Evaluating the Connection between MicroRNAs and Long Non-Coding RNAs for the Establishment of the Major Depressive Disorder Diagnosis
Abstract
1. Introduction
1.1. Major Depressive Disorders
1.1.1. General Considerations
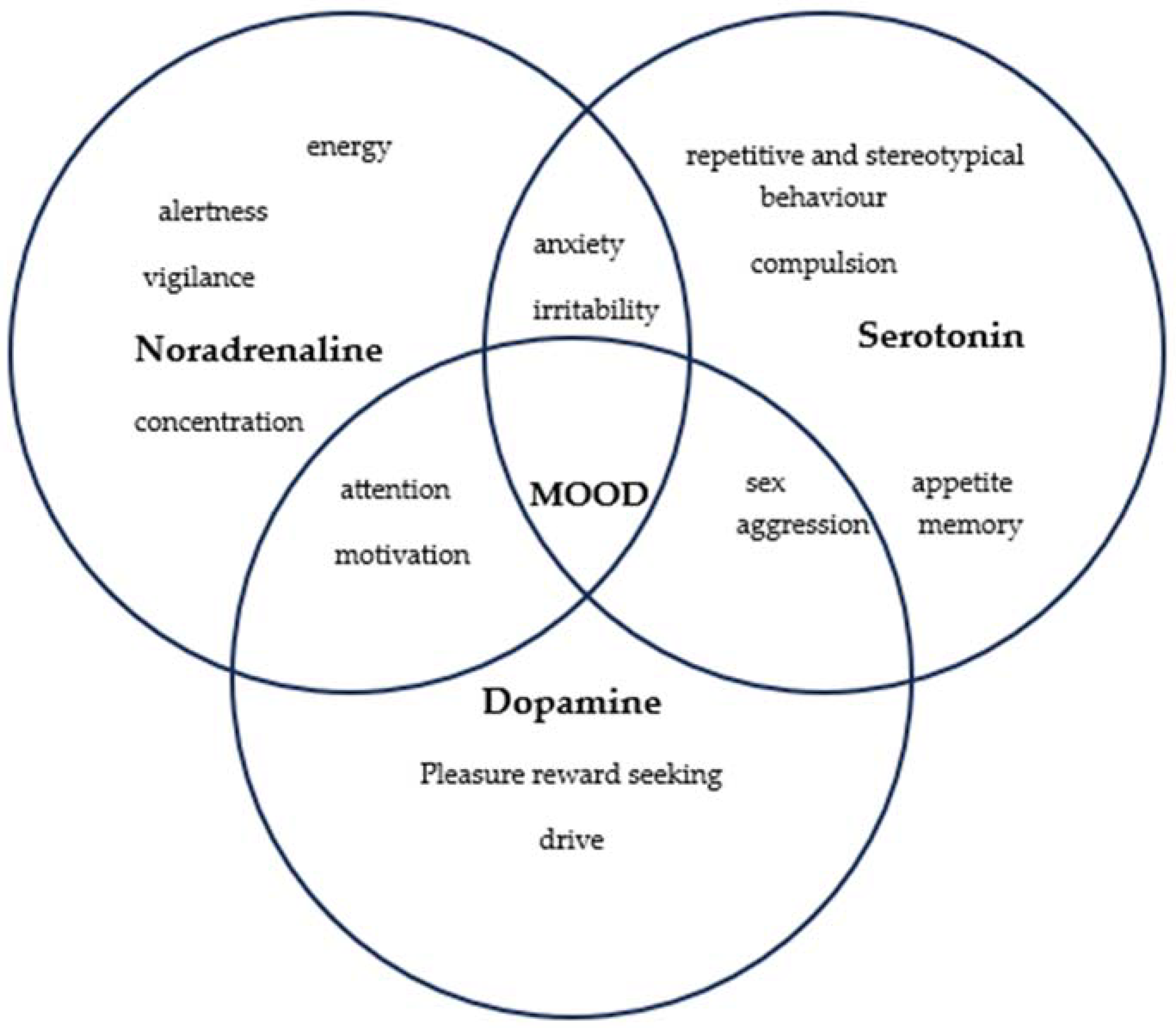
1.1.2. Ethiopathogeny
1.1.3. Treatment of Major Depressive Disorder
1.1.4. Evaluation of Therapeutic Response
1.2. MicroRNA and Long Non-Coding RNA
1.2.1. The Role of MicroRNA in Signaling Pathways
1.2.2. The Influence of MicroRNA on Growth Factors
1.2.3. The Influence of MicroRNAs on Behavioral Phenotypes
1.2.4. Long Non-Coding RNAs
2. Materials and Methods
3. Results
3.1. First Set of Articles Reviewed—Evaluation of Blood RNA Level Variations in Human Subjects with MDD and Healthy Human Subsets
3.2. Second Set of Articles Reviewed—Assessment of RNA Levels in Blood Samples/Brain Biopsy Samples from Human Subjects and Rodent Species
3.3. Third Set of Articles Reviewed—Assessment of RNA Levels in Blood Samples after Antidepressant Medication
4. Discussion
- (1)
- Do human models of MDD reveal miRNA and lncRNA variations? Are there differences in miRNA and lncRNAs at the plasma level? Do samples of brain tissue biopsy show miRNA and lncRNA variants as well?
- (2)
- Do animal models differ in their miRNA and lncRNA composition? Are miRNA and lncRNA variants present in plasma? Do biopsy samples of the brain tissue exhibit miRNA and lncRNA variations?
- (3)
- Do human models with antidepressant therapy exhibit miRNA and lncRNA variations?
- (1)
- Possibility of correlating elements identified in rodents with what applies to human subjects.
- (2)
- The need to understand and clarify these miRNAs and lncRNAs variations, e.g., what is upregulated and what is downregulated and most importantly which of them are associated with a favorable development of depression.
- (3)
- Assess the possibility of using this biomarker in clinical practice.
- (4)
- Quantify the human, technological, and financial resources required.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marx, W.; Penninx, B.W.J.H.; Solmi, M.; Furukawa, T.A.; Firth, J.; Carvalho, A.F.; Berk, M. Major depressive disorder. Nat. Rev. Dis. Primers 2023, 9, 44. [Google Scholar] [CrossRef]
- Institute of Health Metrics and Evaluation. Global Health Data Exchange (GHDx). Available online: https://vizhub.healthdata.org/gbd-results/ (accessed on 4 April 2023).
- Southam-Gerow, M.A.; McLeod, B.D.; Brown, R.C.; Quinoy, A.M. Cognitive-Behavioral Therapy for Adolescents. Encycl. Adolesc. 2011, 100–108. [Google Scholar] [CrossRef]
- Feather, A.; Randall, D.; Waterhouse, M. Kumar and Clark Clinical Medicine, 10th ed.; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Ortega, M.A.; Alvarez-Mon, M.A.; García-Montero, C.; Fraile-Martinez, O.; Lahera, G.; Monserrat, J.; Muñoz-Merida, L.; Mora, F.; Rodríguez-Jiménez, R.; Fernandez-Rojo, S.; et al. MicroRNAs as Critical Biomarkers of Major Depressive Disorder: A Comprehensive Perspective. Biomedicines 2021, 9, 1659. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef]
- Hunsberger, J.G.; Austin, D.R.; Chen, G.; Manji, H.K. MicroRNAs in Mental Health: From Bio-logical Underpinnings to Potential Therapies. Neuromol. Med. 2009, 11, 173–182. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, Y.N.; Zhang, Y.Q.; Wang, H.; Deng, Y.L.; Li, N.M. A New Player in Depression: MiRNAs as Modulators of Altered Synaptic Plasticity. Int. J. Mol. Sci. 2022, 23, 4555. [Google Scholar] [CrossRef]
- Huang, X.; Luo, Y.-L.; Mao, Y.-S.; Ji, J.-L. The link between long noncoding RNAs and depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2017, 73, 73–78. [Google Scholar] [CrossRef]
- Ding, R.; Su, D.; Zhao, Q.; Wang, Y.; Wang, J.Y.; Lv, S.; Ji, X. The role of microRNAs in depression. Front. Pharmacol. 2023, 14, 1129186. [Google Scholar] [CrossRef]
- Popova, N.K.; Tsybko, A.S.; Naumenko, V.S. The Implication of 5-HT Receptor Family Members in Aggression, Depression and Suicide: Similarity and Difference. Int. J. Mol. Sci. 2022, 23, 8814. [Google Scholar] [CrossRef]
- Platania, G.A.; Savia Guerrera, C.; Sarti, P.; Varrasi, S.; Pirrone, C.; Popovic, D.; Ventimiglia, A.; De Vivo, S.; Cantarella, R.A.; Tascedda, F.; et al. Predictors of functional outcome in patients with major depression and bipolar disorder: A dynamic network approach to identify distinct patterns of interacting symptoms. PLoS ONE 2023, 18, e0276822. [Google Scholar] [CrossRef]
- Guerrera, C.S.; Platania, G.A.; Boccaccio, F.M.; Sarti, P.; Varrasi, S.; Colliva, C.; Grasso, M.; De Vivo, S.; Cavallaro, D.; Tascedda, F.; et al. The dynamic interaction between symptoms and pharmacological treatment in patients with major depressive disorder: The role of network intervention analysis. BMC Psychiatry 2023, 23, 885. [Google Scholar] [CrossRef]
- Ferrúa, C.P.; Giorgi, R.; da Rosa, L.C.; do Amaral, C.C.; Ghisleni, G.C.; Pinheiro, R.T.; Nedel, F. MicroRNAs expressed in depression and their associated pathways: A systematic review and a bioinformatics analysis. J. Chem. Neuroanat. 2019, 100, 101650. [Google Scholar] [CrossRef]
- Belzeaux, R.; Bergon, A.; Jeanjean, V.; Loriod, B.; Formisano-Tréziny, C.; Verrier, L.; Loundou, A.; Baumstarck-Barrau, K.; Boyer, L.; Gall, V.; et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl. Psychiatry 2012, 2, e185. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, Y.; Wang, X.; Wu, J.L.; Liu, K.Z.; Zhou, J.; Liu, L.; Zhang, C.H. Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS ONE 2015, 10, e0121975. [Google Scholar] [CrossRef]
- Li, J.; Meng, H.; Cao, W.; Qiu, T. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci. Lett. 2015, 606, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Maffioletti, E.; Cattaneo, A.; Rosso, G.; Maina, G.; Maj, C.; Gennarellia, M.; Tardito, D.; Bocchio-Chiavetto, L. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J. Affect. Disord. 2016, 200, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Lei, L.; Wang, Y.; Yang, C.X.; Liu, Z.F.; Li, X.R.; Zhang, K. Preliminary comparison of plasma notch-associated microRNA-34b and -34c levels in drug naive, first episode depressed patients and healthy controls. J. Affect. Disord. 2016, 194, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Sun, X.; Niu, W.; Kong, L.; He, M.; Zhong, A.; Chen, S.; Jiang, K.; Zhang, L.; Cheng, Z. Long Non-Coding RNA: Potential Diagnostic and Therapeutic Biomarker for Major Depressive Disorder. Med. Sci. Monit. 2016, 22, 5240–5248. [Google Scholar] [CrossRef]
- Hung, Y.Y.; Wu, M.K.; Tsai, M.C.; Huang, Y.L.; Kang, H.Y. Aberrant Expression of Intracellular let-7e, miR-146a, and miR-155 Correlates with Severity of Depression in Patients with Major Depressive Disorder and Is Ameliorated after Antidepressant Treatment. Cells 2019, 8, 647. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, F.; Zheng, Y.; He, S.; Zhang, T.; Guo, Q.; Xu, H.; Chen, H.; Liu, C.; Yu, S.; et al. The role of circulating blood microRNA-374 and microRNA-10 levels in the pathogenesis and therapeutic mechanisms of major depressive disorder. Neurosci. Lett. 2021, 763, 136184. [Google Scholar] [CrossRef]
- Lou, D.; Wang, J.; Wang, X. miR-124 ameliorates depressive-like behavior by targeting STAT3 to regulate microglial activation. Mol. Cell Probes. 2019, 48, 101470. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Dunbar, M.; Shelton, R.; Dwivedi, Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacology 2017, 42, 864–875. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liu, X.; Jiang, K.; Peng, D.; Hong, W.; Fang, Y.; Qian, Y.; Yu, S.; Li, H. Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J. Psychiatr. Res. 2016, 78, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Smalheiser, N.R.; Lugli, G.; Rizavi, H.S.; Torvik, V.I.; Turecki, G.; Dwivedi, Y. MicroRNA expression is down-regulated and reorganized in the prefrontal cortex of depressed suicide subjects. PLoS ONE 2012, 7, e33201. [Google Scholar] [CrossRef] [PubMed]
- Lian, N.; Niu, Q.; Lei, Y.; Li, X.; Li, Y.; Song, X. MiR-221 is involved in depression by regulating Wnt2/CREB/BDNF axis in hippocampal neurons. Cell Cycle 2018, 17, 2745–2755. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.K.; Dong, Z.Q.; Tian, L.T.; Li, J. MicroRNA-451a, microRNA-34a-5p, and microRNA-221-3p as predictors of response to antidepressant treatment. Braz. J. Med. Biol. Res. 2018, 51, e7212. [Google Scholar] [CrossRef]
- Bocchio-Chiavetto, L.; Maffioletti, E.; Bettinsoli, P. Blood microRNA changes in depressed patients during antidepressant treatment. Eur. Neuropsychopharmacol. 2013, 23, 602–611. [Google Scholar] [CrossRef]
- Lin, C.C.; Tsai, M.C.; Lee, C.T.; Sun, M.H.; Huang, T.L. Antidepressant treatment increased serum miR- 183 and miR-212 levels in patients with major depressive disorder. Psychiatry Res. 2018, 270, 232–237. [Google Scholar] [CrossRef]
- Enatescu, V.R.; Papava, I.; Enatescu, I.; Antonescum, M.; Anghel, A.; Seclaman, E.; Sirbu, I.O.; Marian, C. Circulating Plasma Micro RNAs in patients with major depressive disorder treated with antidepressants: A pilot study. Psychiatry Investig. 2016, 13, 549. [Google Scholar] [CrossRef]
- Higuchi, F.; Uchida, S.; Yamagata, H.; Abe-Higuchi, N.; Hobara, T.; Hara, K.; Kobayashi, A.; Shintaku, T.; Itoh, Y.; Suzuki, T.; et al. Hippocampal MicroRNA-124 Enhances Chronic Stress Resilience in Mice. J. Neurosci. 2016, 36, 7253–7267. [Google Scholar] [CrossRef]
- Liu, L.; Wang, H.; Chen, X.; Zhang, Y.; Li, W.; Rao, X.; Liu, Y.; Zhao, L.; Pu, J.; Gui, S.; et al. Integrative Analysis of Long Non-coding RNAs, Messenger RNAs, and MicroRNAs Indicates the Neurodevelopmental Dysfunction in the Hippocampus of Gut Microbiota-Dysbiosis Mice. Front. Mol. Neurosci. 2022, 14, 745437. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, M.; Li, M.; Yang, J.; Jia, J.; Liu, L.; Zhou, J.; Zhang, C.; Wang, X. Serum miR-221-3p as a new potential biomarker for depressed mood in perioperative patients. Brain Res. 2019, 1720, 146296. [Google Scholar] [CrossRef] [PubMed]

| Reference | No. of Patients | Controls | Upregulated lncRNA | Downregulated lncRNAs | Total RNA |
|---|---|---|---|---|---|
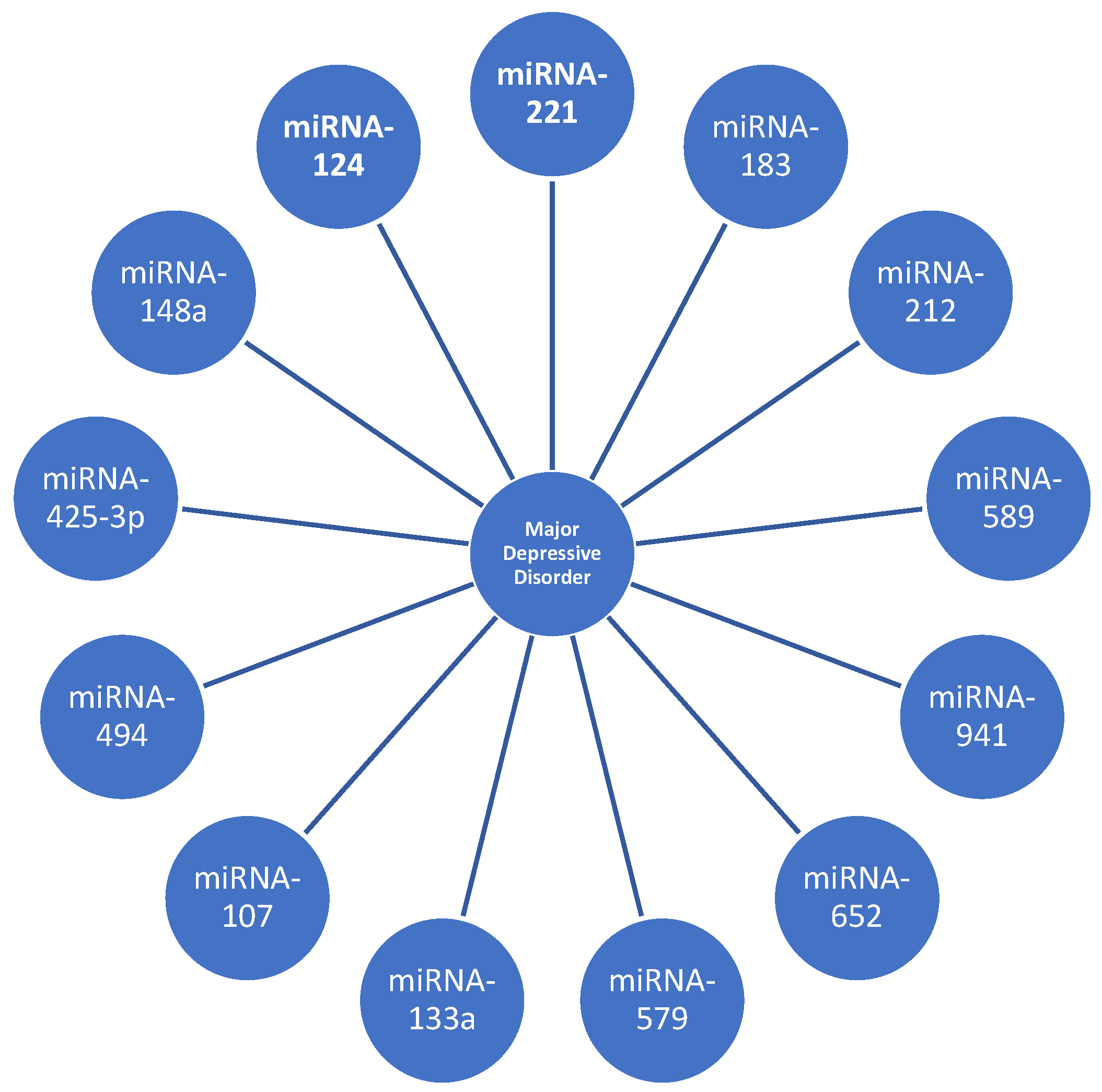
| [15] | 16 | 13 | miRNA-107 miRNA-133a miRNA-148a miRNA-425-3p miRNA-494 miRNA-579 miRNA-652 miRNA-941 miRNA-589 | miRNA-200c miRNA-381 miRNA-571 miRNA-636 miRNA-1243 | 9 upregulated and 5 downregulated |
| [16] | 38 | 27 | miRNA-29b-3p miRNA-10a-5p miRNA-375 miRNA-155-5p miRNA-33a-5p miRNA-139-5p | miRNA-106-5p miRNA-590-5p miRNA-185-5p | 5 downregulated |
| [17] | 18 | 18 | miRNA-644 miRNA-450b miRNA-328 miRNA-182 | miRNA-335 miRNA-583 miRNA-708a miRNA-650 miRNA-654a | 4 upregulated, 5 downregulated, and 3 unchanged |
| [18] | 20 | 20 | miRNA-199a-5p miRNA-24-3p miRNA-425-3p miRNA-29c-5p miRNA-330-3p miRNA-345-5p | let-7a-5p let-7d-5p let-7f-5p hmiRNA-1915-3p | 6 upregulated, 4 downregulated, and 13 unchanged |
| [19] | 32 | 32 | miRNA-34b-5p miRNA-34c-5p | - | 2 upregulated and 3 unchanged |
| [20] | 5 | 5 | 534 upregulated RNA | 2115 downregulated lncRNAs | 534 upregulated RNA, 2115 downregulated lncRNAs |
| [21] | 84 | 43 | miRNA-21-5p miRNA-145 miRNA-223 | miRNA-146a miRNA-155 let-7e | 3 upregulated and 3 downregulated |
| [22] | 5 | 2 | miRNA-4539 miRNA-4281 | miRNA-374b-5p miRNA-98 miRNA-10a-5p | 2 upregulated and 3 downregulated |
| Dysregulations of miRNA-124 in Post-Mortem Brain Tissues and Blood Samples | ||||
| Reference | Species | Sample Type | miRNA | Changes |
| [23] | Rodent | Hippocampus | miRNA-124 | Decrease |
| [24] | Humans | The prefrontal cortex (BA46) | miRNA-124 | Increase |
| [25] | Humans | Peripheral blood mononuclear cells | miRNA-124 | Increase |
| Dysregulations of miRNA-221 in humans and rodent | ||||
| Reference | Species | Sample type | miRNA | Changes |
| [26] | Humans | The prefrontal cortex (BA10) | miRNA-221 | Increase |
| [27] | Humans | Cerebrospinal fluid | miRNA-221 | Increase |
| Rodent | Serum | Increase | ||
| Hippocampus | Increase | |||
| [28] | Humans | Serum | miRNA-221 | Increase |
| Reference | Species | Upregulated miRNA | Downregulated miRNA | Therapeutic Agent | Treatment Duration |
|---|---|---|---|---|---|
| [29] | Whole-blood sample | miRNA-130b miRNA-505 miRNA-29b-2 miRNA-26b miRNA-22 miRNA-26a miRNA-664 miRNA-494 let-7d let-7g let-7f miRNA-629 miRNA-106b miRNA-103 miRNA-191 miRNA-128 miRNA-502-3p miRNA-374b miRNA-132 miRNA-30d miRNA-500 miRNA-589 miRNA-183 miRNA-574-3p miRNA-140-3p miRNA-335 miRNA-361-5p | miRNA-34c-5p miRNA-770-5p | Escitalopram | 12 weeks |
| [30] | Serum | miRNA-16 (only in selective serotonin reuptake inhibitors) miRNA-183 miRNA-212 | - | Selective serotonin reuptake inhibitors or selective serotonin-norepinephrine reuptake inhibitors | 4 weeks |
| [31] | Plasma | miRNA-1193 miRNA-4263 miRNA-3173-3p miRNA-382 miRNA-3154 miRNA-129-5p miRNA-3661 miRNA-1287 miRNA-532-3p miRNA-608 miRNA-3691-5p miRNA-2278 miRNA-3150a-3p miRNA-375 miRNA-3909 miRNA-433 miRNA-937 miRNA-676 miRNA-1298 miRNA-489 miRNA-1909 miRNA-637 miRNA-1471 | miRNA-744 miRNA-301b miRNA-27a miRNA-24 miRNA-146a miRNA-126 miRNA-151-5p miRNA-99b miRNA-151-3p let-7d miRNA-221 miRNA-223 miRNA-181b miRNA-146b-5p miRNA-125a-5p miRNA-26a miRNA-652 | Escitalopram | 12 weeks |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prodan-Bărbulescu, C.; Şeclăman, E.P.; Enătescu, V.; Faur, I.F.; Ghenciu, L.A.; Tuţac, P.; Paşca, P.; Grigoriţă, L.O. Evaluating the Connection between MicroRNAs and Long Non-Coding RNAs for the Establishment of the Major Depressive Disorder Diagnosis. Biomedicines 2024, 12, 516. https://doi.org/10.3390/biomedicines12030516
Prodan-Bărbulescu C, Şeclăman EP, Enătescu V, Faur IF, Ghenciu LA, Tuţac P, Paşca P, Grigoriţă LO. Evaluating the Connection between MicroRNAs and Long Non-Coding RNAs for the Establishment of the Major Depressive Disorder Diagnosis. Biomedicines. 2024; 12(3):516. https://doi.org/10.3390/biomedicines12030516
Chicago/Turabian StyleProdan-Bărbulescu, Cătălin, Edward Paul Şeclăman, Virgil Enătescu, Ionuţ Flaviu Faur, Laura Andreea Ghenciu, Paul Tuţac, Paul Paşca, and Laura Octavia Grigoriţă. 2024. "Evaluating the Connection between MicroRNAs and Long Non-Coding RNAs for the Establishment of the Major Depressive Disorder Diagnosis" Biomedicines 12, no. 3: 516. https://doi.org/10.3390/biomedicines12030516
APA StyleProdan-Bărbulescu, C., Şeclăman, E. P., Enătescu, V., Faur, I. F., Ghenciu, L. A., Tuţac, P., Paşca, P., & Grigoriţă, L. O. (2024). Evaluating the Connection between MicroRNAs and Long Non-Coding RNAs for the Establishment of the Major Depressive Disorder Diagnosis. Biomedicines, 12(3), 516. https://doi.org/10.3390/biomedicines12030516






