CD44 Is Associated with Poor Prognosis of ccRCC and Facilitates ccRCC Cell Migration and Invasion through HAS1/MMP9
Abstract
1. Introduction
2. Material and Methods
2.1. Bioinformatics Analysis
2.2. Clinical ccRCC Samples
2.3. Immunohistochemistry
2.4. ccRCC Cell Lines
2.5. Lentivirus and Cell Transfection
2.6. Colony Formation Assay
2.7. Cell Counting Kit-8 (CCK-8) Assay
2.8. Transwell Migration and Invasion Analysis
2.9. Real-Time Quantitative RT-PCR
2.10. Western Blotting Analysis
2.11. Animal Experiments
2.12. Statistical Analysis
3. Results
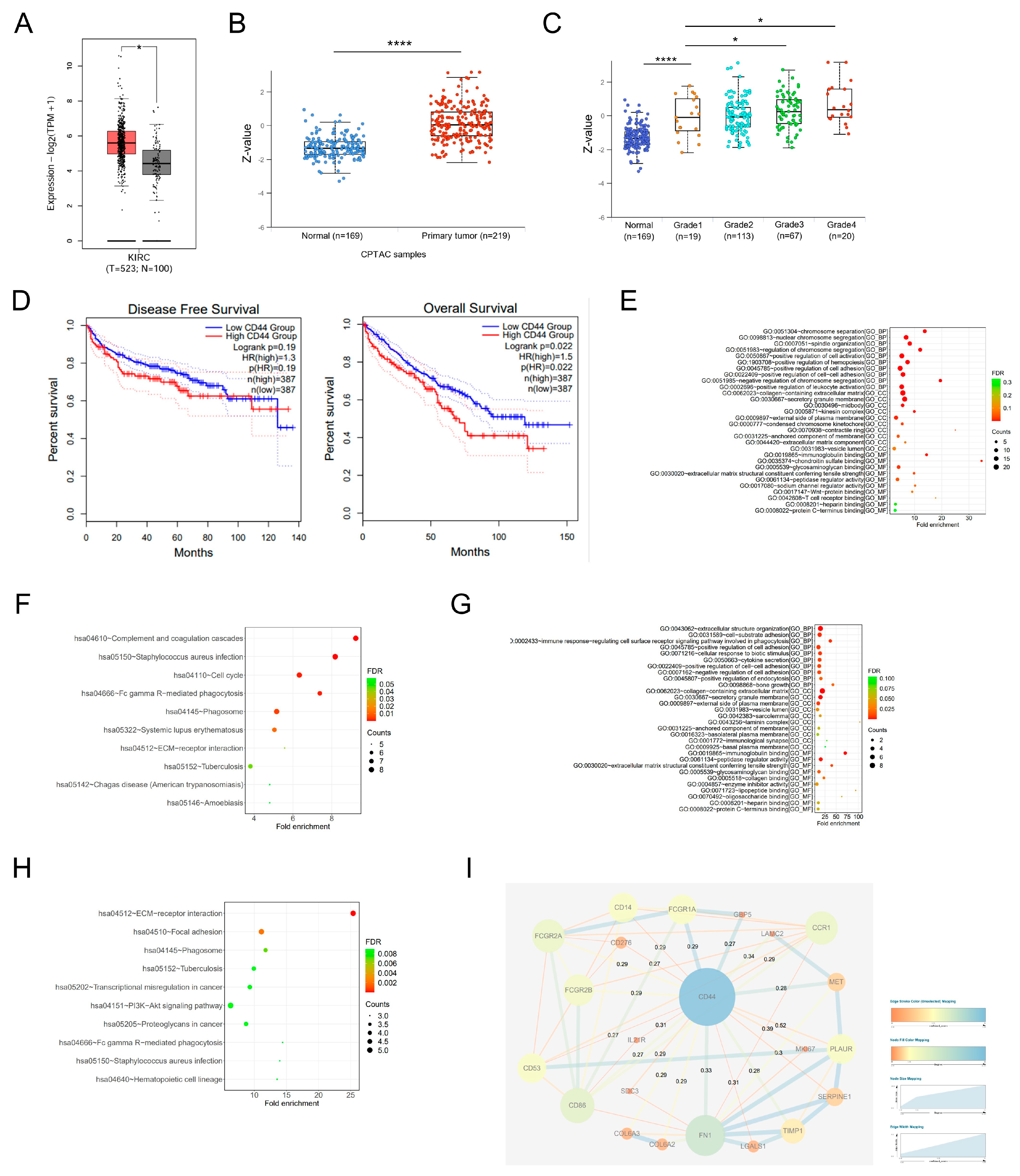
3.1. Elevated Expression of CD44 Is Indicative of an Unfavorable Prognosis among Patients with ccRCC
3.2. CD44 Is Overexpressed in ccRCC Samples and Is Significantly Correlated with Tumor Stage
3.3. CD44 Facilitates ccRCC Cell Proliferation, Migration, and Invasion In Vitro
3.4. CD44 Regulates MMP9 in ccRCC by Up-Regulating HAS1 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lih, T.M.; Dhanasekaran, S.M.; Mannan, R.; Chen, L.; Cieslik, M.; Wu, Y.; Lu, R.J.; Clark, D.J.; Kolodziejczak, I.; et al. Histopathologic and proteogenomic heterogeneity reveals features of clear cell renal cell carcinoma aggressiveness. Cancer Cell 2023, 41, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Jonasch, E.; Walker, C.L.; Rathmell, W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2021, 17, 245–261. [Google Scholar] [CrossRef]
- Motzer, R.J.; Bukowski, R.M.; Figlin, R.A.; Hutson, T.E.; Michaelson, M.D.; Kim, S.T.; Baum, C.M.; Kattan, M.W. Prognostic nomogram for sunitinib in patients with metastatic renal cell carcinoma. Cancer-Am. Cancer Soc. 2008, 113, 1552–1558. [Google Scholar] [CrossRef] [PubMed]
- Nerich, V.; Hugues, M.; Paillard, M.J.; Borowski, L.; Nai, T.; Stein, U.; Nguyen, T.H.T.; Montcuquet, P.; Maurina, T.; Mouillet, G.; et al. Clinical impact of targeted therapies in patients with metastatic clear-cell renal cell carcinoma. Oncotargets Ther. 2014, 7, 365–374. [Google Scholar] [CrossRef]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Sottnik, J.L.; Dancik, G.M.; Sahu, D.; Hansel, D.E.; Theodorescu, D.; Schwartz, M.A. An Osteopontin/CD44 Axis in RhoGDI2-Mediated Metastasis Suppression. Cancer Cell 2016, 30, 432–443. [Google Scholar] [CrossRef]
- Louderbough, J.M.; Schroeder, J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol. Cancer Res. 2011, 9, 1573–1586. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, Y.; Xie, L.; Huang, A.; Xue, C.; Gu, Z.; Wang, K.; Zong, S. The Prognostic and Clinical Value of CD44 in Colorectal Cancer: A Meta-Analysis. Front. Oncol. 2019, 9, 309. [Google Scholar] [CrossRef]
- Desai, B.; Rogers, M.J.; Chellaiah, M.A. Mechanisms of osteopontin and CD44 as metastatic principles in prostate cancer cells. Mol. Cancer 2007, 6, 18. [Google Scholar] [CrossRef]
- Sacks, J.D.; Barbolina, M.V. Expression and Function of CD44 in Epithelial Ovarian Carcinoma. Biomolecules 2015, 5, 3051–3066. [Google Scholar] [CrossRef]
- Yoon, C.; Park, D.J.; Schmidt, B.; Thomas, N.J.; Lee, H.J.; Kim, T.S.; Janjigian, Y.Y.; Cohen, D.J.; Yoon, S.S. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin. Cancer Res. 2014, 20, 3974–3988. [Google Scholar] [CrossRef] [PubMed]
- Nallasamy, P.; Nimmakayala, R.K.; Karmakar, S.; Leon, F.; Seshacharyulu, P.; Lakshmanan, I.; Rachagani, S.; Mallya, K.; Zhang, C.; Ly, Q.P.; et al. Pancreatic Tumor Microenvironment Factor Promotes Cancer Stemness via SPP1-CD44 Axis. Gastroenterology 2021, 161, 1998–2013. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Won, M.; Kim, J.H.; Jung, E.; Min, K.; Jangili, P.; Kim, J.S. Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem. Soc. Rev. 2020, 49, 7856–7878. [Google Scholar] [CrossRef]
- Weng, X.; Maxwell-Warburton, S.; Hasib, A.; Ma, L.; Kang, L. The membrane receptor CD44: Novel insights into metabolism. Trends Endocrinol. Metab. 2022, 33, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Lee, Y.M.; Kim, J.M.; Lee, H.J.; Seong, I.O.; Kim, K.H. Immunohistochemical expression of CD44, matrix metalloproteinase2 and matrix metalloproteinase9 in renal cell carcinomas. Urology 2019, 37, 742–748. [Google Scholar] [CrossRef]
- Ma, J.; Li, M.; Chai, J.; Wang, K.; Li, P.; Liu, Y.; Zhao, D.; Xu, J.; Yu, K.; Yan, Q.; et al. Expression of RSK4, CD44 and MMP-9 is upregulated and positively correlated in metastatic ccRCC. Diag. Pathol. 2020, 15, 28. [Google Scholar] [CrossRef]
- Wang, X.M.; Lu, Y.; Song, Y.M.; Dong, J.; Li, R.Y.; Wang, G.L.; Wang, X.; Zhang, S.D.; Dong, Z.H.; Lu, M.; et al. Integrative genomic study of Chinese clear cell renal cell carcinoma reveals features associated with thrombus. Nat. Commun. 2020, 11, 739. [Google Scholar] [CrossRef]
- Siiskonen, H.; Oikari, S.; Pasonen-Seppanen, S.; Rilla, K. Hyaluronan synthase 1: A mysterious enzyme with unexpected functions. Front. Immunol. 2015, 6, 43. [Google Scholar] [CrossRef]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R.; Stach, K. MMP9: A Tough Target for Targeted Therapy for Cancer. Cancers 2022, 14, 1847. [Google Scholar] [CrossRef]
- Zhang, Y.; Thant, A.A.; Machida, K.; Ichigotani, Y.; Naito, Y.; Hiraiwa, Y.; Senga, T.; Sohara, Y.; Matsuda, S.; Hamaguchi, M. Hyaluronan-CD44s signaling regulates matrix metalloproteinase-2 secretion in a human lung carcinoma cell line QG90. Cancer Res. 2002, 62, 3962–3965. [Google Scholar]
- Psutka, S.P.; Leibovich, B.C. Management of inferior vena cava tumor thrombus in locally advanced renal cell carcinoma. Ther. Adv. Urol. 2015, 7, 216–229. [Google Scholar] [CrossRef]
- Hassn, M.M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef]
- Wang, F.; Scoville, D.; He, X.C.; Mahe, M.M.; Box, A.; Perry, J.M.; Smith, N.R.; Lei, N.Y.; Davies, P.S.; Fuller, M.K.; et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology 2013, 145, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell Physiol. Biochem. 2018, 46, 860–872. [Google Scholar] [CrossRef]
- Vermeulen, L.; de Sousa, E.M.F.; Richel, D.J.; Medema, J.P. The developing cancer stem-cell model: Clinical challenges and opportunities. Lancet Oncol. 2012, 13, e83–e89. [Google Scholar] [CrossRef]
- Liu, T.; Jiang, L.; Tavana, O.; Gu, W. The Deubiquitylase OTUB1 Mediates Ferroptosis via Stabilization of SLC7A11. Cancer Res. 2019, 79, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef]
- Zanjani, L.S.; Madjd, Z.; Abolhasani, M.; Rasti, A.; Fodstad, O.; Andersson, Y.; Asgari, M. Increased expression of CD44 is associated with more aggressive behavior in clear cell renal cell carcinoma. Biomark. Med. 2018, 12, 45–61. [Google Scholar] [CrossRef]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Gotte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef]
- Skandalis, S.S.; Karalis, T.; Heldin, P. Intracellular hyaluronan: Importance for cellular functions. Semin. Cancer Biol. 2020, 62, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, G.E.; Akim, A.; Naveed, Z.M.; Safdar, N.; Sung, Y.Y.; Muhammad, T. Understanding Hyaluronan Receptor (CD44) Interaction, HA-CD44 Activated Potential Targets in Cancer Therapeutics. Adv. Pharm. Bull. 2021, 11, 426–438. [Google Scholar] [CrossRef]
- Liu, S.; Cheng, C. Akt Signaling Is Sustained by a CD44 Splice Isoform-Mediated Positive Feedback Loop. Cancer Res. 2017, 77, 3791–3801. [Google Scholar] [CrossRef]
- Golshani, R.; Hautmann, S.H.; Estrella, V.; Cohen, B.L.; Kyle, C.C.; Manoharan, M.; Jorda, M.; Soloway, M.S.; Lokeshwar, V.B. HAS1 expression in bladder cancer and its relation to urinary HA test. Int. J. Cancer 2007, 120, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Huang, J.; Yao, J.; Li, N.; Yang, Z. miR-125a regulates HAS1 and inhibits the proliferation, invasion and metastasis by targeting STAT3 in non-small cell lung cancer cells. J. Cell Biochem. 2020, 121, 3197–3207. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef]
- Niland, S.; Riscanevo, A.X.; Eble, J.A. Matrix Metalloproteinases Shape the Tumor Microenvironment in Cancer Progression. Int. J. Mol. Sci. 2021, 23, 146. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, T.; Wu, Z.; Wu, Y.; Liu, Y.; Song, Y.; Ma, L. CD44 Is Associated with Poor Prognosis of ccRCC and Facilitates ccRCC Cell Migration and Invasion through HAS1/MMP9. Biomedicines 2023, 11, 2077. https://doi.org/10.3390/biomedicines11072077
Du T, Wu Z, Wu Y, Liu Y, Song Y, Ma L. CD44 Is Associated with Poor Prognosis of ccRCC and Facilitates ccRCC Cell Migration and Invasion through HAS1/MMP9. Biomedicines. 2023; 11(7):2077. https://doi.org/10.3390/biomedicines11072077
Chicago/Turabian StyleDu, Tan, Zonglong Wu, Yaqian Wu, Yunchong Liu, Yimeng Song, and Lulin Ma. 2023. "CD44 Is Associated with Poor Prognosis of ccRCC and Facilitates ccRCC Cell Migration and Invasion through HAS1/MMP9" Biomedicines 11, no. 7: 2077. https://doi.org/10.3390/biomedicines11072077
APA StyleDu, T., Wu, Z., Wu, Y., Liu, Y., Song, Y., & Ma, L. (2023). CD44 Is Associated with Poor Prognosis of ccRCC and Facilitates ccRCC Cell Migration and Invasion through HAS1/MMP9. Biomedicines, 11(7), 2077. https://doi.org/10.3390/biomedicines11072077





