SCFFBXW11 Complex Targets Interleukin-17 Receptor A for Ubiquitin–Proteasome-Mediated Degradation
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Inhibition of Proteasome and Cullin-Ring Ligase (CRL) Complex Increases IL-17RA Protein Stability
3.2. F-Box and WD Repeat Domain-Containing (FBXW) Proteins Are Predicted to Recognize IL-17RA
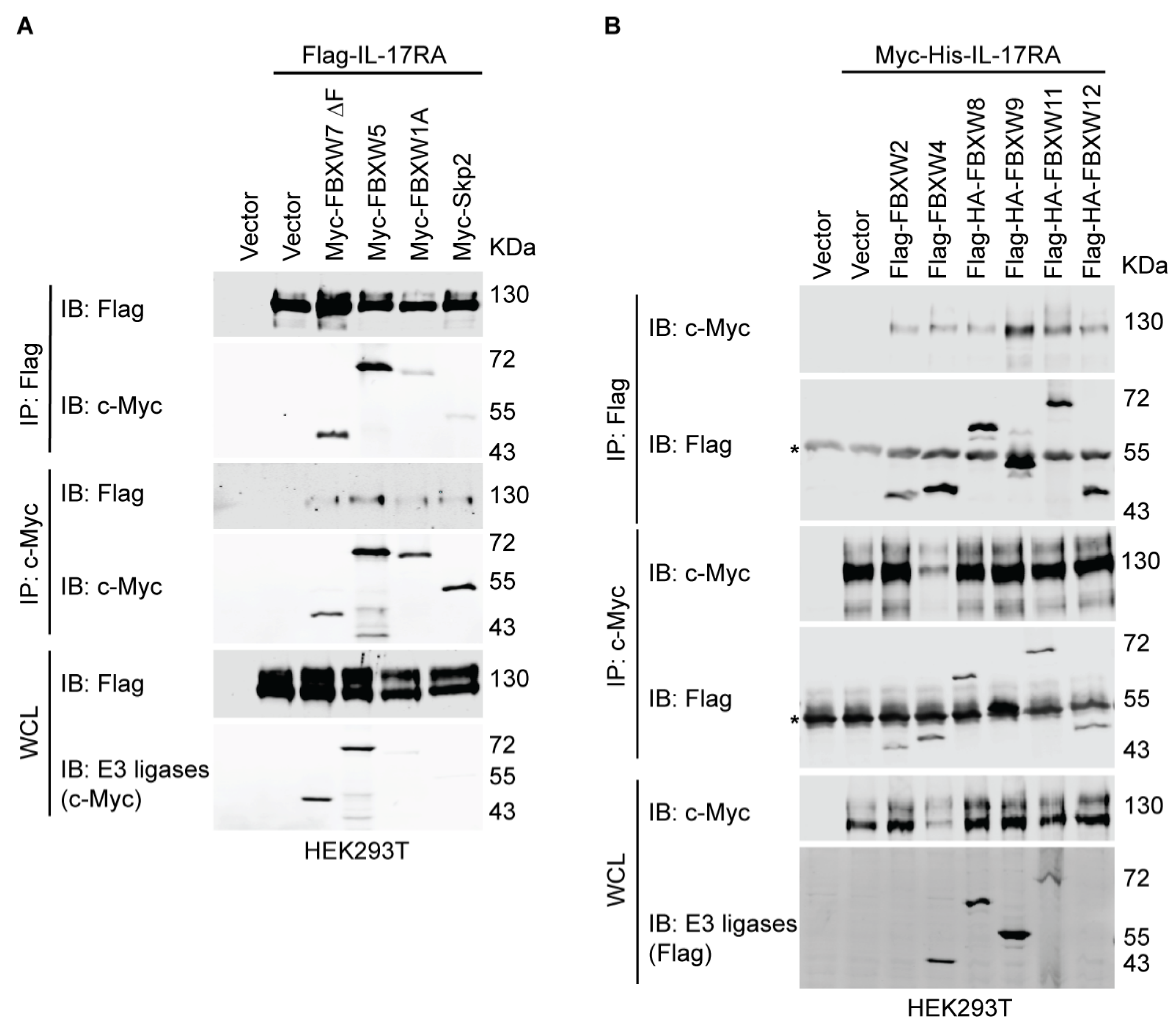
3.3. Several SCF E3 Ligases Are Associated with IL-17RA
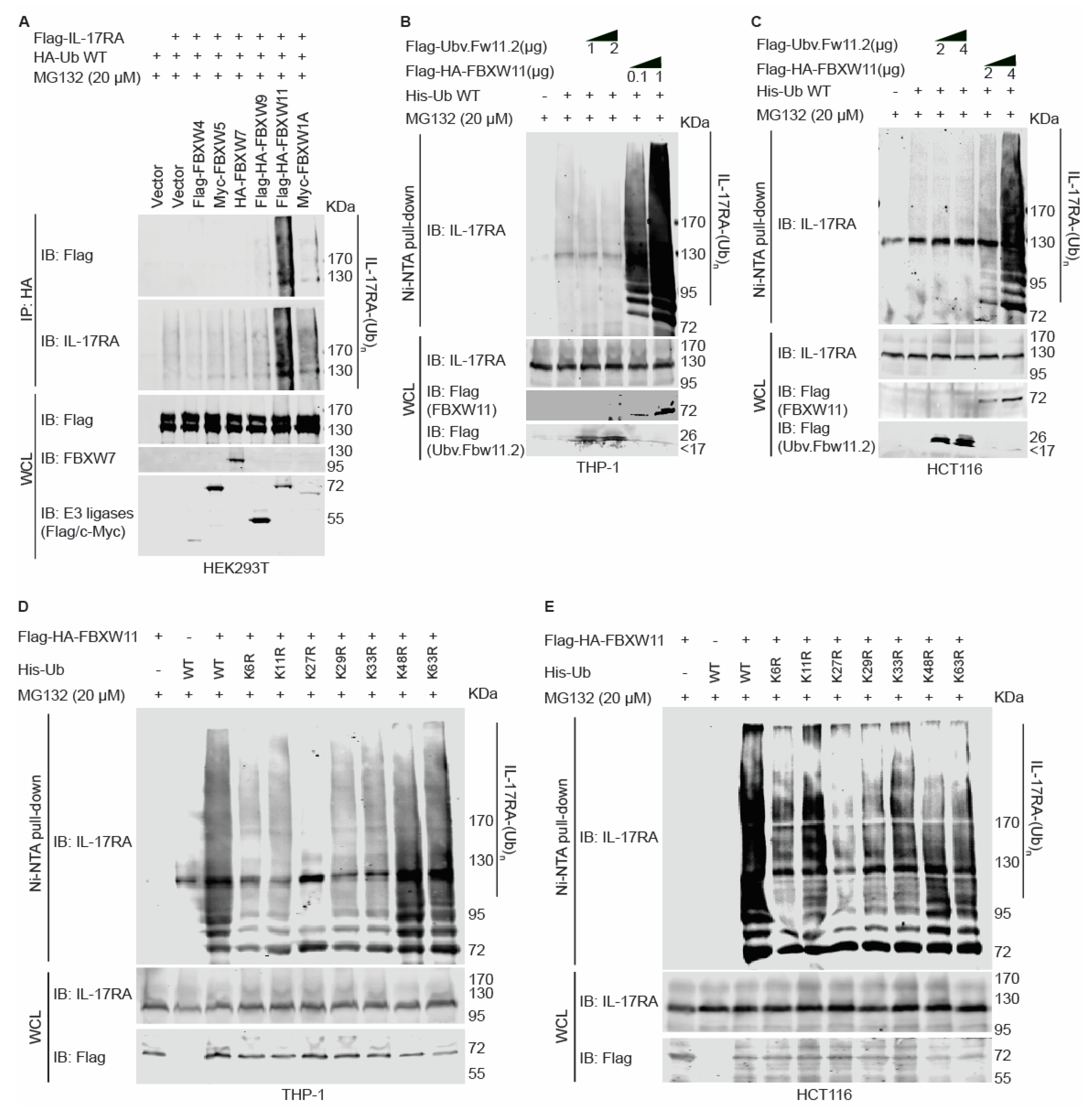
3.4. FBXW11 Ubiquitylates IL-17RA via K27-Linked Polyubiquitin in a Dose-Dependent Way
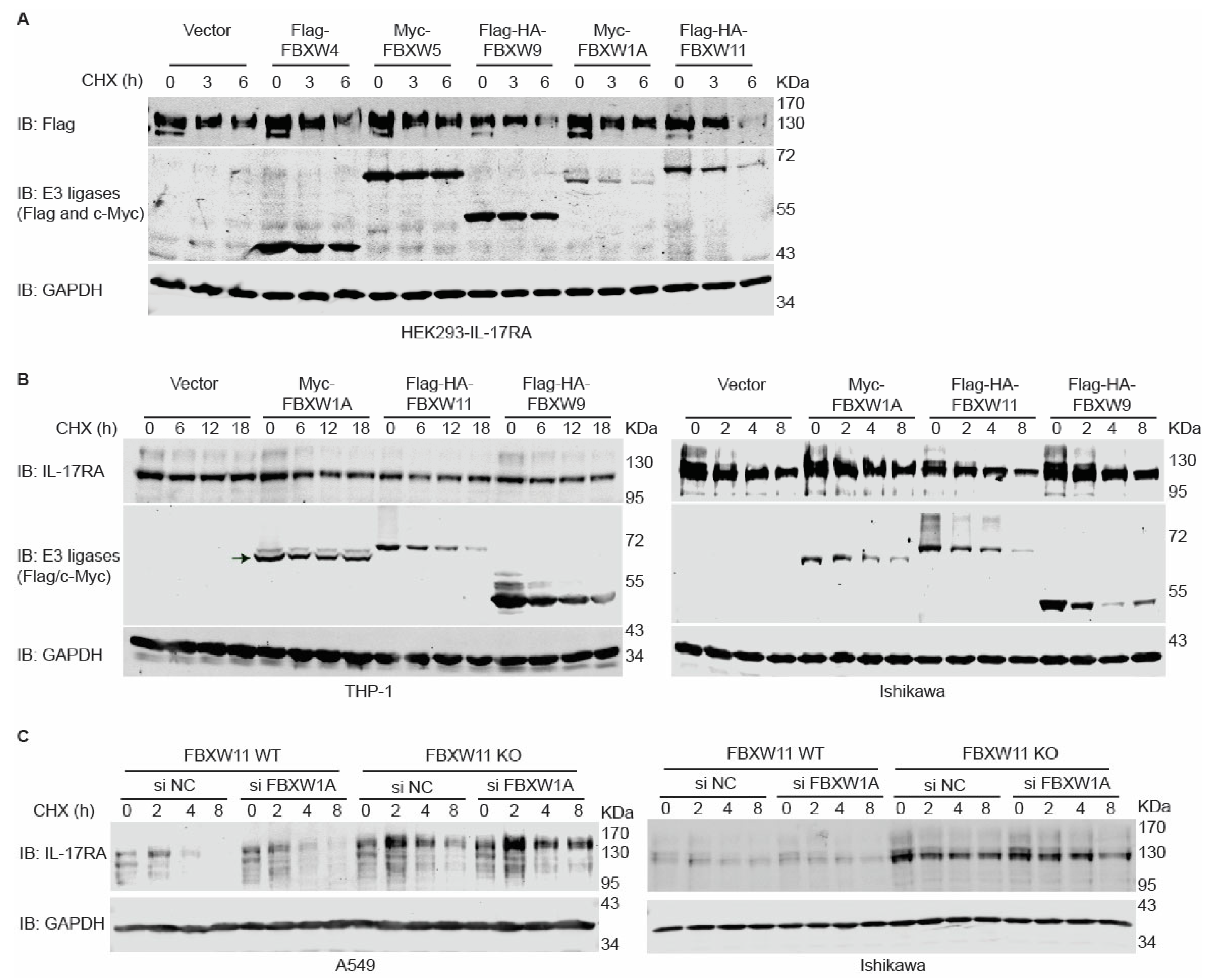
3.5. Overexpression of FBXW11 Accelerates Degradation of IL-17RA, While Knock-Out of FBXW11 Increases Protein Stability of IL-17RA
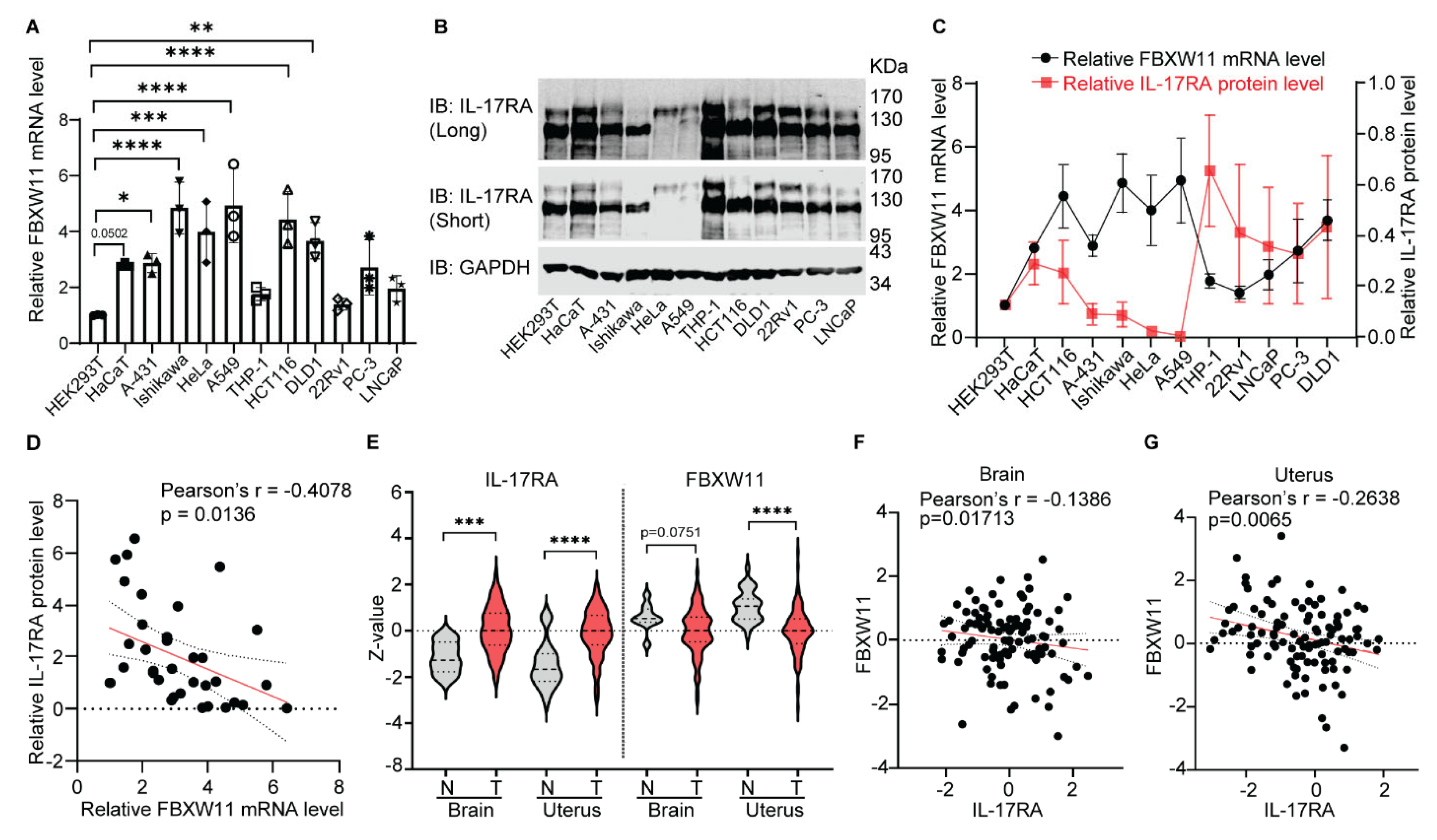
3.6. Expression Levels of FBXW11 and IL-17RA Are Inversely Correlated
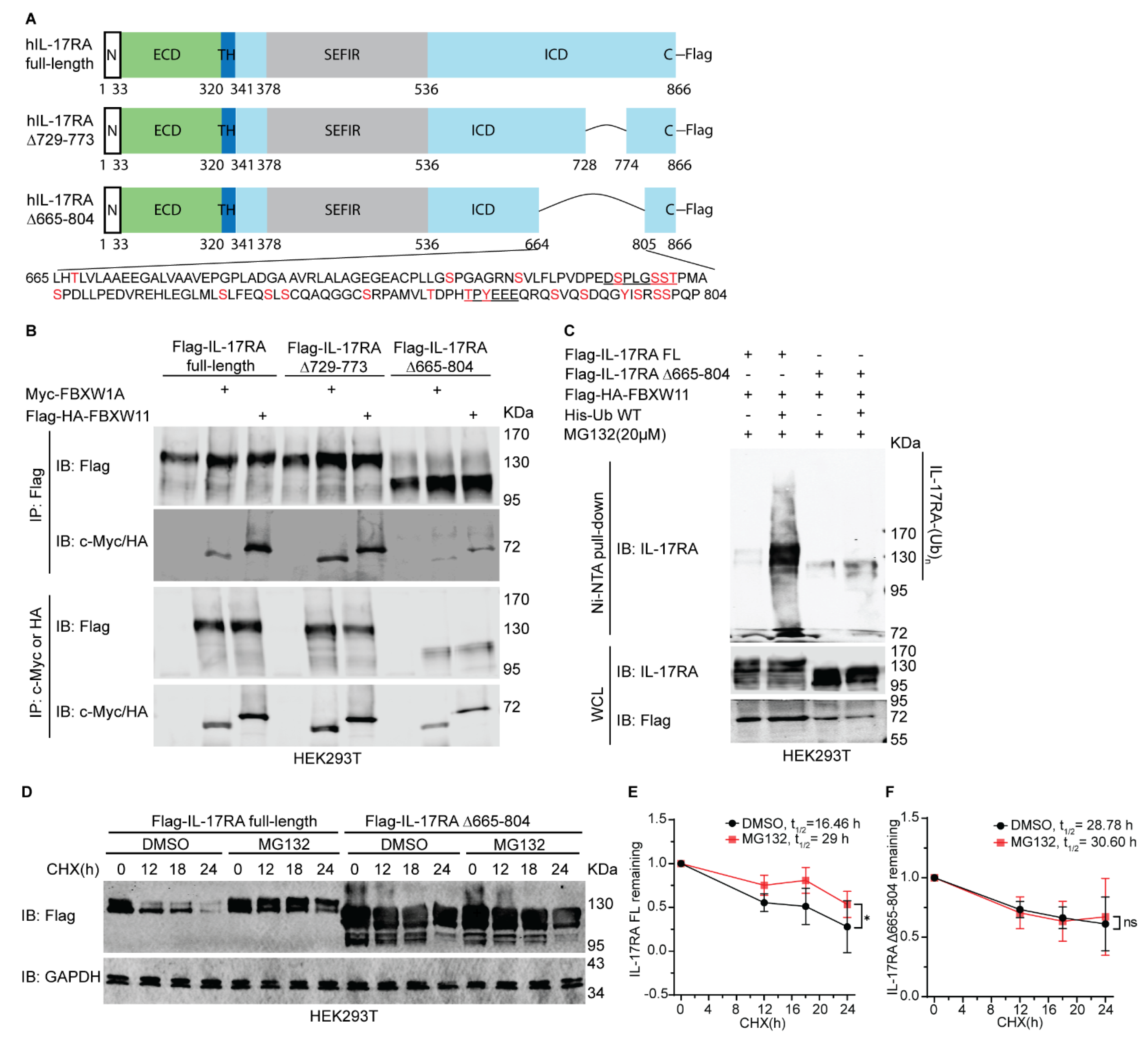
3.7. 665-804 Domain of IL-17RA Determines Its Protein Stability and Ubiquitylation Mediated by FBXW11
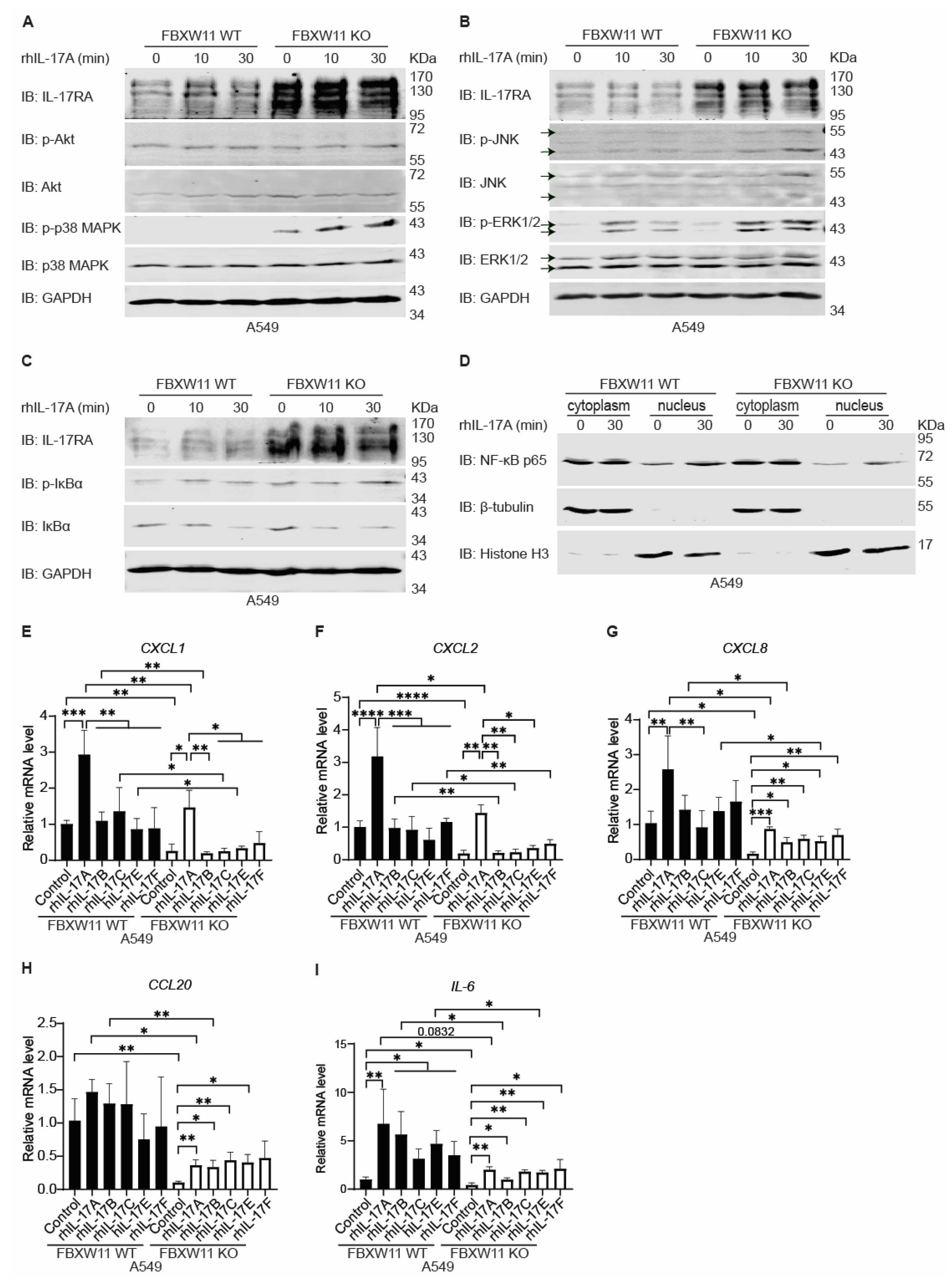
3.8. Knock-Out of FBXW11 Suppresses Expression of IL-17-Downstream Genes through Inhibiting Nuclear Entry of NF-κB p65
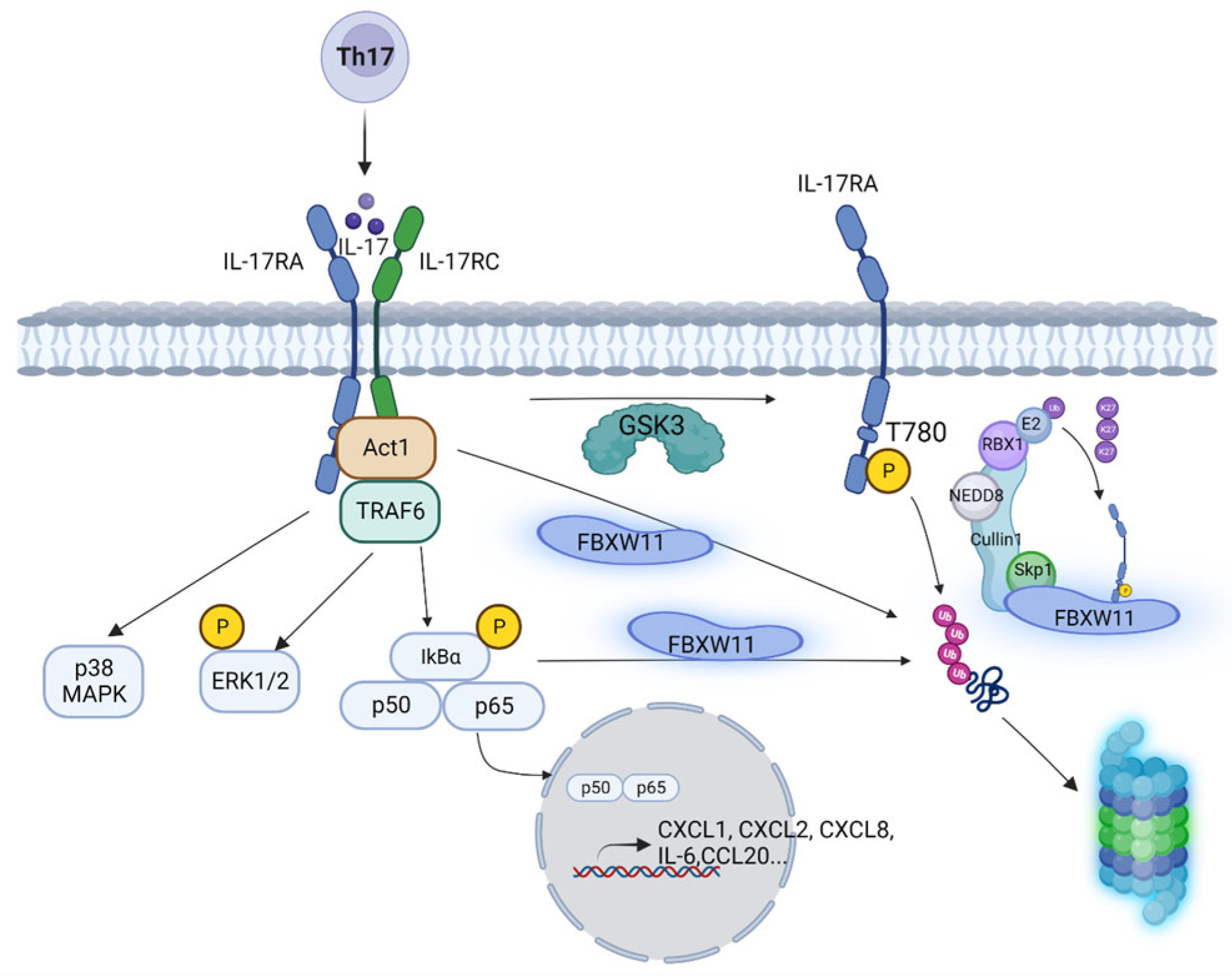
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monin, L.; Gaffen, S.L. Interleukin 17 Family Cytokines: Signaling Mechanisms, Biological Activities, and Therapeutic Implications. Cold Spring Harb. Perspect. Biol. 2018, 10, a028522. [Google Scholar] [CrossRef] [PubMed]
- Gaffen, S.L. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 2009, 9, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lee, H.-Y.; Zhao, X.; Han, J.; Su, Y.; Sun, Q.; Shao, J.; Ge, J.; Zhao, Y.; Bai, X.; et al. Interleukin-17D regulates group 3 innate lymphoid cell function through its receptor CD93. Immunity 2021, 54, 673–686.e4. [Google Scholar] [CrossRef]
- Wright, J.F.; Bennett, F.; Li, B.; Brooks, J.; Luxenberg, D.P.; Whitters, M.J.; Tomkinson, K.N.; Fitz, L.J.; Wolfman, N.M.; Collins, M.; et al. The Human IL-17F/IL-17A Heterodimeric Cytokine Signals through the IL-17RA/IL-17RC Receptor Complex. J. Immunol. 2008, 181, 2799–2805. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Huang, J.; Zhao, X.; Lu, H.; Wang, W.; Yang, X.O.; Shi, Y.; Wang, X.; Lai, Y.; Dong, C. Interleukin-17 receptor D constitutes an alternative receptor for interleukin-17A important in psoriasis-like skin inflammation. Sci. Immunol. 2019, 4, eaau9657. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ho, W.-H.; Maruoka, M.; Corpuz, R.T.; Baldwin, D.T.; Foster, J.S.; Goddard, A.D.; Yansura, D.G.; Vandlen, R.L.; Wood, W.I.; et al. IL-17E, a Novel Proinflammatory Ligand for the IL-17 Receptor Homolog IL-17Rh1. J. Biol. Chem. 2001, 276, 1660–1664. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.C.; Caveney, N.A.; Yen, M.; Pollmann, C.; Xiang, X.; Jude, K.M.; Hafer, M.; Tsutsumi, N.; Piehler, J.; Garcia, K.C. Organizing structural principles of the IL-17 ligand–receptor axis. Nature 2022, 609, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Reynolds, J.M.; Pappu, B.P.; Chen, G.; Martinez, G.J.; Dong, C. Interleukin-17C Promotes Th17 Cell Responses and Autoimmune Disease via Interleukin-17 Receptor E. Immunity 2011, 35, 611–621. [Google Scholar] [CrossRef]
- Goepfert, A.; Lehmann, S.; Blank, J.; Kolbinger, F.; Rondeau, J.-M. Structural Analysis Reveals that the Cytokine IL-17F Forms a Homodimeric Complex with Receptor IL-17RC to Drive IL-17RA-Independent Signaling. Immunity 2020, 52, 499–512.e5. [Google Scholar] [CrossRef]
- Onishi, R.M.; Park, S.J.; Hanel, W.; Ho, A.W.; Maitra, A.; Gaffen, S.L. SEF/IL-17R (SEFIR) is not enough: An extended SEFIR domain is required for il-17RA-mediated signal transduction. J. Biol. Chem. 2010, 285, 32751–32759. [Google Scholar] [CrossRef]
- Maitra, A.; Shen, F.; Hanel, W.; Mossman, K.; Tocker, J.; Swart, D.; Gaffen, S.L. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc. Natl. Acad. Sci. USA 2007, 104, 7506–7511. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, B.; Kumar, M.; Gibson, T.J.; Uyar, B.; Dosztányi, Z. Degrons in cancer. Sci. Signal. 2017, 10, eaak9982. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Sarikas, A.; Hartmann, T.; Pan, Z.-Q. The cullin protein family. Genome Biol. 2011, 12, 220. [Google Scholar] [CrossRef]
- Baek, K.; Krist, D.T.; Prabu, J.R.; Hill, S.; Klügel, M.; Neumaier, L.M.; von Gronau, S.; Kleiger, G.; Schulman, B.A. NEDD8 nucleates a multivalent cullin–RING–UBE2D ubiquitin ligation assembly. Nature 2020, 578, 461–466. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, J. K27-linked noncanonic ubiquitination in immune regulation. J. Leukoc. Biol. 2022, 111, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.V.; Ahmed, M.; Vallejo, A.N.; Ma, A.; Gaffen, S.L. The Deubiquitinase A20 Mediates Feedback Inhibition of Interleukin-17 Receptor Signaling. Sci. Signal. 2013, 6, ra44. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Zhu, S.; Lin, Y.; Liu, Y.; Liu, Y.; Chen, Z.; Shi, Y.; Qian, Y. Persistent stimulation with interleukin-17 desensitizes cells through SCFβ-TrCP-mediated degradation of Act1. Sci. Signal. 2011, 4, ra73. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Gao, H.; Zhu, S.; Shi, P.; Zhang, Y.; Liu, Y.; Jallal, B.; Yao, Y.; Shi, Y.; Qian, Y. TRAF6-dependent Act1 phosphorylation by the IκB kinase-related kinases suppresses interleukin-17-induced NF-κB activation. Mol. Cell. Biol. 2012, 32, 3925–3937. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Q.; Chen, C.; Ge, D.; Qu, Y.; Chen, R.; Fan, Y.M.; Li, N.; Tang, W.W.; Zhang, W.; et al. Hyperinsulinemia enhances interleukin-17-induced inflammation to promote prostate cancer development in obese mice through inhibiting glycogen synthase kinase 3-mediated phosphorylation and degradation of interleukin-17 receptor. Oncotarget 2016, 7, 13651–13666. [Google Scholar] [CrossRef] [PubMed]
- Winston, J.T.; Strack, P.; Beer-Romero, P.; Chu, C.Y.; Elledge, S.J.; Harper, J.W. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999, 13, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Kainulainen, M.; Lau, S.; Samuel, C.E.; Hornung, V.; Weber, F. NSs Virulence Factor of Rift Valley Fever Virus Engages the F-Box Proteins FBXW11 and β-TRCP1 To Degrade the Antiviral Protein Kinase PKR. J. Virol. 2016, 90, 6140–6147. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, Q.; Wang, J.; Li, J.; Huang, C.; Zhang, Y.; Wu, X.; Lu, H.; Zhou, X. Ubiquitin ligase TRIM71 suppresses ovarian tumorigenesis by degrading mutant p53. Cell Death Dis. 2019, 10, 737. [Google Scholar] [CrossRef]
- Qian, Y.; Liu, C.; Hartupee, J.; Altuntas, C.Z.; Gulen, M.F.; Jane-Wit, D.; Xiao, J.; Lu, Y.; Giltiay, N.; Liu, J.; et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 2007, 8, 247–256. [Google Scholar] [CrossRef]
- You, Z.; Dong, Y.; Kong, X.; Zhang, Y.; Vessella, R.L.; Melamed, J. Differential Expression of IL-17RC Isoforms in Androgen-Dependent and Androgen-Independent Prostate Cancers. Neoplasia 2007, 9, 464–470. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- van Kley, H.; Hale, S.M. Assay for protein by dye binding. Anal. Biochem. 1977, 81, 485–487. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single–cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Mészáros, B.; Sámano-Sánchez, H.; Zeke, A.; Dobson, L.; Lazar, T.; Örd, M.; Nagpal, A.; et al. The Eukaryotic Linear Motif resource: 2022 release. Nucleic Acids Res. 2022, 50, D497–D508. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Kawaler, E.A.; Zhou, D.C.; Gritsenko, M.A.; Huang, C.; Blumenberg, L.; Karpova, A.; Petyuk, V.A.; Savage, S.R.; Satpathy, S.; et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell 2020, 180, 729–748.e26. [Google Scholar] [CrossRef]
- Wang, L.B.; Karpova, A.; Gritsenko, M.A.; Marina, A.; Kyle, J.E.; Jennifer, E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 2021, 39, 509–528.e20. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Savage, S.R.; Liao, Y.; Dou, Y.; Shi, Z.; Yi, X.; Jiang, W.; Lei, J.T.; Zhang, B. LinkedOmicsKB: A web portal to explore pan-cancer molecular and phenotype associations in AACR. Cancer Res. 2023, 83, 6575. [Google Scholar] [CrossRef]
- Baek, K.; Scott, D.C.; A Schulman, B. NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr. Opin. Struct. Biol. 2021, 67, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Yada, M.; Hatakeyama, S.; Kamura, T.; Nishiyama, M.; Tsunematsu, R.; Imaki, H.; Ishida, N.; Okumura, F.; Nakayama, K.; Nakayama, K.I. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004, 23, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; van der Linden, W.A.; Overkleeft, H.S. Proteasome Inhibitors: An Expanding Army Attacking a Unique Target. Chem. Biol. 2012, 19, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Crawford, L.J.; Walker, B.; Ovaa, H.; Chauhan, D.; Anderson, K.C.; Morris, T.C.; Irvine, A.E. Comparative Selectivity and Specificity of the Proteasome Inhibitors BzLLLCOCHO, PS-341, and MG-132. Cancer Res. 2006, 66, 6379–6386. [Google Scholar] [CrossRef]
- Yim, W.W.-Y.; Mizushima, N. Lysosome biology in autophagy. Cell Discov. 2020, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short Linear Motifs: Ubiquitous and Functionally Diverse Protein Interaction Modules Directing Cell Regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, T.; Pagano, M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004, 5, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Lan, R.; Jin, B.; Liu, Y.-Z.; Zhang, K.; Niu, T.; You, Z. Genome and transcriptome profiling of FBXW family in human prostate cancer. Am. J. Clin. Exp. Urol. 2020, 8, 116–128. [Google Scholar] [PubMed]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Draberova, H.; Janusova, S.; Knizkova, D.; Semberova, T.; Pribikova, M.; Ujevic, A.; Harant, K.; Knapkova, S.; Hrdinka, M.; Fanfani, V.; et al. Systematic analysis of the IL -17 receptor signalosome reveals a robust regulatory feedback loop. EMBO J. 2020, 39, e104202. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Avvakumov, G.; Tong, J.; Fan, Y.; Zhao, Y.; Alberts, P.; Persaud, A.; Walker, J.R.; Neculai, A.-M.; Neculai, D.; et al. A Strategy for Modulation of Enzymes in the Ubiquitin System. Science 2013, 339, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, M.; Orlicky, S.; Sartori, M.A.; Tang, X.; Marcon, E.; Kurinov, I.; Greenblatt, J.F.; Tyers, M.; Moffat, J.; Sicheri, F.; et al. Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1–F-box interface. Proc. Natl. Acad. Sci. USA 2016, 113, 3527–3532. [Google Scholar] [CrossRef]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Pagano, M. Ubiquitin ligases in cancer: Functions and clinical potentials. Cell Chem. Biol. 2021, 28, 918–933. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bechara, R.; Zhao, J.; McGeachy, M.J.; Gaffen, S.L. IL-17 receptor–based signaling and implications for disease. Nat. Immunol. 2019, 20, 1594–1602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, X.; Herjan, T.; Li, X. The role of interleukin-17 in tumor development and progression. J. Exp. Med. 2020, 217, e20190297. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, R.; Wang, B.; Lian, J.; Yao, Y.; Sun, H.; Zhang, C.; Fang, L.; Guan, X.; Shi, J.; et al. Blocking IL-17A enhances tumor response to anti-PD-1 immunotherapy in microsatellite stable colorectal cancer. J. Immunother. Cancer 2021, 9, e001895. [Google Scholar] [CrossRef] [PubMed]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef]
- Levin, S.D. IL-17 Receptor Signaling: Ubiquitin Gets In On the Act. Sci. Signal. 2009, 2, pe64. [Google Scholar] [CrossRef]
- Xu, W.; Huang, Y. Regulation of Inflammatory Cell Death by Phosphorylation. Front. Immunol. 2022, 13, 851169. [Google Scholar] [CrossRef] [PubMed]
- Battaglioni, S.; Benjamin, D.; Wälchli, M.; Maier, T.; Hall, M.N. mTOR substrate phosphorylation in growth control. Cell 2022, 185, 1814–1836. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Cheng, L.; Ren, Y.; Li, Z.; Li, Y.; Li, X.; Li, H.; Fu, X.-Y.; Chang, Z. Interleukin-17F signaling requires ubiquitination of interleukin-17 receptor via TRAF6. Cell. Signal. 2007, 19, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Akhoondi, S.; Sun, D.; von der Lehr, N.; Apostolidou, S.; Klotz, K.; Maljukova, A.; Cepeda, D.; Fiegl, H.; Dofou, D.; Marth, C.; et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007, 67, 9006–9012. [Google Scholar] [CrossRef] [PubMed]
- Nateri, A.S.; Riera-Sans, L.; Costa, C.D.; Behrens, A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 2004, 303, 1374–1378. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Li, G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012, 31, 75–87. [Google Scholar] [CrossRef]
- Alan, W.L.; Lau, A.W.; Fukushima, H.; Wei, W. The Fbw7 and BetaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front. Biosci. 2012, 17, 2197. [Google Scholar]
- Knizkova, D.; Pribikova, M.; Draberova, H.; Semberova, T.; Trivic, T.; Synackova, A.; Ujevic, A.; Stefanovic, J.; Drobek, A.; Huranova, M.; et al. CMTM4 is a subunit of the IL-17 receptor and mediates autoimmune pathology. Nat. Immunol. 2022, 23, 1644–1652. [Google Scholar] [CrossRef]
- Kim, T.Y.; Siesser, P.F.; Rossman, K.L.; Goldfarb, D.; Mackinnon, K.; Yan, F.; Yi, X.H.; MacCoss, M.J.; Moon, R.T.; Der, C.J.; et al. Substrate trapping proteomics reveals targets of the βTrCP2/FBXW11 ubiquitin ligase. Mol. Cell. Biol. 2015, 35, 167–181. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1. [Google Scholar] [CrossRef] [PubMed]
- Kathania, M.; Khare, P.; Zeng, M.; Cantarel, B.; Zhang, H.; Ueno, H.; Venuprasad, K. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat. Immunol. 2016, 17, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Nakasone, M.A.; Livnat-Levanon, N.; Glickman, M.H.; Cohen, R.E.; Fushman, D. Mixed-Linkage Ubiquitin Chains Send Mixed Messages. Structure 2013, 21, 727–740. [Google Scholar] [CrossRef]
- Gao, D.; Inuzuka, H.; Tan, M.K.; Fukushima, H.; Locasale, J.W.; Liu, P.; Wan, L.; Zhai, B.; Chin, Y.R.; Shaik, S.; et al. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 2011, 44, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. The Age of Crosstalk: Phosphorylation, Ubiquitination, and Beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef]
- Li, X.; Song, Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 2020, 13, 50. [Google Scholar] [CrossRef]
- Prescott, J.A.; Mitchell, J.P.; Cook, S.J. Inhibitory feedback control of NF-κB signalling in health and disease. Biochem. J. 2021, 478, 2619–2664. [Google Scholar] [CrossRef]

| Gene Name | Primers (Synthesized by Eurofins Genomics) |
|---|---|
| hCXCL1 | Forward: 5′-AACCGAAGTCATAGCCACAC-3′ Reverse: 5-GTTGGATTTGTCACTGTTCAGC-3′ |
| hCXCL2 | Forward: 5′-CTGCGCTGCCAGTGCTT-3′ Reverse: 5′-CCTTCACACTTTGGATGTTCTTGA-3′ |
| hCXCL8 | Forward: 5′-GTGCAGTTTTGCCAAGGAGT-3′ Reverse: 5′-CTCTGCACCCAGTTTTCCTT-3′ |
| hCCL20 | Forward: 5′-TGCTGTACCAAGAGTTTGCTC-3′ Reverse: 5′-CGCACACAGACAACTTTTTCTTT-3′ |
| hIL-6 | Forward: 5′-GGTACATCCTCGACGGCATCT-3′ Reverse: 5′-GTGCCTCTTTGCTGCTTTCAC-3′ |
| hFBXW11 | Forward: 5′-GTGGGATGTGAACACGGGTGA-3′ Reverse: 5′-CGTAAAGTGATGTCGGTCGCAG-3′ |
| hGAPDH | Forward: 5′-CCATGGGGAAGGTGAAGGTC-3′ Reverse: 5′-AGTGATGGCATGGACTGTGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, B.; Moududee, S.A.; Ge, D.; Zhou, P.; Wang, A.R.; Liu, Y.-Z.; You, Z. SCFFBXW11 Complex Targets Interleukin-17 Receptor A for Ubiquitin–Proteasome-Mediated Degradation. Biomedicines 2024, 12, 755. https://doi.org/10.3390/biomedicines12040755
Jin B, Moududee SA, Ge D, Zhou P, Wang AR, Liu Y-Z, You Z. SCFFBXW11 Complex Targets Interleukin-17 Receptor A for Ubiquitin–Proteasome-Mediated Degradation. Biomedicines. 2024; 12(4):755. https://doi.org/10.3390/biomedicines12040755
Chicago/Turabian StyleJin, Ben, Sayed Ala Moududee, Dongxia Ge, Pengbo Zhou, Alun R. Wang, Yao-Zhong Liu, and Zongbing You. 2024. "SCFFBXW11 Complex Targets Interleukin-17 Receptor A for Ubiquitin–Proteasome-Mediated Degradation" Biomedicines 12, no. 4: 755. https://doi.org/10.3390/biomedicines12040755
APA StyleJin, B., Moududee, S. A., Ge, D., Zhou, P., Wang, A. R., Liu, Y.-Z., & You, Z. (2024). SCFFBXW11 Complex Targets Interleukin-17 Receptor A for Ubiquitin–Proteasome-Mediated Degradation. Biomedicines, 12(4), 755. https://doi.org/10.3390/biomedicines12040755






