A, Miljak A, Brzica K, Glavina Durdov M. The Abundance of FOXP3, FOXP3/CD4 and CD8 Cells in the Microenvironment of Nodular Sclerosis and Mixed Cellularity Subtypes Is Associated with the Epstein–Barr Virus Status of Classic Hodgkin Lymphoma. Biomedicines. 2024; 12(8):1680. https://doi.org/10.3390/biomedicines12081680
Chicago/Turabian StylePavlović, Antonia, Antonija Miljak, Katarina Brzica, and Merica Glavina Durdov. 2024. "The Abundance of FOXP3, FOXP3/CD4 and CD8 Cells in the Microenvironment of Nodular Sclerosis and Mixed Cellularity Subtypes Is Associated with the Epstein–Barr Virus Status of Classic Hodgkin Lymphoma" Biomedicines 12, no. 8: 1680. https://doi.org/10.3390/biomedicines12081680
APA StylePavlović, A., Miljak, A., Brzica, K., & Glavina Durdov, M. (2024). The Abundance of FOXP3, FOXP3/CD4 and CD8 Cells in the Microenvironment of Nodular Sclerosis and Mixed Cellularity Subtypes Is Associated with the Epstein–Barr Virus Status of Classic Hodgkin Lymphoma. Biomedicines, 12(8), 1680. https://doi.org/10.3390/biomedicines12081680






