Abstract
Malignant neoplasms are characterized by high molecular heterogeneity due to multilevel deregulation of gene expression and cellular functions. It is known that non-coding RNAs, including long intergenic non-coding RNAs (lincRNAs), can play significant roles in cancer biology. The current review focuses on a systematical analysis of genomic, transcriptomic, epigenomic, interactomic, and literature data on 65 lincRNAs of human chromosome 18 in the context of pan-cancer studies. The entire group of lincRNAs can be conditionally divided into 4 subgroups depending on experimental evidence on direct or indirect involvement in cancers and the biological associations with cancers, which we found during the data-mining process: the most studied (5 lincRNAs), moderately or poorly studied (11 lincRNAs), and understudied (31 lincRNAs). For the remaining 18 lincRNAs, data for analysis were fragmentary or missing. Among the key findings were the following: Of the lincRNAs of human chromosome 18, 40% have tissue-specific expression patterns, 22% of lincRNAs are known to have gene fusions, 40% of lincRNAs are prone to gene amplifications and/or deletions in cancers at a frequency greater than 3%, and 23% of lincRNAs are differentially expressed across cancer types, whereas 7% have subtype-specific expression patterns. LincRNAs’ interactomes consist of ‘master’ microRNAs and 47 proteins (including cancer-associated proteins and microRNAs) that can interact with 3 or more lincRNAs. Functional enrichment analysis of a set of highly co-expressed genes retrieved for 17 lincRNAs in different cancer types indicated the potential associations of these lincRNAs with cellular signaling pathways. Six lincRNAs encoded small open-reading frame (smORF) proteins with emerging roles in cancers, and microRNAs as well as proteins with known functions in molecular carcinogenesis can bind to coding regions of smORFs. We identified seven transcriptomic signatures with potential prognostic value, consisting of two to seven different lincRNAs only. Taken together, the literature, biomedical, and molecular biology data analyzed indicated that only five of all lincRNAs of human chromosome 18 are cancer-associated, while eleven other lincRNAs have the tendency to be associated with cancers.
1. Introduction
Long non-coding RNAs (lncRNAs) have been arbitrarily defined as non-coding transcripts of more than 200 nucleotides (200 nt) [1], but today, the term ‘lncRNA’ defines a more expanded list of different RNAs [2]. With respect to protein-coding genes, lncRNAs can be intergenic, antisense, or intronic. Long intergenic non-coding RNAs (lincRNAs) are autonomously transcribed RNAs whose genes do not trespass on nearby protein-coding loci [2,3]. It is believed that lincRNAs are expressed in a tissue-, cell-, stage-, and disease-specific manner. There are several known universal mechanisms by which all lncRNAs, and lincRNAs in particular, realize their biologic functions, such as control of chromatin architecture, modulation of enhancer activity, formation of biomolecular condensates [2], and epigenetic and transcriptional regulation of gene expression [3]. LincRNAs can influence other biomolecules, acting as signals, decoys, scaffolds, and guides [4] that are mediated by binding with chromatin and chromatin-modifying complexes [5,6], transcription factors [7], RNA-binding proteins (RBPs) [8], and various types of non-coding RNAs [4,9].
It is known that lincRNAs affect biological pathways in autoimmune and neurodegenerative disorders [10], cardiovascular diseases [9], inflammation [11,12], and normal and malignant hematopoiesis [13]. LincRNAs are aberrantly expressed in various malignant tumors [14]. For example, aberrant expression of LINC00173 affects the initiation and progression of human cancers [15], while LINC01094 indirectly stabilizes the brain-derived neurotrophic factor via microRNA miR-577 in glioblastoma cells [16]. Overexpression of LINC01355 significantly inhibits tumorigenesis of breast cancer cells through interaction and stabilization of forkhead box O3 protein (FOXO3), leading to transcriptional repression of the cyclin D1 gene [15]. LINC00680 enhances hepatocellular carcinoma stem cell behavior and chemoresistance by sponging miR-568 to upregulate AKT Ser/Thr protein kinase 3 [17]. Thus, some lincRNAs involved in the pathogenesis of various cancer or non-cancer diseases may be considered as potential molecular targets or prognostic, predictive, and diagnostic biomarkers. Recent studies of human chromosome 18 within the framework of the Russian segment of the international program ‘The Human Proteome Project’ have investigated a detailed proteogenomic landscape of human chromosome 18 genes in the HepG2 cell line and liver tissue [18,19,20]. Taken together, literature data speak in favor of the associations between some lincRNAs of human chromosome 18 and cancers. The goal of the current review is a systematic analysis of genomic, transcriptomic, epigenomic, interactomic, and literature data on 65 lincRNAs of human chromosome 18 in the context of pan-cancer studies.
2. A Spectrum of Genes Encoding lincRNAs of Human Chromosome 18
Sixty-five genes of human chromosome 18 encode lincRNAs-of-interest (Table S1). The lengths of lincRNAs range from 297 to 6201 nucleotides. LincRNAs’ transcripts per gene vary from 1 to 84 due to the alternative splicing. Records on subcellular localization in the RNAlocate database [21] are available only for 22 of 65 lincRNAs. They are localized in the circulating blood exosomes, nucleus, nucleoplasm, membranes, and cytosol. Four linc-genes (LINC00305, LINC00470, LINC00526, and LINC01387) encode open-reading frames of uncharacterized chromosome-specific proteins C18orf20, C18orf2, C18orf18, and C18orf64, respectively. However, their protein existence status remains ‘uncertain’ according to the PepPsy portal [22]. LincRNAs-of-interest in the form of circRNAs, a type of single-stranded RNA forming a covalently closed continuous loop, were not found in the ‘Circular RNA Interactome’ [23] and ‘CircBank’ [24] databases. Knockouts of LINC01387, LINC01899, and LINC01909 genes resulted in significant biological effects in different cell lines (≥four independent sources, according to the BioGRID Open Repository of CRISPR Screens v.1.1.14 [25]). At least 8 lincRNAs are associated with 17 different types of malignancies (Figure 1), which follows from the RNA-Disease Repository v. 4.0 database [26].
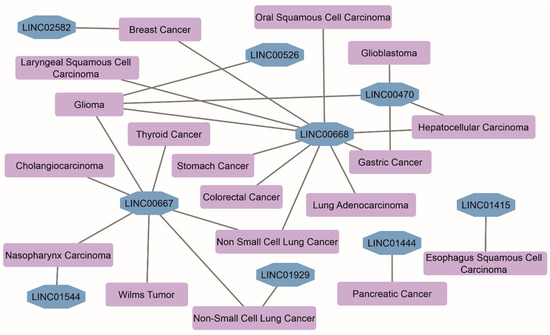
Figure 1.
The network of associations between diseases and lincRNAs of chromosome 18 (data were retrieved from the RNADisease v. 4.0 repository database [26]; selection of disease terms was performed based on a score > 0.9 and experimental evidence).
2.1. Alterations of Genes Encoding lincRNAs of Human Chromosome 18
Single-nucleotide polymorphism (SNP) of linc-genes can lead to the disruption of gene function or production of defective RNAs with altered secondary structures. SNPs are described for 41 of 65 lincRNAs-of-interest (Table S1). For example, 108, 234, and 259 pathogenic variants are known for LINC01477, LINC01917, and LINC02565 genes, respectively (Table S1).
Gene fusions due to the inter- or intra-chromosomal rearrangements are the common gene alterations in tumor cells. They ultimately affect the expression of chimeric RNAs encoding protein products with abnormal activity. Another source of chimeric RNAs is the alternative intergenic splicing [27,28].
The landscape of fusions with participation of linc-genes of human chromosome 18 in cancer cell lines was explored using the ‘Cancer Dependency Map’ database [29]. Slightly more than half (63%) of the found gene fusions are within the same loci as a linc-gene or nearby chromosome loci (Table S2). Fusions of linc-genes and cancer-associated genes, such as RAD54L (RAD54 like), PIK3C3 (phosphatidylinositol 3-kinase catalytic subunit type 3), MBP (myelin basic protein), PFN2 (profilin 2), MBD2 (methyl-CpG binding domain protein 2), and YES1 (YES proto-oncogene 1, Src-family tyrosine kinase), as well as tumor suppressors SMAD4 (SMAD family member 4) and SDHA (succinate dehydrogenase complex flavoprotein subunit A), can be an additional factor in cancer promotion.
The landscape of amplifications and deletions of linc-genes of human chromosome 18 was examined in 11 different cancer types (with >100 cases in each cohort) using the cBioPortal database [30] (Table S3). There are several conventional subgroups of linc-genes with frequency of deletions >3%—D1 (LINC00305 and LINC00907 in metastatic breast cancer (mBRCA), esophageal carcinoma (ESCA), and pancreatic adenocarcinoma (PAAD) and D2 (LINC01538, LINC01541, LINC01544, LINC01924, LINC02582, and LINC02864 in ESCA, head and neck cancer (HNSC), and PAAD), as well as amplifications > 3%—A1 (LINC00470, LINC00526, LINC00667, and LINC00668 in bladder cancer (BLCA), mBRCA, ESCA, PAAD, prostate adenocarcinoma (PRAD), and stomach adenocarcinoma (STAD)) and A2 (LINC01387, LINC01543, and LINC01915 in ovarian cancer (OV), PAAD, PRAD, and STAD). The highest frequency of deletions (16–25%) is observed in PAAD and mBRCA, while the highest frequency of amplifications—9.8% and 8.7%—is observed in PRAD (LINC00907) and PAAD (LINC01915), respectively.
2.2. Promoter Methylation of Genes Encoding lincRNAs of Human Chromosome 18
Differential promoter methylation patterns are found in The Cancer Genome Atlas (TCGA) pan-cancer cohort (n > 30 cases in each cohort) using the web-based tool DNMIVD [31] for LINC00470 in colorectal adenocarcinoma (COAD; Figure S1A), for LINC00305 in head and neck cancer (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC; Figure S1B–E), and for LINC00526 in uterine corpus endometrial carcinoma (UCEC; Figure S1F). At the same time, there are associations of LINC00526 gene promoter methylation with patients’ survival rates in UCEC (46 normal and 432 tumor cases; Figure 2A) and an inverse relationship between an increase in promoter methylation of LINC00526 gene and a decrease in its gene expression levels (Figure 2B).
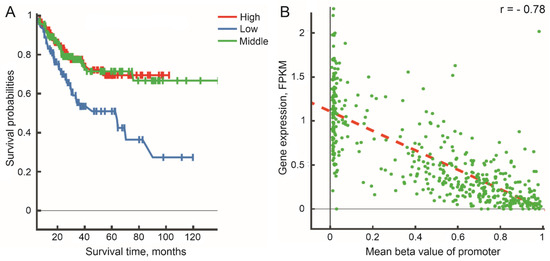
Figure 2.
(A) Associations of promoter methylation of LINC00526 gene with progression-free survival (p-value log-rank test = 6.02 × 10−4) in TCGA_UCEC cohort. (B) A plot of gene expression levels vs. mean beta values of promoter methylation (Spearman correlation coefficient r = −0.78, p-value = 2.46 × 10−95) in TCGA_UCEC cohort. A regression line is indicated by a red dashed line. DNA methylation beta values of zero–0.3, 0.3–0.7, and 0.7–1 were divided into low-, middle-, and high-risk groups of patients, respectively. FPKM—fragments per kilobase million (RNA-seq data). Images are the original outputs from the web-based tool DNA Methylation Interactive Visualization Database (DNMIVD).
2.3. Differentially Expressed Genes Encoding lincRNAs of Human Chromosome 18
A landscape of tissue-specific expression of 47 of 65 lincRNAs of human chromosome 18 was analyzed using the Gene Tissue Expression Portal (GTEx portal, https://www.gtexportal.org, accessed on 5 February 2024; Figure S2). Two lincRNAs, encoded by LINC00526 and LINC00667 genes, are expressed in almost all tissues examined >3 TPM (Transcript per Million). It is also interesting to note that 14 different lincRNAs have tissue-specific expression in testis tissue (>3 TPM). Other lincRNAs with expression levels > 3 TPM are as follows: LINC01544 (brain cerebellum and cerebellar hemisphere), LINC01909 (liver tissue), LINC00668 (colon transverse and testis tissues), LINC01539 (thyroid and testis tissues), LINC01543 (kidney medulla), LINC01926 (prostate and testis tissues), and LINC01444 (thyroid tissue). Thus, it can be seen that the tissue-specific expression patterns are characteristic for almost 30 lincRNAs.
Further, eight differentially expressed linc-genes in cancers were selected using the web-based tool GEPIA2 [32] at |log2fold-change (FC)| > 1.5 tumor/normal tissue and p-value < 0.01 using TCGA as a data source, without taking into account cancer subtypes. All the boxplots, depicting differentially expressed linc-genes of human chromosome 18 in different cancer types, are presented in Figure S3A–S. Of them, downregulation of LINC00305 (Figure S3A), LINC00470 (Figure S3B), LINC00668 (Figure S3C), LINC01255 (Figure S3D), LINC01478 (Figure S3E), and LINC01539 (Figure S3F) genes occurs in testicular germ cell tumors (TGCT), of LINC00470 (Figure S3G), LINC00526 (Figure S3H), and LINC00668 (Figure S3I) genes occurs in acute myeloid leukemia (LAML), and of LINC01539 and LINC00667 genes occurs in thyroid cancer (THCA; Figure S3J) and UCEC (Figure S3K), respectively. Upregulation of LINC00668 gene occurs in COAD, READ, STAD, and LUSC (Figure S3L–O), of LINC00526 and LINC00667 genes occurs in thymoma (THYM; Figure S3P,R), and of LINC01443 gene occurs in skin cutaneous melanoma (SKCM; Figure S3S).
Next, cancer subtype-specific expression patterns of linc-genes were examined using the web-based tool GEPIA2 at |log2FC| > 1 tumor/normal tissue and p-value < 0.05 (Figure S4A–I). Subtype-specific differential expression of six genes is found in eight cancer types. Statistically significant induction of gene expression occurs for the following lincRNAs: LINC00470 in basal and classical subtypes of lung squamous carcinoma (Figure S4A), LINC00526 in the pro-neural subtype of glioblastoma (Figure S4B), and LINC01415 in oligodendroglioma (a subtype of low-grade glioma). The downregulation of gene expression is characteristic for LINC00667 in the papillary subtype of bladder cancer (Figure S4C), LINC00667 in basal-like and HER2 breast cancer (Figure S4D), and LINC00667 in colorectal adenocarcinoma with high microsatellite instability (Figure S4E). LINC00668 is upregulated in microsatellite-stable colorectal and rectal adenocarcinoma with low microsatellite instability (Figures S4F and S4G, respectively). Changes in LINC00470 gene expression occur between classical and primitive subtypes in lung squamous carcinoma (Figure S4A). LINC00668 gene expression varies in rectal adenocarcinoma between subtypes with high and low microsatellite instability (Figure S4G). There are no subtype-specific gene expression changes between seminoma and non-seminoma subtypes of six linc-genes (Figure S3A–F), with differential expression in TGCT as well as between subtypes of skin cutaneous melanoma in the case of LINC01443 (Figure S4I).
Statistically significant changes of stage-specific expression of linc-genes in cancers were not found (F-test, p-value ≤ 0.05).
A list of the top-100 highly co-expressed genes (r ≥ 0.8) for the above-mentioned differentially expressed linc-genes of human chromosome 18 in cancer types was retrieved from TCGA and GTEx portals using the web-based tool GEPIA2. The list, containing co-expressed genes and results of functional enrichment analysis, is presented in Table S4. It is shown that LINC00668, LINC01478, and LINC01539 genes are co-expressed with genes participating in the meiotic cell cycle and cellular process involved in reproduction in multicellular organisms and motile cilium in testis tissue. LINC00526 and LINC00667 genes are co-expressed with genes participating in the processing of capped intron-containing pre-mRNA, RNA processing, and transcriptional and post-transcriptional regulation of gene expression in thymoma.
Analysis of the NCBI Gene Expression Omnibus (GEO; ≥10 cases in each dataset) on condition-specific expression of linc-genes of human chromosome 18 in tumor tissues or cell lines was performed. We analyzed 36 relevant datasets (Table S5) that were previously selected by GEO Profiles to search for differently expressed linc-genes using the GEO2R tool at |log2FC| ≥ 0.8 and p-value < 0.05. Ten differently expressed linc-genes were found (Table S6) with expression patterns specific to certain cancer types, metastasis, and therapy effects [33,34,35,36].
2.4. Transcriptional Regulation of Genes Encoding lincRNAs of Human Chromosome 18
LincRNAs’ accumulation in tumor tissues may be related to transcriptional regulation via different combinations of transcriptional factors (TFs). We analyzed data on 350 potential TFs, whose binding sites were predicted in the promoter or enhancer regions of 30 linc-genes (Table S7) using the GeneHancer Regulatory Elements in the frame of the GeneCards database [37]. All findings on potential TFs found for genes, encoding lincRNAs of human chromosome 18, are shown in Table S8. Non-overlapping groups I and II include TFs, each of which interacts with DNA regions of >15 and >10 linc-genes, respectively (Table S9). Some genes, encoding TFs, were upregulated in cancers, but functional enrichment analysis of TF sets did not show their over-representation in cancer-associated pathways from both groups. It should be noted that the aspects of transcriptional regulation of genes encoding lincRNAs of human chromosome 18 are practically not studied.
3. Interactomics of lincRNAs of Human Chromosome 18
3.1. Interactions of lincRNAs with microRNAs
The post-transcriptional regulation can be realized via interactions of lincRNAs with microRNAs. The miRDB database [38] was used to predict a number of interacting microRNAs based on the lincRNAs’ sequences. As a result, 1004 microRNA/lincRNA interactions were found (Table S10). Of them, 57% of microRNAs interact with only one lincRNA, 27.5% with 2 different lincRNAs, 11% with 3 lincRNAs, and 4% with 4 lincRNAs. One microRNA, hsa-miR-670-3p, was predicted to interact with five different lincRNAs. Figure 3 shows a core part of a microRNAs/lincRNAs subnetwork, from which it follows that the highest connectivity was observed for LINC02864 (degree = 9), LINC01902 (degree = 7), and LINC03035 (degree = 6). Of 1004 predicted interactions, 33 have been previously verified in experiments recorded in the LncBook v. 2.0 database [39] (Table S10).
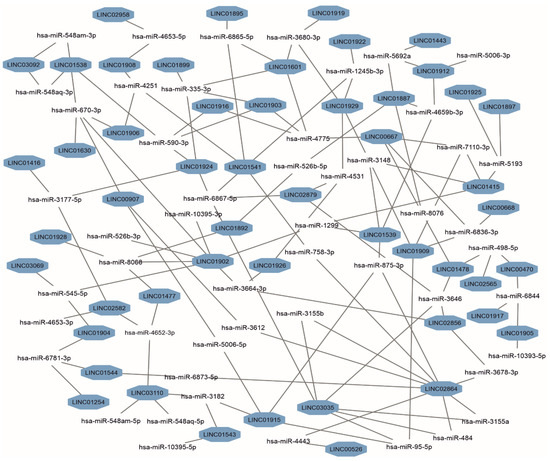
Figure 3.
A subnetwork of predicted miRNAs interacting with three or more lincRNAs of human chromosome 18. The subnetwork was generated based on data retrieved from the miRDB portal.
The RNA–RNA interaction network is quite complex to find disease-specific associations, and the number of predicted hypotheses may be excessive, despite the stringent selection criteria. We performed a functional enrichment analysis with the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology terms of experimentally verified microRNA/lincRNA interactions with each of six lincRNAs of human chromosome 18 (Table S11) using the web-based tool miEAA v.2.1 [40].
The potential for participation in cancer-associated processes decreases in a set of LINC00667, LINC00668 > LINC00470 > LINC01926, LINC01544, LINC01909, depending on the number of functional terms. Further, using the PlasmiR database [41], we additionally searched records on the diagnostic value of 17 microRNAs interacting with these lincRNAs (excluding LINC01909) in plasma and serum blood samples (Table S12). Since lincRNAs act as molecular sponges for microRNAs [42], tissue-specific accumulation of lincRNAs may indirectly influence microRNA content in tissues and, hence, secretion to the biological fluids.
Generally speaking, microRNAs are capable of regulating the half-life of lncRNAs. The latter affects molecular processes and biological functions, and changes in microRNAs’ content directly alter the cellular responses in pathological conditions [43]. However, a reverse phenomenon is also known. Thus, a high negative correlation between the levels of lncRNA OIP5-AS1 and miR-7 was associated with OIP5-AS1-mediated miR-7 degradation, which promoted myotube formation by stimulating a myogenic fusion program [44]. Opposite expression directions of three linc-genes and interacting microRNAs (Table 1) may imply the existence of cancer-dependent regulation of microRNA transcripts’ half-life. For example, LINC00668 and miR-236-3p are up- and down-regulated in READ, respectively. Otherwise, LINC01539 and miR-34a-5p are down- and up-regulated in THCA, respectively.

Table 1.
Differential expression of lincRNAs and microRNAs in tumor tissues.
3.2. Interactions of lincRNAs of Human Chromosome 18 with Other lncRNAs
The analysis of the landscape of co-expressed genes allows us to predict the functional processes and molecular pathways, with which lincRNAs of human chromosome 18 can be associated under normal and disease conditions, in particular, cancer. For this, lists of the top-100 co-expressed genes for each of eleven differentially expressed lincRNAs in a set of cancer types were retrieved from TCGA database using the web-based tool GEPIA2, with a Pearson correlation coefficient ≥ 0.7 as a cut-off (Table S13). We also analyzed lists of highly co-expressed genes retrieved regarding lincRNAs without differential expression in cancer types. Since plenty of co-expressed genes represented non-coding RNAs and pseudogenes, we used the NCpath web-based tool [46] adapted for functional enrichment analysis of gene sets containing non-coding RNAs. All the pathway terms are shown in Table S14, and the most frequent pathway terms are summarized in Table S15. It shows that some lincRNAs of human chromosome 18 may be associated with signaling pathways in cancers under conditions of probable interactions with other non-coding RNAs having similar expression patterns. Thus, LINC00526 and LINC00667 are represented in the majority of pathways in different cancer types. The more cancer-specific lincRNAs are LINC00470, LINC00668, LINC00907, LINC01254, LINC01415, LINC01478, and LINC01539. Finally, there is a subset of LINC01378, LINC01443, LINC01477, and LINC01544, which are typically represented in one or two pathways in only one cancer type. The most common pathways, with which lincRNAs of human chromosome 18 are associated, are ‘adherent junction’, ‘focal adhesion’, ‘FoxO’, and ‘mTOR’.
3.3. Interactions of lincRNAs of Human Chromosome 18 with Cellular Proteins
A list of 613 binary interactions between lincRNAs of human chromosome 18 and cellular proteins was retrieved from four different databases (LncTarD v. 2.0 [47], RNAinter [48], NPInter v.5.0 [49], and Biogrid v.4.4 [25]; Table S16). Further, 47 proteins, which bind with at least ≥3 lincRNAs, were selected (Table S16). Of them, histones H3 methylated at lysine 4 or lysine 27 or acetylated at lysine 27 are the most common interactors for almost all 46 lincRNAs (Table S16). A functional enrichment analysis and protein–protein interaction analysis of 42 non-histone proteins using the tool WebGestalt [50] showed that lincRNA-binding proteins are involved in the epigenetic regulation of gene expression and mRNA processing (Table 2).

Table 2.
Functional enrichment analysis of cellular proteins interacting with lincRNAs of human chromosome 18.
There are also a number of proteins: AR, ESR1, EWSR1, FOXA1, IGF2BP3, HNF4A, POU5F1, SMARCA4, and SOX2, that are associated with epithelial cancers, urogenital neoplasms, and adenocarcinomas, and capable of forming a highly connected subnetwork of protein–protein interactions (Figure S5). In addition, genes encoding these proteins, excluding IGF2BP3 and HNF4A, are causally implicated in cancer promotion [51] due to increased rates of driver mutations.
Generally, the direct interactions of lncRNAs with histone proteins, and especially TFs, are a type of transcriptional regulation through activation or recruitment of TFs [52,53], and these events may have a cancer-specific pattern [54]. Moreover, in the LncBase v.3.0 database [55], we found that experimentally supported interactions of a number of genes: LINC00470, LINC00526, LINC00667, LINC00668, and LINC02582, with cellular proteins are associated with increased growth, proliferation, migration, and invasion of tumor cells, as well as tumor progression through apoptosis suppression and a decrease in radio-sensitivity (Table S17). An interesting fact is that TF FOXA1 (forkhead box protein A1) interacts with 21 lincRNAs-of-interest, while LINC00907, LINC01255, and LINC01910 gene expressions are predicted to be regulated by FOXA1 (GeneCards database). The same coincidence was observed in three other cases: LINC00526 and LINC01910 gene expression may be regulated by HNF4A (hepatocyte nuclear factor 4α), LINC01919 by POU5F1 (POU domain, class 5, transcription factor 1), and LINC00470 by SMARCA4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4).
3.4. Small Open-Reading Frame Proteins (smORF-Proteins)
As a rule, lincRNAs have no protein-coding potential, but some of them are annotated as protein-coding small open-reading frames (smORFs) and thus can be considered as coding RNAs. Eukaryotic proteins encoded by such RNAs are known as ‘small proteins’ that are usually from 15 to 100 amino acid residues in length [56,57]. As an example, a small protein derived from LINC00675 was detected only in tumor tissues [58], and participates in cancer-associated processes [58]. The protein-coding or peptide-coding potential of lincRNAs of human chromosome 18 was examined using the LncBook 2.0 database linked with the SmProt database [59]. SmProt is an online repository with annotation of small proteins derived from ribosome profiling that is based on high-throughput sequencing of mRNAs interacting with active ribosomes in cells [60]. Table S18 shows a list of 28 potential smORF proteins encoded by six lincRNAs of chromosome 18, though records on their mass spectrometry identification are absent in the Peptide Atlas database [61].
Theoretically, interference of RNAs’ open-reading frames, while interacting with other types of RNAs or RNA-binding proteins (RBPs), modulates the translation of small proteins during the neoplastic transformation of cells. microRNAs and RBPs, interacting with lincRNA sequences, encoding smORF proteins, were predicted using the miRDB [38] and RBPmap [62] tools, respectively. Table S18 shows that predicted interactors of lincRNAs include both cancer-associated microRNAs and RBPs. As it can be seen, RBPs (SRSF2 and SRSF10 (serine- and arginine-rich splicing factor 2 and 10), RBM4 and RBM25 (RNA-binding motif protein 4 and 25), MBNL1 (muscle-blind-like splicing regulator 1), CNOT4 (CCR4-NOT transcription complex subunit 4), TRA2A (transformer 2 alpha homolog), and HNRNPs (heterogeneous nuclear ribonucleoproteins) may act as potential interactors for multiple lincRNAs of chromosome 18. The largest number of interactors (3 microRNAs and 17 RNA-binding proteins) was predicted for SPROHSA300118, which is one of the smORF proteins encoded by LINC00667.
4. Prognostic and Predictive Value of Genes Encoding lincRNAs of Human Chromosome 18
Transcriptomic signatures with participation of non-coding RNAs, in particular lincRNAs, can be associated with disease prognosis or prediction of therapy responses. In this regard, Nie and co-authors [63] found that high tissue expression levels of MNX1-AS1, LINC00330, and LSAMP-AS1 genes in laryngeal cancer correlated with low survival rates, and the high-risk group was sensitive to AKT (protein kinase B) inhibitors. Hence, we searched for associations between gene expression levels of lincRNAs of human chromosome 18 and survival rates of patients with cancer using the Kaplan–Meier plotter [64,65]. All findings regarding lincRNAs of human chromosome 18 are shown in Table S19. Several transcriptomic signatures, containing from four to seven linc-genes, are specific solely for one cancer type (PAAD, STAD, HNSC, or LIHC), as shown in Table 3.

Table 3.
Transcriptomic signatures of lincRNAs of human chromosome 18 with potential prognostic significance.
In groups with low and high expression of linc-genes, the calculated difference in survival rates reached at least a 2-fold value at a follow-up period of 24–48 months. Figure 4A demonstrates the Kaplan–Meier plot of the overall survival of patients with melanoma treated with immune checkpoint inhibitors (ICIs), which correlates with LINC00305 gene expression levels. In addition, Figure 4B shows the post-progression survival of patients with gastric cancer, which correlates with LINC01539 and LINC01541 expression levels. At a follow-up period of 36 months, median survival rates were 21 and 8.5 months in low and high expression groups, respectively.
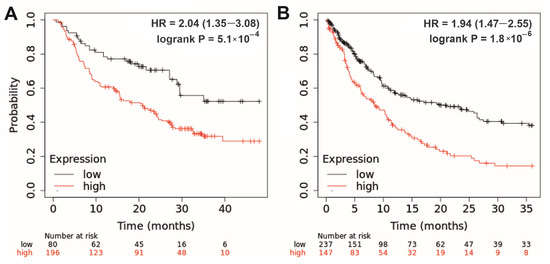
Figure 4.
(A) Overall survival of patients with melanoma correlating with LINC00305 gene expression: restrict analysis to ‘anti-PD-1, anti-PD-L1, anti-CTLA-4 treatment’. Upper quartile survivals were 18.1 and 7.2 months in low and high expression cohorts, respectively. Follow-up threshold = 48 months; FDR = 5%. (B) Post-progression survival of patients with gastric cancer correlating with LINC01539 and LINC01541 gene expression. Median survival rates were 21 and 8.5 months in low and high expression cohorts, respectively. Follow-up threshold = 36 months; FDR = 1%. HR (hazard ratio) is indicated for the high expression group. Kaplan–Meier analysis was performed using the KMplotter web-based tool [64,65].
Immunotherapy is an effective option for treatment of malignant neoplasms. However, only a small portion of patients with cancer achieve positive responses to ICIs, mainly, to inhibitors of PD-1 (programmed cell death protein 1), PD-L1 (programmed death-ligand 1), and CTLA-4 (cytotoxic T-lymphocyte-associated protein 4). Therefore, the identification of predictive biomarkers of tumor susceptibility or resistance to ICIs will help to overcome this clinical complication. The ROC-plotter tool [66] was used to explore associations between expression levels of linc-genes and responses to ICIs. Downregulation of LINC01415 in metastatic melanoma indicated responders to anti-PD-1 treatment, and the area under the curve value (AUC value) of the model and gene expression fold-change were equal to 0.725 and 2.0, respectively (Figure 5A,B). First, we selected the potential markers based on lincRNAs’ gene expression with a predictive value and model quality that correspond to AUC values > 0.7 (good quality). Second, regarding metastatic melanoma, LINC00667 and LINC00526 gene expression levels were also associated with responses to inhibitor of CTLA-4 (ipilimumab), but with AUC values < 0.7 (Figure S6A,B) and a smaller number of clinical cases compared to LINC01415.
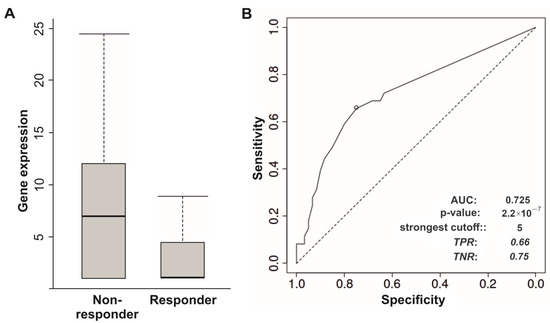
Figure 5.
Association of LINC01415 gene expression levels in metastatic melanoma (81 responders and 105 non-responders) with response to anti-PD-1 therapy. (A) Gene expression levels of LINC01415. (B) Sensitivity and specificity of the model. Mann–Whitney test p-value = <0.001, fold-change = 2. Analysis was performed using the web-based tool ROC-plotter [66].
5. Pharmacologic Aspects of Genes Expression of lincRNAs
Using the PanDrugs2, adapted for personalized treatment selection of patients with cancer through analysis of gene–drug interactions [67], we found that deletion of LINC02864 was associated with resistance to entinostat (a histone deacetylase inhibitor, clinical trials NCT01349959, NCT01038778, and NCT01305499) and AZD8186 (inhibitor of PI3Kβ- and δ-mediated AKT signaling, clinical trials NCT04001569 and NCT01884285).
A search for transcriptomic signatures of chemotherapy response in the NCBI GEO repository using the ncRNADrug tool [68] (at |log2FC| ≥ 1 and FDR < 0.05) allowed us to find the following records. First, downregulation of LINC00470, LINC00526, and LINC00667 genes in MCF-7 cell lines (breast cancer) was associated with sensitivity to doxorubicin. Second, upregulation of LINC00667, LINC01416, and LINC01929 genes in LN229 and U87 cell lines (glioblastoma) was associated with resistance to temozolomide, while downregulation of LINC00668 and LINC00907 genes pointed to sensitivity to this drug.
The SigCom-Library tool [69] helps to provide a signature similarity search for mimickers and reversers, as well as gene–drug associations based on transcriptomic profiling data of cancer cell lines being exposed to various concentrations of drugs. Table S20 presents a summary of the anticancer activity of several candidate drugs as ‘reversers’ that were predicted by the transcriptomic signatures of eight linc-genes with differential expression (LINC00305, LINC00526, LINC00667, LINC00668, LINC00907, LINC01254, LINC01443, and LINC01478) in relation to breast and prostate cancers, as well as lymphoma and leukemia. These drugs were applied in nanomolar concentrations in cell-based assays. Among them, there are receptor tyrosine kinase inhibitors (ibrutinib, lapatinib, lucitanib, quizartinib, rebastinib, and tozasertib), histone deacetylase inhibitor (givinostat), and repurposing drugs (e.g., talinolol and tizanidine; Table S20). Thus, we also demonstrated that the gene expression patterns of lincRNAs of human chromosome 18 might have relevance for prediction of anticancer drugs and immunotherapy responses.
6. Discussion
At least 12 lincRNAs of human chromosome 18 may play certain roles in the malignant transformation of cells, which are mediated via regulation of RNA–RNA and RNA–protein interactions. The main theses from 20 articles addressing these 12 lincRNAs are presented in Table 4. LincRNAs affect pro-tumorigenic processes, such as cell proliferation, migration, invasion, apoptosis, cell senescence, regulation of epithelial–mesenchymal transition, and angiogenesis, which can be applied to a small group of the most studied lincRNAs (LINC00470, LINC00667, LINC00668), while for the remaining ten lincRNAs mentioned in Table 4, investigations in the cancer field are still only occasional. Hence, the majority of lincRNAs of human chromosome 18 have been poorly studied not only in the pan-cancer context, but also in the basic functional aspect. We conducted additional analysis of transcriptomic, interactomic, and other available biomedical data on this group of lincRNAs, but excluding comparative analysis with any other groups of cancer-associated non-coding RNAs. To systematize the collected data on potential associations of each of the studied lincRNAs of human chromosome 18 with cancer, positive (+1) or negative (−1) values were assigned to 16 different hallmarks depending on their presence or absence, respectively. The clustered heat map of hallmarks’ distribution, including a set of 47 lincRNAs of human chromosome 18, is presented in Figure S7. No relevant data were found for the remaining 18 lincRNAs. Among 47 lincRNAs, the first cluster, shown in the far-left part of Figure S7, can be distinguished. This cluster consists of LINC00470, LINC00667, and LINC00668, as well as LINC00305 and LINC00526, which have positive values of most hallmarks, except for hallmarks ‘A’ (somatic gene mutations), ‘B’ (gene fusions), and ‘C’ (gene knockouts effects). Therefore, based on the literature evidence and biomedical data mining, these five lincRNAs are cancer-associated and seem to be directly involved in malignant transformation of cells. The second cluster, shown in the far-right part of Figure S7, is represented by eleven lincRNAs (LINC00907, LINC01254, LINC01387, LINC01415, LINC01416, LINC01443, LINC01477, LINC01478, LINC01538, LINC01539, and LINC01544), which are characterized by hallmarks ‘D’ (copy number variations), ‘G’ (cancer-type-specific differential expression), ‘M’ (predicted relations in cancer-associated pathways), ‘O’ (potential prognostic value), and ‘Q’ (gene expression signatures for drug prediction). This cluster of lincRNAs is characterized by mediocre or hypothetical data on their involvement in cancer-associated processes, so they can be considered as molecular entities moderately or poorly studied in the pan-cancer context. The third cluster (the central part of Figure S7) includes 31 lincRNAs that also form many smaller sub-clusters, pointing to the significant heterogeneity of existing data, which does not yet allow us to hypothesize about their associations with cancer (understudied lincRNAs). The most common hallmarks for all three clusters of lincRNAs are ‘D’ (copy number variations), ‘F’ (tissue-specific expression), and ‘O’ (potential prognostic value).

Table 4.
Literature evidence on participation of lincRNAs of chromosome 18 in cancer-associated processes.
Although this review was primarily focused on synthesizing literature data and the current state-of-the-art of genomic, transcriptomic, epigenomic, and interactomic data on 65 lincRNAs of human chromosome 18, we also touched on some structural aspects. Using data on nucleotide sequences presented in Table S21, we predicted secondary structures for all 65 lincRNAs (Table S22). All sequences were also pairwise-aligned, and the matrix of sequence identity of lincRNAs is shown in Table S23. Only eight pairs of lincRNAs demonstrated high overall sequence identity (≥70%). Among them, we selected four pairs (LINC01916/LINC01415, LINC02564/LINC01919, LINC02879/LINC01926, and LINC02564/LINC01925) having similar secondary structure motifs (p-values < 0.1). Therefore, we tried to predict protein interactors for such motifs (Table S24). The common protein interactors found may indicate similar functions of each pair of compared lincRNAs. This complements our findings on a pool of common protein interactors of lincRNAs of human chromosome 18 (Table 2), as well as proteins with potential to interact with small protein open-reading frames (smORFs) encoded by some lincRNAs (Table S18).
7. Conclusions
The group of long intergenic non-coding RNAs (lincRNAs) of human chromosome 18 are poorly characterized molecular entities both in functional and disease-associated contexts. We have systematized the up-to-date literature data indicating the emerging functional roles of lincRNAs in cancer biology, but this applies to a relatively small subgroup of lincRNAs of human chromosome 18. However, for most lincRNAs, there is a significant gap in the understanding of their contribution to the molecular pathogenesis of diseases, in particular, widely spread solid cancers and malignant proliferative diseases. Therefore, we conducted a search for biomedical data and systems biologic analysis to create a panoramic view, mainly focusing on gene expression and interactomic data for the whole group of lincRNAs of human chromosome 18, which allowed us to consider some lincRNAs as potential candidates for future cancer investigations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines12030544/s1. Table S1: Main characteristics of lincRNAs encoded by genes of human chromosome 18. Table S2: Gene fusions involving linc-genes. Table S3: Copy number variations of linc-genes in cancers. Table S4: A list (#1) of the top-100 co-expressed genes in cancers. Table S5: GEO datasets used to search for differentially expressed linc-genes. Table S6: Condition-specific expression of linc-genes. Table S7: Datasets for analysis of transcriptional regulators of linc-genes. Table S8: Predicted transcriptional regulators of linc-genes. Table S9: Gene expression and functional terms of transcriptional regulators. Table S10: Prediction of microRNAs interacting with lincRNAs. Table S11: Functional enrichment analysis of microRNAs interacting with lincRNAs. Table S12: Diagnostic value* of predicted microRNAs interacting with lincRNAs. Table S13: A list (#2) of the top-100 co-expressed genes in cancers. Table S14: Pathway enrichment analysis of co-expressed genes. Table S15: Associations of individual lincRNAs with signaling pathways in cancers. Table S16: A list of lincRNA–protein interactions. Table S17: Biologic processes associated with interactions between lincRNAs and proteins. Table S18: Predicted interactors of lincRNAs’ open-reading frames small proteins. Table S19: Potential prognostic value of lincRNAs’ expression in different cancer types. Table S20: Anticancer drug candidates predicted by transcriptomic signatures of linc-genes. Table S21: Nucleotide sequences of lincRNAs of human chromosome 18. Table S22: Predicted secondary structures of lincRNAs of human chromosome 18. Table S23: A matrix of pairwise sequence alignment of lincRNAs of human chromosome 18. Table S24: Secondary structure motifs of lincRNAs with high identity. Figure S1: Differentially methylated promoter regions of linc-genes in cancers. Figure S2: Transcriptional landscape of lincRNAs of human chromosome 18. Figure S3: Differentially expressed linc-genes in cancer types. Figure S4: Differentially expressed linc-genes in cancer subtypes. Figure S5: The subnetwork of protein–protein interactions of lincRNA-binding proteins. Figure S6: Gene expression levels of two linc-genes associated with clinical responses to ipilimumab. Figure S7: Panoramic view of characteristics of lincRNAs of human chromosome 18 (clustering and visualization). Supporting information include references [21,32,37,38,39,40,41,51,55,62,90,91,92,93,94,95,96,97].
Author Contributions
Conceptualization, P.V.E.; methodology, P.V.E.; investigation, P.V.E., E.O.Y. and Y.V.M.; writing—original draft preparation, P.V.E.; writing—review and editing, Y.V.M., E.O.Y. and A.S.I.; supervision, A.S.I. All authors have read and agreed to the published version of the manuscript.
Funding
The work was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021–2030; No. 122030100168-2).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Predicted secondary structures of lincRNAs of human chromosome 18 (json format) are deposited in FigShare, https://doi.org/10.6084/m9.figshare.25257343 (accessed on 21 February 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Jayasuriya, R.; Ganesan, K.; Xu, B.; Ramkumar, K.M. Emerging Role of Long Non-Coding RNAs in Endothelial Dysfunction and Their Molecular Mechanisms. Biomed. Pharmacother. 2022, 145, 112421. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, D.; Cai, Y.; Han, X.; Tang, L.; Gao, F.; Qi, Y.; Cai, D.; Wang, H.; Ri, M.; et al. Very Long Intergenic Non-Coding (Vlinc) RNAs Directly Regulate Multiple Genes in Cis and Trans. BMC Biol. 2021, 19, 108. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Chan, S.-J.; Liu, X.; Wei, A.-C.; Jian, R.-I.; Huang, K.-W.; Lang, Y.-D.; Shih, J.-H.; Liao, C.-C.; Luan, C.-L.; et al. Long Noncoding RNA Smyca Coactivates TGF-β/Smad and Myc Pathways to Drive Tumor Progression. J. Hematol. Oncol. 2022, 15, 85. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic Discovery of Xist RNA Binding Proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- Wahba, A.S.; Ibrahim, M.E.; Mesbah, N.M.; Saleh, S.M.; Abo-Elmatty, D.M.; Mehanna, E.T. Serum LINC00305 Expression and Its Genetic Variant Rs2850711 Are Associated with Clinical and Laboratory Features of Rheumatoid Arthritis. Br. J. Biomed. Sci. 2020, 77, 142–147. [Google Scholar] [CrossRef]
- Plewka, P.; Raczynska, K.D. Long Intergenic Noncoding RNAs Affect Biological Pathways Underlying Autoimmune and Neurodegenerative Disorders. Mol. Neurobiol. 2022, 59, 5785–5808. [Google Scholar] [CrossRef]
- Zhang, D.-D.; Wang, W.-T.; Xiong, J.; Xie, X.-M.; Cui, S.-S.; Zhao, Z.-G.; Li, M.J.; Zhang, Z.-Q.; Hao, D.-L.; Zhao, X.; et al. Long Noncoding RNA LINC00305 Promotes Inflammation by Activating the AHRR-NF-κB Pathway in Human Monocytes. Sci. Rep. 2017, 7, 46204. [Google Scholar] [CrossRef]
- Li, X.; Yu, M.; Han, L.; Chen, L.; Zhang, D.; Zhou, G.; Zhao, Q.; Sun, T. LINC00305 Represses miR-124 Expression to Trigger Inflammatory Insults in the Presence of Lipopolysaccharide. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2352–2360. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Lodish, H.F. Emerging Mechanisms of Long Noncoding RNA Function during Normal and Malignant Hematopoiesis. Blood 2017, 130, 1965–1975. [Google Scholar] [CrossRef]
- Mao, W.; Liao, Y.; Tang, L. Long Intergenic Non-Protein Coding RNA 173 in Human Cancers. Cancers 2022, 14, 5923. [Google Scholar] [CrossRef]
- Ai, B.; Kong, X.; Wang, X.; Zhang, K.; Yang, X.; Zhai, J.; Gao, R.; Qi, Y.; Wang, J.; Wang, Z.; et al. LINC01355 Suppresses Breast Cancer Growth through FOXO3-Mediated Transcriptional Repression of CCND1. Cell Death Dis. 2019, 10, 502. [Google Scholar] [CrossRef]
- Dong, X.; Fu, X.; Yu, M.; Li, Z. Long Intergenic Non-Protein Coding RNA 1094 Promotes Initiation and Progression of Glioblastoma by Promoting microRNA-577-Regulated Stabilization of Brain-Derived Neurotrophic Factor. Cancer Manag. Res. 2020, 12, 5619–5631. [Google Scholar] [CrossRef] [PubMed]
- Shu, G.; Su, H.; Wang, Z.; Lai, S.; Wang, Y.; Liu, X.; Dai, L.; Bi, Y.; Chen, W.; Huang, W.; et al. LINC00680 Enhances Hepatocellular Carcinoma Stemness Behavior and Chemoresistance by Sponging miR-568 to Upregulate AKT3. J. Exp. Clin. Cancer Res. 2021, 40, 45. [Google Scholar] [CrossRef] [PubMed]
- Archakov, A.I.; Aseev, A.L.; Bykov, V.A.; Grigoriev, A.I.; Govorun, V.M.; Ilgisonis, E.V.; Ivanov, Y.D.; Ivanov, V.T.; Kiseleva, O.I.; Kopylov, A.T.; et al. Challenges of the Human Proteome Project: 10-Year Experience of the Russian Consortium. J. Proteome Res. 2019, 18, 4206–4214. [Google Scholar] [CrossRef] [PubMed]
- Radko, S.P.; Poverennaya, E.V.; Kurbatov, L.K.; Ponomarenko, E.A.; Lisitsa, A.V.; Archakov, A.I. The “Missing” Proteome: Undetected Proteins, Not-Translated Transcripts, and Untranscribed Genes. J. Proteome Res. 2019, 18, 4273–4276. [Google Scholar] [CrossRef] [PubMed]
- Poverennaya, E.V.; Kopylov, A.T.; Ponomarenko, E.A.; Ilgisonis, E.V.; Zgoda, V.G.; Tikhonova, O.V.; Novikova, S.E.; Farafonova, T.E.; Kiseleva, Y.Y.; Radko, S.P.; et al. State of the Art of Chromosome 18-Centric HPP in 2016: Transcriptome and Proteome Profiling of Liver Tissue and HepG2 Cells. J. Proteome Res. 2016, 15, 4030–4038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tan, P.; Wang, L.; Jin, N.; Li, Y.; Zhang, L.; Yang, H.; Hu, Z.; Zhang, L.; Hu, C.; et al. RNALocate: A Resource for RNA Subcellular Localizations. Nucleic Acids Res. 2017, 45, D135–D138. [Google Scholar] [CrossRef] [PubMed]
- Sallou, O.; Duek, P.D.; Darde, T.A.; Collin, O.; Lane, L.; Chalmel, F. PepPSy: A Web Server to Prioritize Gene Products in Experimental and Biocuration Workflows. Database 2016, 2016, baw070. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A Web Tool for Exploring Circular RNAs and Their Interacting Proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A Comprehensive Database for circRNA with Standard Nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.-J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Chen, J.; Lin, J.; Hu, Y.; Ye, M.; Yao, L.; Wu, L.; Zhang, W.; Wang, M.; Deng, T.; Guo, F.; et al. RNADisease v4.0: An Updated Resource of RNA-Associated Diseases, Providing RNA-Disease Analysis, Enrichment and Prediction. Nucleic Acids Res. 2023, 51, D1397–D1404. [Google Scholar] [CrossRef]
- Taniue, K.; Akimitsu, N. Fusion Genes and RNAs in Cancer Development. Noncoding RNA 2021, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.P. The Ever-Changing World of Gene Fusions in Cancer: A Secondary Gene Fusion and Progression. Oncogene 2019, 38, 7197–7199. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Ding, W.; Chen, J.; Feng, G.; Chen, G.; Wu, J.; Guo, Y.; Ni, X.; Shi, T. DNMIVD: DNA Methylation Interactive Visualization Database. Nucleic Acids Res. 2020, 48, D856–D862. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Koti, M.; Gooding, R.J.; Nuin, P.; Haslehurst, A.; Crane, C.; Weberpals, J.; Childs, T.; Bryson, P.; Dharsee, M.; Evans, K.; et al. Identification of the IGF1/PI3K/NF κB/ERK Gene Signalling Networks Associated with Chemotherapy Resistance and Treatment Response in High-Grade Serous Epithelial Ovarian Cancer. BMC Cancer 2013, 13, 549. [Google Scholar] [CrossRef] [PubMed]
- McMullin, R.P.; Wittner, B.S.; Yang, C.; Denton-Schneider, B.R.; Hicks, D.; Singavarapu, R.; Moulis, S.; Lee, J.; Akbari, M.R.; Narod, S.A.; et al. A BRCA1 Deficient-like Signature Is Enriched in Breast Cancer Brain Metastases and Predicts DNA Damage-Induced Poly (ADP-Ribose) Polymerase Inhibitor Sensitivity. Breast Cancer Res. 2014, 16, R25. [Google Scholar] [CrossRef]
- Bowen, N.J.; Walker, L.D.; Matyunina, L.V.; Logani, S.; Totten, K.A.; Benigno, B.B.; McDonald, J.F. Gene Expression Profiling Supports the Hypothesis That Human Ovarian Surface Epithelia Are Multipotent and Capable of Serving as Ovarian Cancer Initiating Cells. BMC Med. Genom. 2009, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Snipstad, K.; Fenton, C.G.; Kjaeve, J.; Cui, G.; Anderssen, E.; Paulssen, R.H. New Specific Molecular Targets for Radio-Chemotherapy of Rectal Cancer. Mol. Oncol. 2010, 4, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-Wide Integration of Enhancers and Target Genes in GeneCards. Database 2017, 2017, bax028. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: Integrating Human Long Non-Coding RNAs with Multi-Omics Annotations. Nucleic Acids Res. 2023, 51, D186–D191. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Hirsch, P.; Schmartz, G.P.; Kern, F.; Fehlmann, T.; Keller, A. miEAA 2023: Updates, New Functional microRNA Sets and Improved Enrichment Visualizations. Nucleic Acids Res. 2023, 51, W319–W325. [Google Scholar] [CrossRef] [PubMed]
- Tastsoglou, S.; Miliotis, M.; Kavakiotis, I.; Alexiou, A.; Gkotsi, E.C.; Lambropoulou, A.; Lygnos, V.; Kotsira, V.; Maroulis, V.; Zisis, D.; et al. PlasmiR: A Manual Collection of Circulating microRNAs of Prognostic and Diagnostic Value. Cancers 2021, 13, 3680. [Google Scholar] [CrossRef] [PubMed]
- Pasieka, R.; Zasoński, G.; Raczyńska, K.D. Role of Long Intergenic Noncoding RNAs in Cancers with an Overview of MicroRNA Binding. Mol. Diagn. Ther. 2023, 27, 29–47. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Functional Interactions among microRNAs and Long Noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Chang, M.-W.; Tsitsipatis, D.; Yang, X.; Martindale, J.L.; Munk, R.; Cheng, A.; Izydore, E.; Pandey, P.R.; Piao, Y.; et al. LncRNA OIP5-AS1-Directed miR-7 Degradation Promotes MYMX Production during Human Myogenesis. Nucleic Acids Res. 2022, 50, 7115–7133. [Google Scholar] [CrossRef]
- Pan, C.-Y.; Lin, W.-C. miR-TV: An Interactive microRNA Target Viewer for microRNA and Target Gene Expression Interrogation for Human Cancer Studies. Database 2020, 2020, baz148. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Fang, J.; Xu, Z.; Zhang, H.; Mao, M.; Chen, Y.; Zhang, L.; Pian, C. NcPath: A Novel Platform for Visualization and Enrichment Analysis of Human Non-Coding RNA and KEGG Signaling Pathways. Bioinformatics 2023, 39, btac812. [Google Scholar] [CrossRef]
- Zhao, H.; Yin, X.; Xu, H.; Liu, K.; Liu, W.; Wang, L.; Zhang, C.; Bo, L.; Lan, X.; Lin, S.; et al. LncTarD 2.0: An Updated Comprehensive Database for Experimentally-Supported Functional lncRNA-Target Regulations in Human Diseases. Nucleic Acids Res. 2023, 51, D199–D207. [Google Scholar] [CrossRef]
- Kang, J.; Tang, Q.; He, J.; Li, L.; Yang, N.; Yu, S.; Wang, M.; Zhang, Y.; Lin, J.; Cui, T.; et al. RNAInter v4.0: RNA Interactome Repository with Redefined Confidence Scoring System and Improved Accessibility. Nucleic Acids Res. 2022, 50, D326–D332. [Google Scholar] [CrossRef]
- Zheng, Y.; Luo, H.; Teng, X.; Hao, X.; Yan, X.; Tang, Y.; Zhang, W.; Wang, Y.; Zhang, P.; Li, Y.; et al. NPInter v5.0: ncRNA Interaction Database in a New Era. Nucleic Acids Res. 2023, 51, D232–D239. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing Genetic Dysfunction across All Human Cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How Do lncRNAs Regulate Transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Wang, G.; Li, H.; Hou, Y. LncRNA MAGI2-AS3 Inhibits Tumor Progression and Angiogenesis by Regulating ACY1 via Interacting with Transcription Factor HEY1 in Clear Cell Renal Cell Carcinoma. Cancer Gene Ther. 2022, 29, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing Experimentally Supported miRNA Targets on Non-Coding Transcripts. Nucleic Acids Res. 2020, 48, D101–D110. [Google Scholar] [CrossRef]
- Steinberg, R.; Koch, H.-G. The Largely Unexplored Biology of Small Proteins in Pro- and Eukaryotes. FEBS J. 2021, 288, 7002–7024. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Ling, Y.; Yu, J.; Wu, J.; Xiao, J. Small Proteins: Untapped Area of Potential Biological Importance. Front. Genet. 2013, 4, 286. [Google Scholar] [CrossRef]
- Li, X.L.; Pongor, L.; Tang, W.; Das, S.; Muys, B.R.; Jones, M.F.; Lazar, S.B.; Dangelmaier, E.A.; Hartford, C.C.; Grammatikakis, I.; et al. A Small Protein Encoded by a Putative lncRNA Regulates Apoptosis and Tumorigenicity in Human Colorectal Cancer Cells. eLife 2020, 9, e53734. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.; Chen, X.; Zheng, Y.; Kang, Q.; Hao, D.; Zhang, L.; Song, T.; Luo, H.; Hao, Y.; et al. SmProt: A Reliable Repository with Comprehensive Annotation of Small Proteins Identified from Ribosome Profiling. Genom. Proteom. Bioinform. 2021, 19, 602–610. [Google Scholar] [CrossRef]
- Bagheri, A.; Astafev, A.; Al-Hashimy, T.; Jiang, P. Tracing Translational Footprint by Ribo-Seq: Principle, Workflow, and Applications to Understand the Mechanism of Human Diseases. Cells 2022, 11, 2966. [Google Scholar] [CrossRef]
- Desiere, F.; Deutsch, E.W.; King, N.L.; Nesvizhskii, A.I.; Mallick, P.; Eng, J.; Chen, S.; Eddes, J.; Loevenich, S.N.; Aebersold, R. The PeptideAtlas Project. Nucleic Acids Res. 2006, 34, D655–D658. [Google Scholar] [CrossRef]
- Paz, I.; Argoetti, A.; Cohen, N.; Even, N.; Mandel-Gutfreund, Y. RBPmap: A Tool for Mapping and Predicting the Binding Sites of RNA-Binding Proteins Considering the Motif Environment. Methods Mol. Biol. 2022, 2404, 53–65. [Google Scholar] [CrossRef]
- Nie, Q.; Cao, H.; Yang, J.; Liu, T.; Wang, B. PI3K/Akt Signalling Pathway-Associated Long Noncoding RNA Signature Predicts the Prognosis of Laryngeal Cancer Patients. Sci. Rep. 2023, 13, 14764. [Google Scholar] [CrossRef]
- Győrffy, B. Discovery and Ranking of the Most Robust Prognostic Biomarkers in Serous Ovarian Cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Fekete, J.T.; Győrffy, B. ROCplot.Org: Validating Predictive Biomarkers of Chemotherapy/Hormonal Therapy/Anti-HER2 Therapy Using Transcriptomic Data of 3,104 Breast Cancer Patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef]
- Jiménez-Santos, M.J.; Nogueira-Rodríguez, A.; Piñeiro-Yáñez, E.; López-Fernández, H.; García-Martín, S.; Gómez-Plana, P.; Reboiro-Jato, M.; Gómez-López, G.; Glez-Peña, D.; Al-Shahrour, F. PanDrugs2: Prioritizing Cancer Therapies Using Integrated Individual Multi-Omics Data. Nucleic Acids Res. 2023, 51, W411–W418. [Google Scholar] [CrossRef]
- Cao, X.; Zhou, X.; Hou, F.; Huang, Y.-E.; Yuan, M.; Long, M.; Chen, S.; Lei, W.; Zhu, J.; Chen, J.; et al. ncRNADrug: A Database for Validated and Predicted ncRNAs Associated with Drug Resistance and Targeted by Drugs. Nucleic Acids Res. 2023, 52, gkad1042. [Google Scholar] [CrossRef]
- Evangelista, J.E.; Clarke, D.J.B.; Xie, Z.; Lachmann, A.; Jeon, M.; Chen, K.; Jagodnik, K.M.; Jenkins, S.L.; Kuleshov, M.V.; Wojciechowicz, M.L.; et al. SigCom LINCS: Data and Metadata Search Engine for a Million Gene Expression Signatures. Nucleic Acids Res. 2022, 50, W697–W709. [Google Scholar] [CrossRef] [PubMed]
- Luan, P.-B.; Sun, X.-M.; Yao, J. LINC00355 Inhibits Apoptosis and Promotes Proliferation of Gastric Cancer Cells by Regulating Wnt/β-Catenin Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8377–8383. [Google Scholar] [CrossRef]
- Yan, J.; Huang, X.; Zhang, X.; Chen, Z.; Ye, C.; Xiang, W.; Huang, Z. LncRNA LINC00470 Promotes the Degradation of PTEN mRNA to Facilitate Malignant Behavior in Gastric Cancer Cells. Biochem. Biophys. Res. Commun. 2020, 521, 887–893. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; She, X.; Fan, L.; Li, P.; Feng, J.; Fu, H.; Liu, Q.; Liu, Q.; Zhao, C.; et al. A Cytoplasmic Long Noncoding RNA LINC00470 as a New AKT Activator to Mediate Glioblastoma Cell Autophagy. J. Hematol. Oncol. 2018, 11, 77. [Google Scholar] [CrossRef]
- Huang, W.; Liu, J.; Yan, J.; Huang, Z.; Zhang, X.; Mao, Y.; Huang, X. LncRNA LINC00470 Promotes Proliferation through Association with NF45/NF90 Complex in Hepatocellular Carcinoma. Hum. Cell 2020, 33, 131–139. [Google Scholar] [CrossRef]
- Yan, J.; Li, Y.; Xu, C.; Tang, B.; Xie, S.; Hong, T.; Zeng, E. Long Noncoding RNA LINC00526 Represses Glioma Progression via Regulating miR-5581-3p/BEX1. J. Oncol. 2021, 2021, 8171250. [Google Scholar] [CrossRef]
- Yu, J.; Wang, F.; Zhang, J.; Li, J.; Chen, X.; Han, G. LINC00667/miR-449b-5p/YY1 Axis Promotes Cell Proliferation and Migration in Colorectal Cancer. Cancer Cell Int. 2020, 20, 322. [Google Scholar] [CrossRef]
- Yang, H.; Yang, W.; Dai, W.; Ma, Y.; Zhang, G. LINC00667 Promotes the Proliferation, Migration, and Pathological Angiogenesis in Non-Small Cell Lung Cancer through Stabilizing VEGFA by EIF4A3. Cell Biol. Int. 2020, 44, 1671–1680. [Google Scholar] [CrossRef]
- Pan, J.; Zang, Y. LINC00667 Promotes Progression of Esophageal Cancer Cells by Regulating miR-200b-3p/SLC2A3 Axis. Dig. Dis. Sci. 2022, 67, 2936–2947. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Liu, X.; Li, Z.; Wang, G.; Feng, Z.; Liu, Y.; Yang, H.; Tan, C.; Zhang, Z.; Li, K. LncRNA LINC00667 Aggravates the Progression of Hepatocellular Carcinoma by Regulating Androgen Receptor Expression as a miRNA-130a-3p Sponge. Cell Death Discov. 2021, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Dong, J.; Zhao, Z.; Li, J.; Cai, X. LncRNA LINC00668 Promotes the Progression of Breast Cancer by Inhibiting Apoptosis and Accelerating Cell Cycle. Onco Targets Ther. 2019, 12, 5615–5625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Yin, D.; Han, L.; He, X.; Si, X.; Chen, W.; Xia, R.; Xu, T.; Gu, D.; De, W.; et al. E2F1-Induced Upregulation of Long Noncoding RNA LINC00668 Predicts a Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation through Epigenetically Silencing of CKIs. Oncotarget 2016, 7, 23212–23226. [Google Scholar] [CrossRef] [PubMed]
- An, Y.-X.; Shang, Y.-J.; Xu, Z.-W.; Zhang, Q.-C.; Wang, Z.; Xuan, W.-X.; Zhang, X.-J. STAT3-Induced Long Noncoding RNA LINC00668 Promotes Migration and Invasion of Non-Small Cell Lung Cancer via the miR-193a/KLF7 Axis. Biomed. Pharmacother. 2019, 116, 109023. [Google Scholar] [CrossRef]
- Liu, Q.; Dong, J.; Li, J.; Duan, Y.; Wang, K.; Kong, Q.; Zhang, H. LINC01255 Combined with BMI1 to Regulate Human Mesenchymal Stromal Senescence and Acute Myeloid Leukemia Cell Proliferation through Repressing Transcription of MCP-1. Clin. Transl. Oncol. 2021, 23, 1105–1116. [Google Scholar] [CrossRef]
- Qiao, D.; Qin, X.; Yang, H.; Liu, X.; Liu, L.; Liu, S.; Jia, Z. Estradiol Mediates the Interaction of LINC01541 and miR-429 to Promote Angiogenesis of G1/G2 Endometrioid Adenocarcinoma in-Vitro: A Pilot Study. Front. Oncol. 2022, 12, 951573. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, F.; Hu, L.; Zhang, F.; Wang, J.; Huang, K.; Wang, Y. lncRNA RP11-838N2.3 Promoted Cisplatin Resistance in Lung Adenocarcinoma. BioMed. Res. Int. 2020, 2020, 2806042. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, S.; Liu, D.; Zhou, J. LINC01915 Facilitates the Conversion of Normal Fibroblasts into Cancer-Associated Fibroblasts Induced by Colorectal Cancer-Derived Extracellular Vesicles through the miR-92a-3p/KLF4/CH25H Axis. ACS Biomater. Sci. Eng. 2021, 7, 5255–5268. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Deng, W.; Fu, F.; Yan, S.; Yang, H.; Liu, R.; Geng, J.; Xu, J.; Wu, Y.; et al. Viral Integration in BK Polyomavirus-Associated Urothelial Carcinoma in Renal Transplant Recipients: Multistage Carcinogenesis Revealed by next-Generation Virome Capture Sequencing. Oncogene 2020, 39, 5734–5742. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Pang, M.; Du, Y.; Yu, X.; Yuan, J.; Liu, W.; Wang, L.; Liu, X. The LINC01929/miR-6875-5p/ADAMTS12 Axis in the ceRNA Network Regulates the Development of Advanced Bladder Cancer. Front. Oncol. 2022, 12, 856560. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Wang, H.; Wang, S.; Liu, F. Long Non-Coding RNA LINC01929 Facilitates Cell Proliferation and Metastasis as a Competing Endogenous RNA Against MicroRNA miR-1179 in Non-Small Cell Lung Carcinoma. Br. J. Biomed. Sci. 2022, 79, 10598. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zheng, J.; Li, R.; Tian, Y.; Lin, J.; Liang, Y.; Sun, Q.; Xu, A.; Zheng, R.; Liu, M.; et al. Long Noncoding RNA LINC02582 Acts Downstream of miR-200c to Promote Radioresistance through CHK1 in Breast Cancer Cells. Cell Death Dis. 2019, 10, 764. [Google Scholar] [CrossRef]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef]
- Ogris, C.; Castresana-Aguirre, M.; Sonnhammer, E.L.L. PathwAX II: Network-based pathway analysis with interactive visualization of network crosstalk. Bioinformatics 2022, 38, 2659–2660. [Google Scholar] [CrossRef] [PubMed]
- Sundfeld, D.; Havgaard, J.H.; de Melo, A.C.; Gorodkin, J. Foldalign 2.5: Multithreaded implementation for pairwise structural RNA alignment. Bioinformatics 2016, 32, 1238–1240. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnaiah, Y.; Morris, A.P.; Dhaliwal, J.; Philip, M.; Kuhlmann, L.; Tyagi, S. Linc2function: A Comprehensive Pipeline and Webserver for Long Non-Coding RNA (lncRNA) Identification and Functional Predictions Using Deep Learning Approaches. Epigenomes 2023, 7, 22. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- RNAcentral Consortium. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).