Influence of Genetic Polymorphisms on the Age at Cancer Diagnosis in a Homogenous Lynch Syndrome Cohort of Individuals Carrying the MLH1:c.1528C>T South African Founder Variant
Abstract
1. Introduction
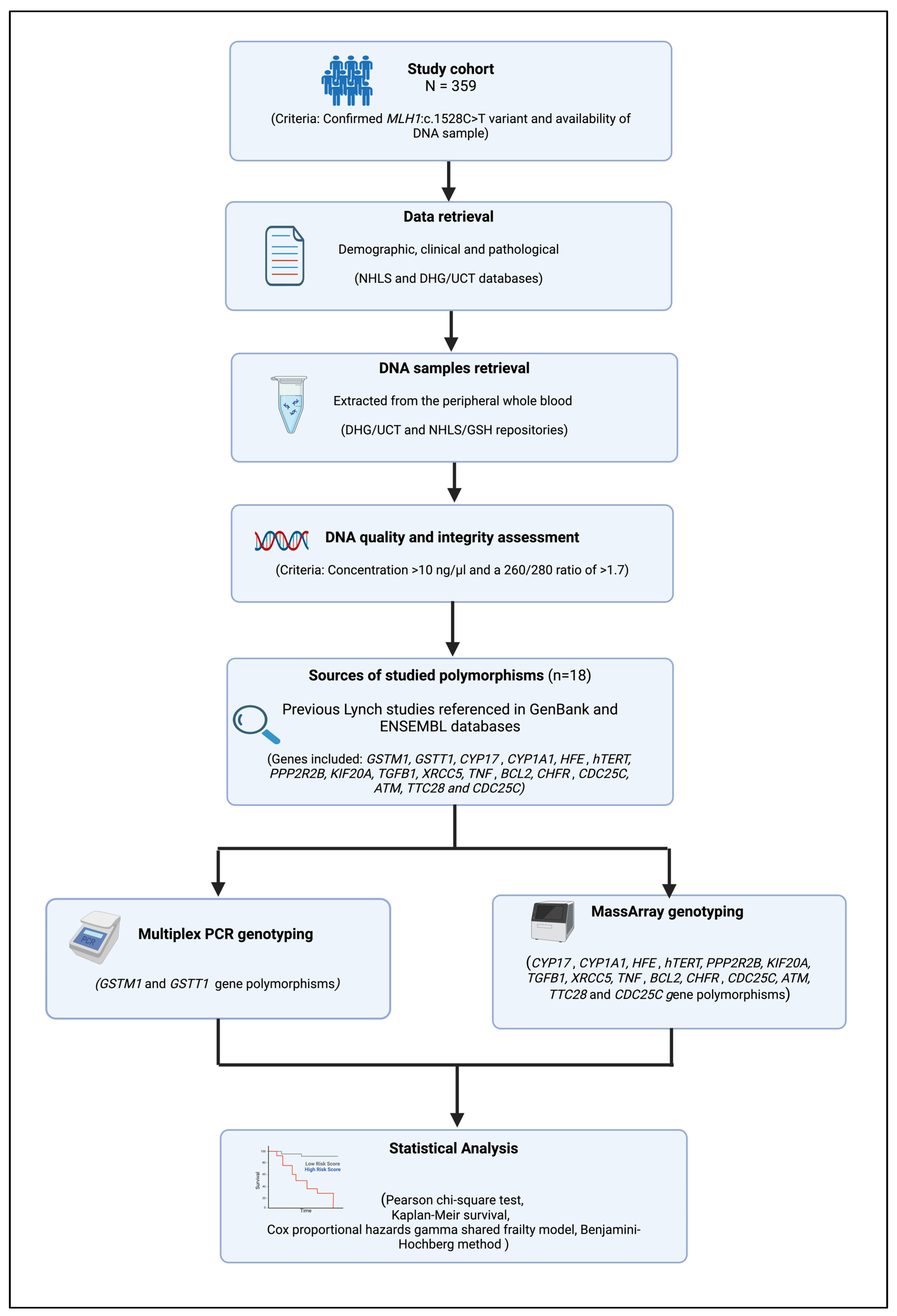
2. Materials and Methods
2.1. Patient Selection
- Confirmed genetic diagnosis of LS due to MLH1: c.1528C>T PV;
- Availability of a blood DNA sample with good quality and quantity according to the minimum requirements of the Multiplex PCR and MassArray genotyping assays (i.e., intact DNA with a concentration of >10 ng/µL and a 260/280 ratio of >1.7) [57].
2.2. Demographic and Pathological Data
2.3. DNA Samples
2.4. DNA Quality and Integrity Assessment
2.5. Sources of Studied Polymorphisms
2.6. Multiplex PCR
2.7. MassArray Genotyping
2.8. Statistical Analysis
3. Results
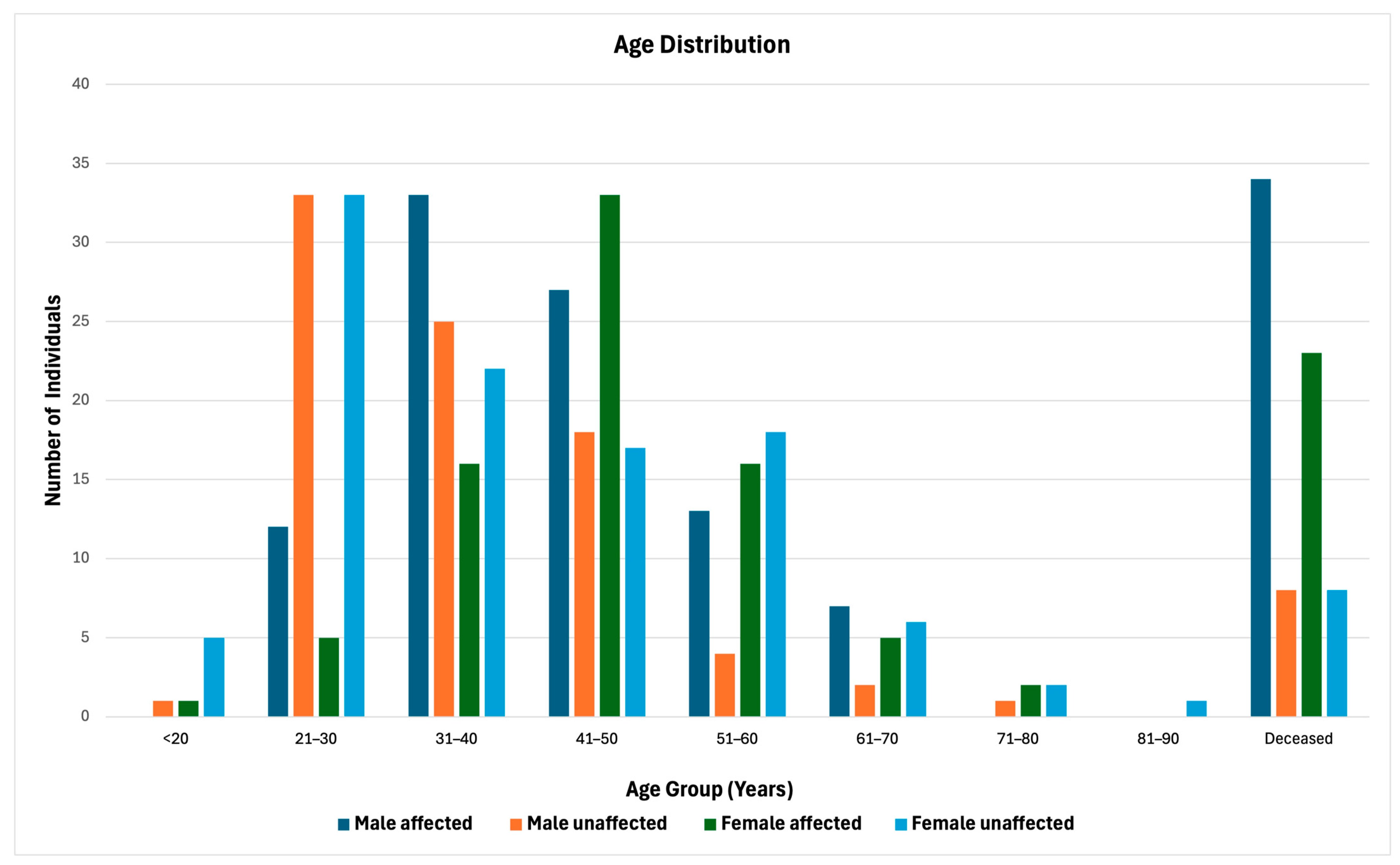
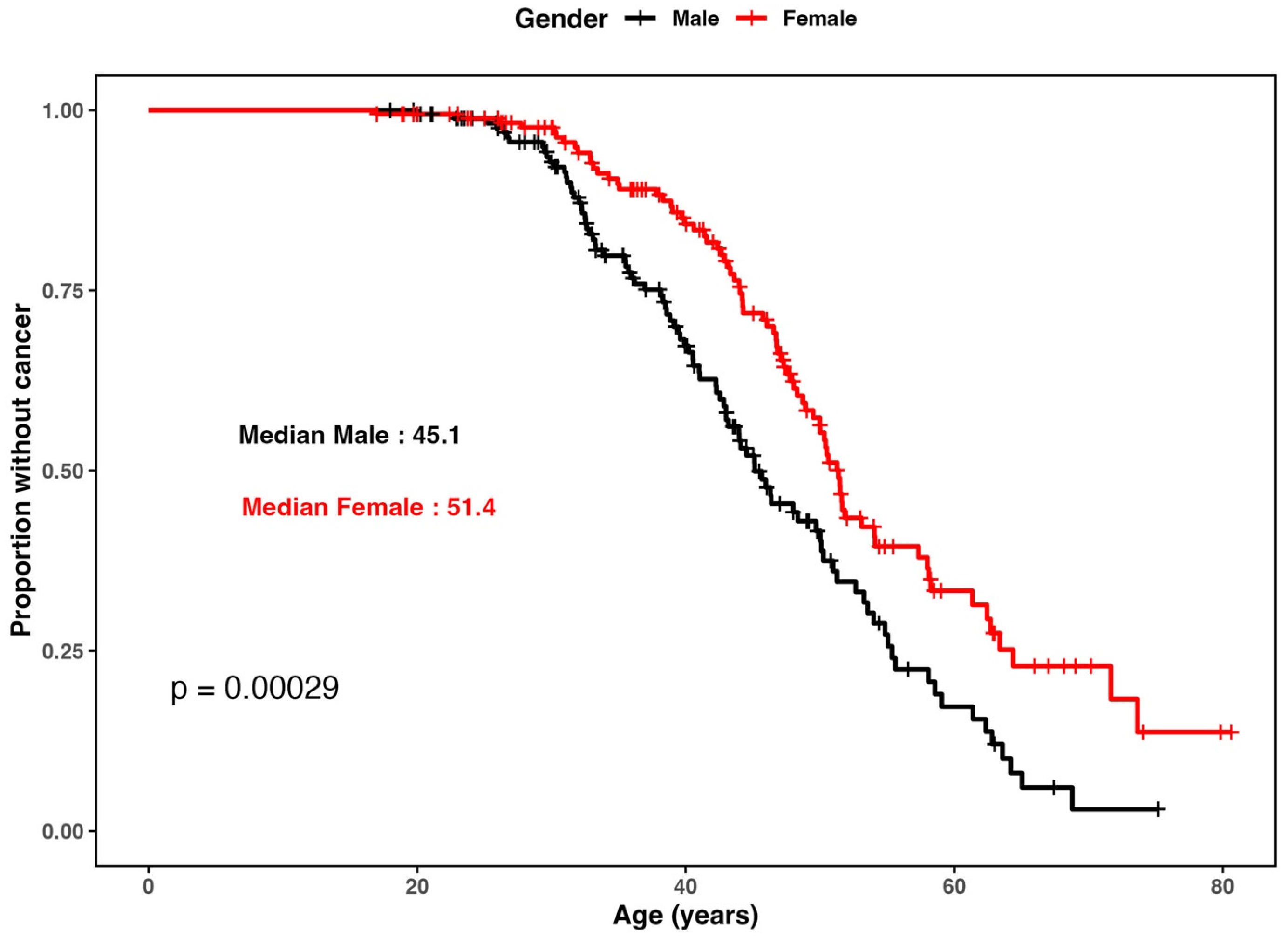
3.1. Patient Cohort and Demographics
3.2. Single Nucleotide Polymorphisms Genotype Frequency Distribution and Hardy–Weinberg Equilibrium Analysis
3.3. Evaluation of the Impact of Polymorphism Genotypes on the Age at First Any-Cancer Diagnosis
- i.
- Identified risk genotypes for age at first any cancer diagnosis.
- ii.
- Identified protective genotypes for age at first any-cancer diagnosis.
3.4. Evaluation of the Impact of Polymorphism Genotype on the Age at First CRC Diagnosis
- i.
- Identified risk genotypes for age at CRC diagnosis.
- ii.
- Identified protective genotypes for age at CRC diagnosis.
3.5. Aggregated Effect of Combined Risk Genotypes
3.6. Aggregated Effect of Combined Protective Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mishra, N.; Hall, J. Identification of patients at risk for hereditary colorectal cancer. Clin. Colon. Rectal Surg. 2012, 25, 67–82. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valle, L.; Vilar, E.; Tavtigian, S.V.; Stoffel, E.M. Genetic predisposition to colorectal cancer: Syndromes, genes, classification of genetic variants and implications for precision medicine. J. Pathol. 2019, 247, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Peltomäki, P.; Nyström, M.; Mecklin, J.P.; Seppälä, T.T. Lynch Syndrome Genetics and Clinical Implications. Gastroenterology 2023, 164, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Krush, A.J. Cancer family “G” revisited: 1895–1970. Cancer 1971, 27, 1505–1511. [Google Scholar] [CrossRef]
- Lynch, H.T.; de la Chapelle, A. Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet. 1999, 36, 801–818. [Google Scholar] [PubMed Central]
- Kempers, M.J.; Kuiper, R.P.; Ockeloen, C.W.; Chappuis, P.O.; Hutter, P.; Rahner, N.; Schackert, H.K.; Steinke, V.; Holinski-Feder, E.; Morak, M.; et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: A cohort study. Lancet Oncol. 2011, 12, 49–55. [Google Scholar] [CrossRef]
- Ligtenberg, M.J.; Kuiper, R.P.; Chan, T.L.; Goossens, M.; Hebeda, K.M.; Voorendt, M.; Lee, T.Y.; Bodmer, D.; Hoenselaar, E.; Hendriks-Cornelissen, S.J.; et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009, 41, 112–117. [Google Scholar] [CrossRef]
- Chen, E.; Xu, X.; Liu, T. Hereditary Nonpolyposis Colorectal Cancer and Cancer Syndromes: Recent Basic and Clinical Discoveries. J. Oncol. 2018, 2018, 3979135. [Google Scholar] [CrossRef]
- Moreira, L.; Balaguer, F.; Lindor, N.; de la Chapelle, A.; Hampel, H.; Aaltonen, L.A.; Hopper, J.L.; Le Marchand, L.; Gallinger, S.; Newcomb, P.A.; et al. Identification of Lynch syndrome among patients with colorectal cancer. Jama 2012, 308, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Baglietto, L.; Lindor, N.M.; Dowty, J.G.; White, D.M.; Wagner, A.; Gomez Garcia, E.B.; Vriends, A.H.; Cartwright, N.R.; Barnetson, R.A.; Farrington, S.M.; et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J. Natl. Cancer Inst. 2010, 102, 193–201. [Google Scholar] [CrossRef]
- Engel, C.; Ahadova, A.; Seppälä, T.T.; Aretz, S.; Bigirwamungu-Bargeman, M.; Bläker, H.; Bucksch, K.; Büttner, R.; de Vos Tot Nederveen Cappel, W.T.; Endris, V.; et al. Associations of Pathogenic Variants in MLH1, MSH2, and MSH6 With Risk of Colorectal Adenomas and Tumors and With Somatic Mutations in Patients With Lynch Syndrome. Gastroenterology 2020, 158, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Bonadona, V.; Bonaïti, B.; Olschwang, S.; Grandjouan, S.; Huiart, L.; Longy, M.; Guimbaud, R.; Buecher, B.; Bignon, Y.J.; Caron, O.; et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. Jama 2011, 305, 2304–2310. [Google Scholar] [CrossRef] [PubMed]
- Talseth-Palmer, B.A.; McPhillips, M.; Groombridge, C.; Spigelman, A.; Scott, R.J. MSH6 and PMS2 mutation positive Australian Lynch syndrome families: Novel mutations, cancer risk and age of diagnosis of colorectal cancer. Hered. Cancer Clin. Pract. 2010, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, F.; Lasset, C.; Carayol, J.; Bonadona, V.; Perdry, H.; Desseigne, F.; Wang, Q.; Bonaïti-Pellié, C. Estimating cancer risk in HNPCC by the GRL method. Eur. J. Hum. Genet. 2007, 15, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Ramsoekh, D.; Wagner, A.; van Leerdam, M.E.; Dooijes, D.; Tops, C.M.; Steyerberg, E.W.; Kuipers, E.J. Cancer risk in MLH1, MSH2 and MSH6 mutation carriers; different risk profiles may influence clinical management. Hered. Cancer Clin. Pract. 2009, 7, 17. [Google Scholar] [CrossRef]
- Win, A.K.; Dowty, J.G.; Reece, J.C.; Lee, G.; Templeton, A.S.; Plazzer, J.P.; Buchanan, D.D.; Akagi, K.; Aksoy, S.; Alonso, A.; et al. Variation in the risk of colorectal cancer in families with Lynch syndrome: A retrospective cohort study. Lancet Oncol. 2021, 22, 1014–1022. [Google Scholar] [CrossRef]
- Cronjé, L.; Paterson, A.C.; Becker, P.J. Colorectal cancer in South Africa: A heritable cause suspected in many young black patients. S. Afr. Med. J. 2009, 99, 103–106. [Google Scholar]
- Katsidzira, L.; Vorster, A.; Gangaidzo, I.T.; Makunike-Mutasa, R.; Govender, D.; Rusakaniko, S.; Thomson, S.; Matenga, J.A.; Ramesar, R. Investigation on the hereditary basis of colorectal cancers in an African population with frequent early onset cases. PLoS ONE 2019, 14, e0224023. [Google Scholar] [CrossRef] [PubMed]
- Liccardo, R.; De Rosa, M.; Duraturo, F. Same MSH2 Gene Mutation But Variable Phenotypes in 2 Families With Lynch Syndrome: Two Case Reports and Review of Genotype-Phenotype Correlation. Clin. Med. Insights Case Rep. 2018, 11, 1179547617753943. [Google Scholar] [CrossRef]
- Felix, R.; Bodmer, W.; Fearnhead, N.S.; van der Merwe, L.; Goldberg, P.; Ramesar, R.S. GSTM1 and GSTT1 polymorphisms as modifiers of age at diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC) in a homogeneous cohort of individuals carrying a single predisposing mutation. Mutat. Res. 2006, 602, 175–181. [Google Scholar] [CrossRef]
- Talseth-Palmer, B.A.; Wijnen, J.T.; Grice, D.M.; Scott, R.J. Genetic modifiers of cancer risk in Lynch syndrome: A review. Fam. Cancer 2013, 12, 207–216. [Google Scholar] [CrossRef]
- Watson, P.; Ashwathnarayan, R.; Lynch, H.T.; Roy, H.K. Tobacco use and increased colorectal cancer risk in patients with hereditary nonpolyposis colorectal cancer (Lynch syndrome). Arch. Intern. Med. 2004, 164, 2429–2431. [Google Scholar] [CrossRef]
- Rahit, K.; Tarailo-Graovac, M. Genetic Modifiers and Rare Mendelian Disease. Genes 2020, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Pandya, D.; Elston, R.C.; Ferlini, C. Defining “mutation” and “polymorphism” in the era of personal genomics. BMC Med. Genom. 2015, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J. Modifier genes and Lynch syndrome: Some considerations. Hered. Cancer Clin. Pract. 2022, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Ndou, L.; Chambuso, R.; Valley-Omar, Z.; Rebello, G.; Algar, U.; Goldberg, P.; Boutall, A.; Ramesar, R. Human Leukocyte Antigen-Allelic Variations May Influence the Age at Cancer Diagnosis in Lynch Syndrome. J. Pers. Med. 2024, 14, 575. [Google Scholar] [CrossRef]
- Thompson, B.A.; Spurdle, A.B.; Plazzer, J.P.; Greenblatt, M.S.; Akagi, K.; Al-Mulla, F.; Bapat, B.; Bernstein, I.; Capellá, G.; den Dunnen, J.T.; et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014, 46, 107–115. [Google Scholar] [CrossRef]
- Goldberg, P.A.; Madden, M.V.; Harocopos, C.; Felix, R.; Westbrook, C.; Ramesar, R.S. In a resource-poor country, mutation identification has the potential to reduce the cost of family management for hereditary nonpolyposis colorectal cancer. Dis. Colon Rectum 1998, 41, 1250–1253; discussion 1253–1255. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Win, A.K.; Hopper, J.L.; Buchanan, D.D.; Young, J.P.; Tenesa, A.; Dowty, J.G.; Giles, G.G.; Goldblatt, J.; Winship, I.; Boussioutas, A.; et al. Are the common genetic variants associated with colorectal cancer risk for DNA mismatch repair gene mutation carriers? Eur. J. Cancer 2013, 49, 1578–1587. [Google Scholar] [CrossRef]
- Bellido, F.; Guinó, E.; Jagmohan-Changur, S.; Seguí, N.; Pineda, M.; Navarro, M.; Lázaro, C.; Blanco, I.; Vasen, H.F.; Moreno, V.; et al. Genetic variant in the telomerase gene modifies cancer risk in Lynch syndrome. Eur. J. Hum. Genet. 2013, 21, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Wijnen, J.T.; Brohet, R.M.; van Eijk, R.; Jagmohan-Changur, S.; Middeldorp, A.; Tops, C.M.; van Puijenbroek, M.; Ausems, M.G.; Gómez García, E.; Hes, F.J.; et al. Chromosome 8q23.3 and 11q23.1 variants modify colorectal cancer risk in Lynch syndrome. Gastroenterology 2009, 136, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Pande, M.; Huang, Y.J.; Wei, C.; Amos, C.I.; Talseth-Palmer, B.A.; Meldrum, C.J.; Chen, W.V.; Gorlov, I.P.; Lynch, P.M.; et al. Cell cycle-related genes as modifiers of age of onset of colorectal cancer in Lynch syndrome: A large-scale study in non-Hispanic white patients. Carcinogenesis 2013, 34, 299–306. [Google Scholar] [CrossRef]
- Loktionov, A.; Watson, M.A.; Gunter, M.; Stebbings, W.S.; Speakman, C.T.; Bingham, S.A. Glutathione-S-transferase gene polymorphisms in colorectal cancer patients: Interaction between GSTM1 and GSTM3 allele variants as a risk-modulating factor. Carcinogenesis 2001, 22, 1053–1060. [Google Scholar] [CrossRef]
- Pande, M.; Amos, C.I.; Osterwisch, D.R.; Chen, J.; Lynch, P.M.; Broaddus, R.; Frazier, M.L. Genetic variation in genes for the xenobiotic-metabolizing enzymes CYP1A1, EPHX1, GSTM1, GSTT1, and GSTP1 and susceptibility to colorectal cancer in Lynch syndrome. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2393–2401. [Google Scholar] [CrossRef]
- Shi, Z.; Johnstone, D.; Talseth-Palmer, B.A.; Evans, T.J.; Spigelman, A.D.; Groombridge, C.; Milward, E.A.; Olynyk, J.K.; Suchy, J.; Kurzawski, G.; et al. Haemochromatosis HFE gene polymorphisms as potential modifiers of hereditary nonpolyposis colorectal cancer risk and onset age. Int. J. Cancer 2009, 125, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Etzel, C.J.; Amos, C.I.; Zhang, Q.; Viscofsky, N.; Lindor, N.M.; Lynch, P.M.; Frazier, M.L. Genetic variants in the cell cycle control pathways contribute to early onset colorectal cancer in Lynch syndrome. Cancer Causes Control 2009, 20, 1769–1777. [Google Scholar] [CrossRef]
- Wiik, M.U.; Negline, M.; Beisvåg, V.; Clapham, M.; Holliday, E.; Dueñas, N.; Brunet, J.; Pineda, M.; Bonifaci, N.; Aretz, S.; et al. MTHFR C677T and A1298C polymorphism’s effect on risk of colorectal cancer in Lynch syndrome. Sci. Rep. 2023, 13, 18783. [Google Scholar] [CrossRef]
- Wiik, M.U.; Evans, T.J.; Belhadj, S.; Bolton, K.A.; Dymerska, D.; Jagmohan-Changur, S.; Capellá, G.; Kurzawski, G.; Wijnen, J.T.; Valle, L.; et al. A genetic variant in telomerase reverse transcriptase (TERT) modifies cancer risk in Lynch syndrome patients harbouring pathogenic MSH2 variants. Sci. Rep. 2021, 11, 11401. [Google Scholar] [CrossRef]
- Talseth, B.A.; Meldrum, C.; Suchy, J.; Kurzawski, G.; Lubinski, J.; Scott, R.J. Genetic polymorphisms in xenobiotic clearance genes and their influence on disease expression in hereditary nonpolyposis colorectal cancer patients. Cancer Epidemiol. Biomark. Prev. 2006, 15, 2307–2310. [Google Scholar] [CrossRef][Green Version]
- Shin, J.H.; Ku, J.L.; Shin, K.H.; Shin, Y.K.; Kang, S.B.; Park, J.G. Glutathione S-transferase M1 associated with cancer occurrence in Korean HNPCC families carrying the hMLH1/hMSH2 mutation. Oncol. Rep. 2003, 10, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.T.; Edwards, L.; McLaughlin, J.R.; Green, J.; Younghusband, H.B.; Woods, M.O. Cytochrome P450 17A1 and catechol O-methyltransferase polymorphisms and age at Lynch syndrome colon cancer onset in Newfoundland. Clin. Cancer Res. 2007, 13, 3783–3788. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Stanley, L.A.; Sim, E.; Strange, R.C.; Wolf, C.R. Metabolic polymorphisms and cancer susceptibility. Cancer Surv. 1995, 25, 27–65. [Google Scholar]
- Singh, R.R.; Reindl, K.M. Glutathione S-Transferases in Cancer. Antioxidants 2021, 10, 701. [Google Scholar] [CrossRef]
- Khan, A.; Jahan, F.; Zahoor, M.; Ullah, R.; Albadrani, G.M.; Mohamed, H.R.H.; Khisroon, M. Association of genetic polymorphism of glutathione S-transferases with colorectal cancer susceptibility in snuff (Naswar) addicts. Braz. J. Biol. 2022, 84, e261509. [Google Scholar] [CrossRef]
- Stojkovic Lalosevic, M.L.; Coric, V.M.; Pekmezovic, T.D.; Simic, T.P.; Pljesa Ercegovac, M.S.; Pavlovic Markovic, A.R.; Krivokapic, Z.V. Deletion and Single Nucleotide Polymorphisms in Common Glutathione-S Transferases Contribute to Colorectal Cancer Development. Pathol. Oncol. Res. 2019, 25, 1579–1587. [Google Scholar] [CrossRef]
- Kamiza, A.B.; You, J.F.; Wang, W.C.; Tang, R.; Chang, C.Y.; Chien, H.T.; Lai, C.H.; Chiu, L.L.; Lo, T.P.; Hung, K.Y.; et al. Polymorphisms of xenobiotic-metabolizing genes and colorectal cancer risk in patients with lynch syndrome: A retrospective cohort study in Taiwan. Environ. Mol. Mutagen. 2018, 59, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Mesic, A.; Rogar, M.; Hudler, P.; Bilalovic, N.; Eminovic, I.; Komel, R. Genetic variations in AURORA cell cycle kinases are associated with glioblastoma multiforme. Sci. Rep. 2021, 11, 17444. [Google Scholar] [CrossRef]
- Köberle, B.; Koch, B.; Fischer, B.M.; Hartwig, A. Single nucleotide polymorphisms in DNA repair genes and putative cancer risk. Arch. Toxicol. 2016, 90, 2369–2388. [Google Scholar] [CrossRef]
- Shieu, M.K.; Ho, H.Y.; Lin, S.H.; Lo, Y.S.; Lin, C.C.; Chuang, Y.C.; Hsieh, M.J.; Chen, M.K. Association of KMT2C Genetic Variants with the Clinicopathologic Development of Oral Cancer. Int. J. Environ. Res. Public Health 2022, 19, 3974. [Google Scholar] [CrossRef] [PubMed]
- Kamiza, A.B.; Hsieh, L.L.; Tang, R.; Chien, H.T.; Lai, C.H.; Chiu, L.L.; Lo, T.P.; Hung, K.Y.; You, J.F.; Wang, W.C.; et al. Polymorphisms of DNA repair genes are associated with colorectal cancer in patients with Lynch syndrome. Mol. Genet. Genom. Med. 2018, 6, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Forciniti, S.; Greco, L.; Grizzi, F.; Malesci, A.; Laghi, L. Iron Metabolism in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2257. [Google Scholar] [CrossRef]
- Huang, L.; Li, W.; Lu, Y.; Ju, Q.; Ouyang, M. Iron metabolism in colorectal cancer. Front. Oncol. 2023, 13, 1098501. [Google Scholar] [CrossRef]
- Lv, Y.F.; Chang, X.; Hua, R.X.; Yan, G.N.; Meng, G.; Liao, X.Y.; Zhang, X.; Guo, Q.N. The risk of new-onset cancer associated with HFE C282Y and H63D mutations: Evidence from 87,028 participants. J. Cell. Mol. Med. 2016, 20, 1219–1233. [Google Scholar] [CrossRef]
- Gaspar, T.B.; Sá, A.; Lopes, J.M.; Sobrinho-Simões, M.; Soares, P.; Vinagre, J. Telomere Maintenance Mechanisms in Cancer. Genes 2018, 9, 241. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.A.; Ong, B. The MassARRAY(®) System for Targeted SNP Genotyping. Methods Mol. Biol. 2017, 1492, 77–94. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Krnajski, Z.; Geering, S.; Steadman, S. Performance verification of the Maxwell 16 Instrument and DNA IQ Reference Sample Kit for automated DNA extraction of known reference samples. Forensic Sci. Med. Pathol. 2007, 3, 264–269. [Google Scholar] [CrossRef]
- Moisio, A.L.; Sistonen, P.; Mecklin, J.P.; Järvinen, H.; Peltomäki, P. Genetic polymorphisms in carcinogen metabolism and their association to hereditary nonpolyposis colon cancer. Gastroenterology 1998, 115, 1387–1394. [Google Scholar] [CrossRef]
- Bell, D.A.; Taylor, J.A.; Paulson, D.F.; Robertson, C.N.; Mohler, J.L.; Lucier, G.W. Genetic risk and carcinogen exposure: A common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J. Natl. Cancer Inst. 1993, 85, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Pemble, S.; Schroeder, K.R.; Spencer, S.R.; Meyer, D.J.; Hallier, E.; Bolt, H.M.; Ketterer, B.; Taylor, J.B. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem. J. 1994, 300 Pt 1, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.W. G. H. Hardy (1908) and Hardy-Weinberg equilibrium. Genetics 2008, 179, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Graffelman, J.; Jain, D.; Weir, B. A genome-wide study of Hardy-Weinberg equilibrium with next generation sequence data. Hum. Genet. 2017, 136, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Seppälä, T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Evans, D.G.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.; et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: First report from the prospective Lynch syndrome database. Gut 2017, 66, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Androutsopoulos, V.P.; Tsatsakis, A.M.; Spandidos, D.A. Cytochrome P450 CYP1A1: Wider roles in cancer progression and prevention. BMC Cancer 2009, 9, 187. [Google Scholar] [CrossRef]
- Bag, A.; Jyala, N.S.; Bag, N. Cytochrome P450 1A1 genetic polymorphisms as cancer biomarkers. Indian J. Cancer 2015, 52, 479–489. [Google Scholar] [CrossRef]
- Ibrahem, S.Q.; Al-Dalawi, Z.T.; Bahaaldin, A.S. Sequence Polymorphism in Xenobiotic Metabolising Genes in Iraqi Colorectal Cancer Patients. Asian Pac. J. Cancer Prev. 2021, 22, 1203–1210. [Google Scholar] [CrossRef]
- Kang, D.; Chen, J.; Wong, J.; Fang, G. The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J. Cell Biol. 2002, 156, 249–259. [Google Scholar] [CrossRef]
- Landi, S. Mammalian class theta GST and differential susceptibility to carcinogens: A review. Mutat. Res. 2000, 463, 247–283. [Google Scholar] [CrossRef]
- Kim, W.J.; Kim, H.; Kim, C.H.; Lee, M.S.; Oh, B.R.; Lee, H.M.; Katoh, T. GSTT1-null genotype is a protective factor against bladder cancer. Urology 2002, 60, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Reeves, S.G.; Meldrum, C.; Groombridge, C.; Spigelman, A.; Suchy, J.; Kurzawski, G.; Lubinski, J.; Scott, R.J. DNA repair gene polymorphisms and risk of early onset colorectal cancer in Lynch syndrome. Cancer Epidemiol. 2012, 36, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Wei, Z.; Yang, W.; Huang, J.; Yang, Y.; Wang, J. The role of BCL-2 family proteins in regulating apoptosis and cancer therapy. Front. Oncol. 2022, 12, 985363. [Google Scholar] [CrossRef]
- Hayden, P.J.; Tewari, P.; Morris, D.W.; Staines, A.; Crowley, D.; Nieters, A.; Becker, N.; de Sanjosé, S.; Foretova, L.; Maynadié, M.; et al. Variation in DNA repair genes XRCC3, XRCC4, XRCC5 and susceptibility to myeloma. Hum. Mol. Genet. 2007, 16, 3117–3127. [Google Scholar] [CrossRef]
- Li, H.; Zimmerman, S.E.; Weyemi, U. Genomic instability and metabolism in cancer. Int. Rev. Cell Mol. Biol. 2021, 364, 241–265. [Google Scholar] [CrossRef] [PubMed]
- Moretton, A.; Loizou, J.I. Interplay between Cellular Metabolism and the DNA Damage Response in Cancer. Cancers 2020, 12, 2051. [Google Scholar] [CrossRef]
- Houlle, S.; Charbonnier, F.; Houivet, E.; Tinat, J.; Buisine, M.P.; Caron, O.; Benichou, J.; Baert-Desurmont, S.; Frebourg, T. Evaluation of Lynch syndrome modifier genes in 748 MMR mutation carriers. Eur. J. Hum. Genet. 2011, 19, 887–892. [Google Scholar] [CrossRef]
- Shah, P.P.; Saurabh, K.; Pant, M.C.; Mathur, N.; Parmar, D. Evidence for increased cytochrome P450 1A1 expression in blood lymphocytes of lung cancer patients. Mutat. Res. 2009, 670, 74–78. [Google Scholar] [CrossRef]
- Agundez, J.A. Cytochrome P450 gene polymorphism and cancer. Curr. Drug Metab. 2004, 5, 211–224. [Google Scholar] [CrossRef]
- Zhang, Z.; Ji, M.; Li, J.; Wu, Q.; Huang, Y.; He, G.; Xu, J. Molecular Classification Based on Prognostic and Cell Cycle-Associated Genes in Patients With Colon Cancer. Front. Oncol. 2021, 11, 636591. [Google Scholar] [CrossRef]
- Heinimann, K.; Scott, R.J.; Chappuis, P.; Weber, W.; Müller, H.; Dobbie, Z.; Hutter, P. N-acetyltransferase 2 influences cancer prevalence in hMLH1/hMSH2 mutation carriers. Cancer Res. 1999, 59, 3038–3040. [Google Scholar] [PubMed]
- Cibeira, M.T.; de Larrea, C.F.; Navarro, A.; Díaz, T.; Fuster, D.; Tovar, N.; Rosiñol, L.; Monzó, M.; Bladé, J. Impact on response and survival of DNA repair single nucleotide polymorphisms in relapsed or refractory multiple myeloma patients treated with thalidomide. Leuk. Res. 2011, 35, 1178–1183. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, L.; Wang, Y.; Yin, J.; Li, X.; Wang, Z.; Li, H.; Zou, T.; Qian, C.; Li, C.; et al. Effect of transporter and DNA repair gene polymorphisms to lung cancer chemotherapy toxicity. Tumour Biol. 2016, 37, 2275–2284. [Google Scholar] [CrossRef]
- Zou, T.; Liu, J.Y.; Qin, Q.; Guo, J.; Zhou, W.Z.; Li, X.P.; Zhou, H.H.; Chen, J.; Liu, Z.Q. Role of rs873601 Polymorphisms in Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy. Biomedicines 2023, 11, 3133. [Google Scholar] [CrossRef]
- Talseth-Palmer, B.A.; Wijnen, J.T.; Brenne, I.S.; Jagmohan-Changur, S.; Barker, D.; Ashton, K.A.; Tops, C.M.; Evans, T.J.; McPhillips, M.; Groombridge, C.; et al. Combined analysis of three Lynch syndrome cohorts confirms the modifying effects of 8q23.3 and 11q23.1 in MLH1 mutation carriers. Int. J. Cancer 2013, 132, 1556–1564. [Google Scholar] [CrossRef]
- Zhu, T.; Gao, Y.; Wang, J.; Li, X.; Shang, S.; Wang, Y.; Guo, S.; Zhou, H.; Liu, H.; Sun, D.; et al. CancerClock: A DNA Methylation Age Predictor to Identify and Characterize Aging Clock in Pan-Cancer. Front. Bioeng. Biotechnol. 2019, 7, 388. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.E.; Clendenning, M.; Wong, E.M.; Rosty, C.; Mahmood, K.; Georgeson, P.; Winship, I.M.; Preston, S.G.; Win, A.K.; Dugué, P.A.; et al. DNA Methylation Signatures and the Contribution of Age-Associated Methylomic Drift to Carcinogenesis in Early-Onset Colorectal Cancer. Cancers 2021, 13, 2589. [Google Scholar] [CrossRef]
- Sarno, F.; Benincasa, G.; List, M.; Barabasi, A.L.; Baumbach, J.; Ciardiello, F.; Filetti, S.; Glass, K.; Loscalzo, J.; Marchese, C.; et al. Clinical epigenetics settings for cancer and cardiovascular diseases: Real-life applications of network medicine at the bedside. Clin. Epigenetics 2021, 13, 66. [Google Scholar] [CrossRef]
- Pastor, D.M.; Schlom, J. Immunology of Lynch Syndrome. Curr. Oncol. Rep. 2021, 23, 96. [Google Scholar] [CrossRef]
- Kloor, M.; Michel, S.; von Knebel Doeberitz, M. Immune evasion of microsatellite unstable colorectal cancers. Int. J. Cancer 2010, 127, 1001–1010. [Google Scholar] [CrossRef]
- Chambuso, R.; Mthembu, M.; Kaambo, E.; Robertson, B.; Ramesar, R. Immunogenomic Biomarkers and Validation in Lynch Syndrome. Cells 2023, 12, 491. [Google Scholar] [CrossRef] [PubMed]
| Character | N (%) N = 359 | Male (176) | Female (183) | p-Value |
|---|---|---|---|---|
| Status | ||||
| Cancer unaffected | 189 | 84 (47.7%) | 105 (57.4%) | 0.085 |
| Cancer affected | 170 | 92 (52.3%) | 78 (42.6%) | |
| Cancer-Affected | ||||
| Age at cancer diagnosis | ||||
| Mean (SD) | 170 | 41.8 (10.9) | 45.5 (10.6) | 0.028 |
| Median [Min, Max] | 170 | 40.5 [19.9, 68.8] | 46.6 [17.0, 73.6] | |
| Cancer type | ||||
| CRC | 136 | 86 (93.5%) | 50 (64.1%) | <0.001 |
| Extra-colonic 1 | 34 | 6 (6.5%) | 28 (35.9%) | |
| CRC histological grade | ||||
| Poorly differentiated | 23 | 20 (23.3%) | 3 (6.0%) | 0.009 |
| Moderately differentiated | 113 | 66 (76.7%) | 47 (94.0%) | |
| CRC histological type | ||||
| Adenocarcinoma | 116 | 70 (81.4%) | 46 (92.0%) | 0.132 |
| Mucinous adenocarcinoma | 20 | 16 (18.6%) | 4 (8.0%) | |
| Cancer recurrence | ||||
| Yes | 36 | 15 (16.3%) | 21 (26.9%) | 0.134 |
| No | 134 | 77 (83.7%) | 57 (73.1%) | |
| Number of Genotypes | Unadjusted HR (95% CI) | p-Value | # p-Value | * Adjusted HR (95% CI) | p-Value | # p-Value |
|---|---|---|---|---|---|---|
| No cancer risk genotype | Ref | Ref | ||||
| One to Two any cancer risk genotypes | 1.44 (1.03–2.00) | 0.031 | 0.050 | 1.49 (1.06–2.09) | 0.020 | 0.030 |
| Number of Genotypes | Unadjusted HR (95% CI) | p-Value | # p-Value | * Adjusted HR (95% CI) | p-Value | # p-Value |
|---|---|---|---|---|---|---|
| No cancer risk genotype | Ref | Ref | ||||
| One to two any cancer protective genotypes | 0.49 (0.35–0.69) | <0.001 | <0.001 | 0.52 (0.37–0.73) | <0.001 | <0.001 |
| One to two CRC protective genotypes | 0.50 (0.34–0.72) | <0.001 | <0.001 | 0.51 (0.36–0.74) | <0.001 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndou, L.; Chambuso, R.; Algar, U.; Goldberg, P.; Boutall, A.; Ramesar, R. Influence of Genetic Polymorphisms on the Age at Cancer Diagnosis in a Homogenous Lynch Syndrome Cohort of Individuals Carrying the MLH1:c.1528C>T South African Founder Variant. Biomedicines 2024, 12, 2201. https://doi.org/10.3390/biomedicines12102201
Ndou L, Chambuso R, Algar U, Goldberg P, Boutall A, Ramesar R. Influence of Genetic Polymorphisms on the Age at Cancer Diagnosis in a Homogenous Lynch Syndrome Cohort of Individuals Carrying the MLH1:c.1528C>T South African Founder Variant. Biomedicines. 2024; 12(10):2201. https://doi.org/10.3390/biomedicines12102201
Chicago/Turabian StyleNdou, Lutricia, Ramadhani Chambuso, Ursula Algar, Paul Goldberg, Adam Boutall, and Raj Ramesar. 2024. "Influence of Genetic Polymorphisms on the Age at Cancer Diagnosis in a Homogenous Lynch Syndrome Cohort of Individuals Carrying the MLH1:c.1528C>T South African Founder Variant" Biomedicines 12, no. 10: 2201. https://doi.org/10.3390/biomedicines12102201
APA StyleNdou, L., Chambuso, R., Algar, U., Goldberg, P., Boutall, A., & Ramesar, R. (2024). Influence of Genetic Polymorphisms on the Age at Cancer Diagnosis in a Homogenous Lynch Syndrome Cohort of Individuals Carrying the MLH1:c.1528C>T South African Founder Variant. Biomedicines, 12(10), 2201. https://doi.org/10.3390/biomedicines12102201





