Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Chemicals and Reagents
2.3. Immunofluorescence Staining and Microscopy Imaging
2.4. Western Blotting
2.5. Ribonucleoprotein Immunoprecipitation and RNA Immunoprecipitation
2.6. Virus and Virus Infection
2.7. Animal Study
3. Results
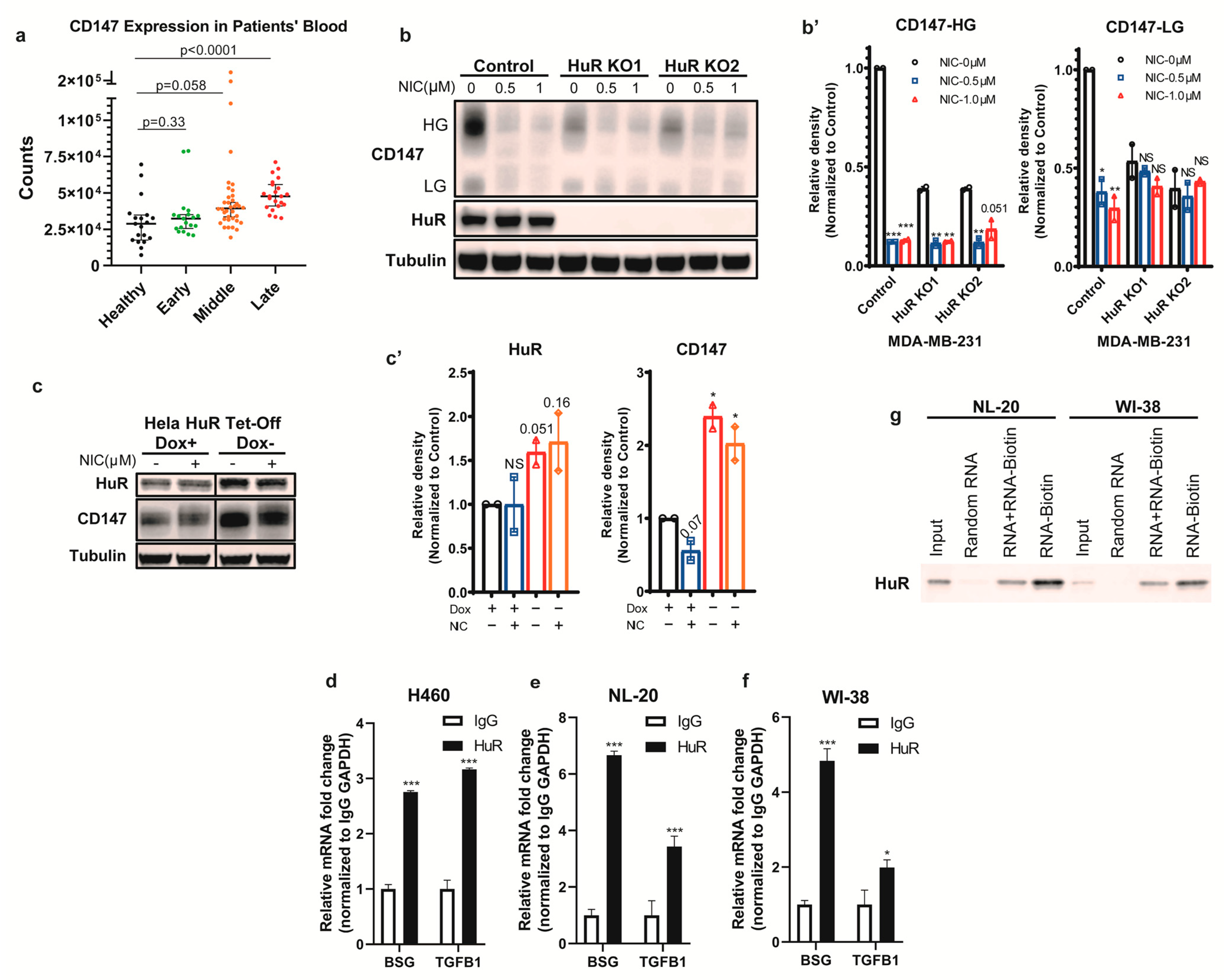
3.1. HuR Binds BSG mRNA and Regulates CD147 Translation
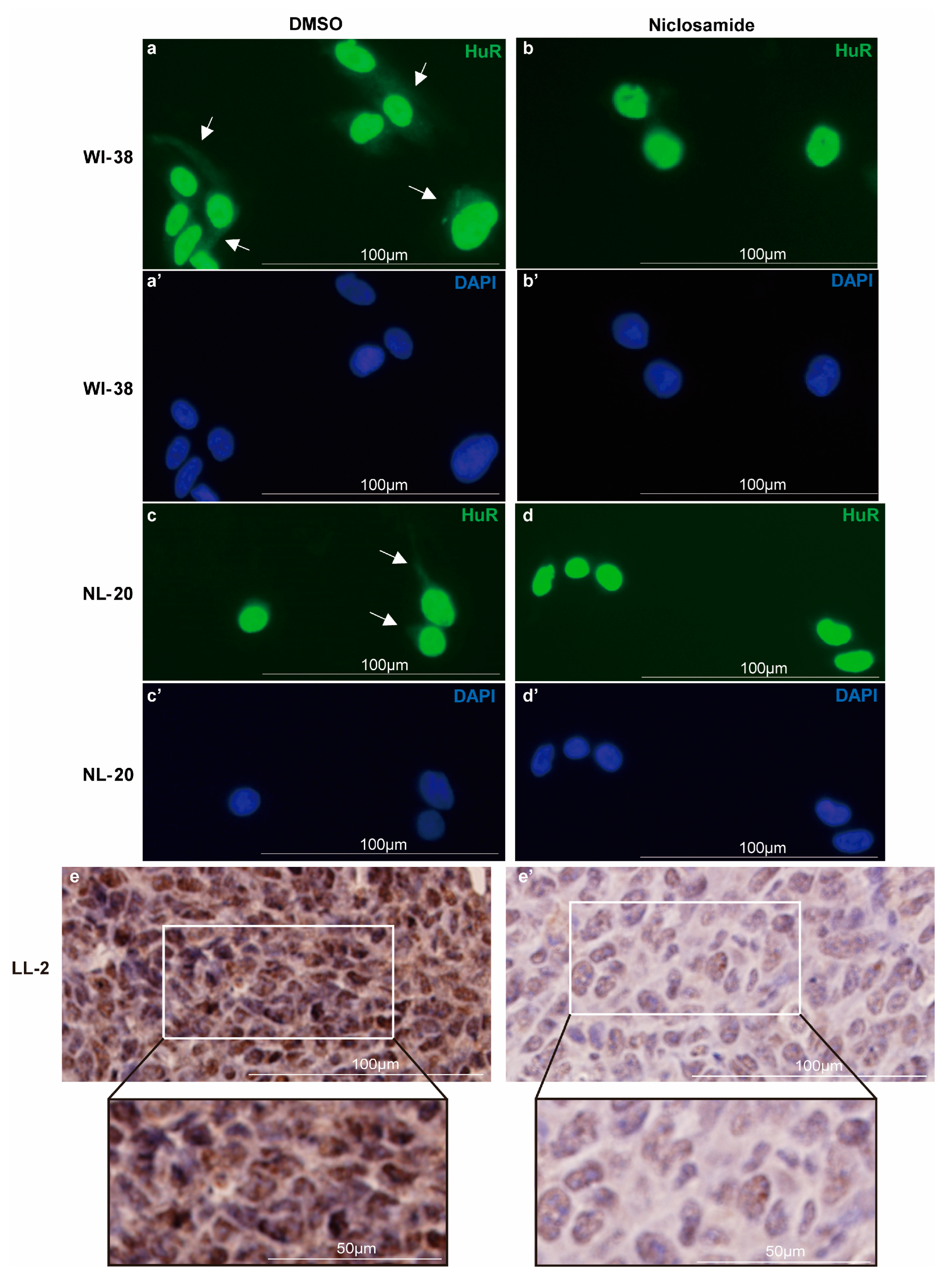
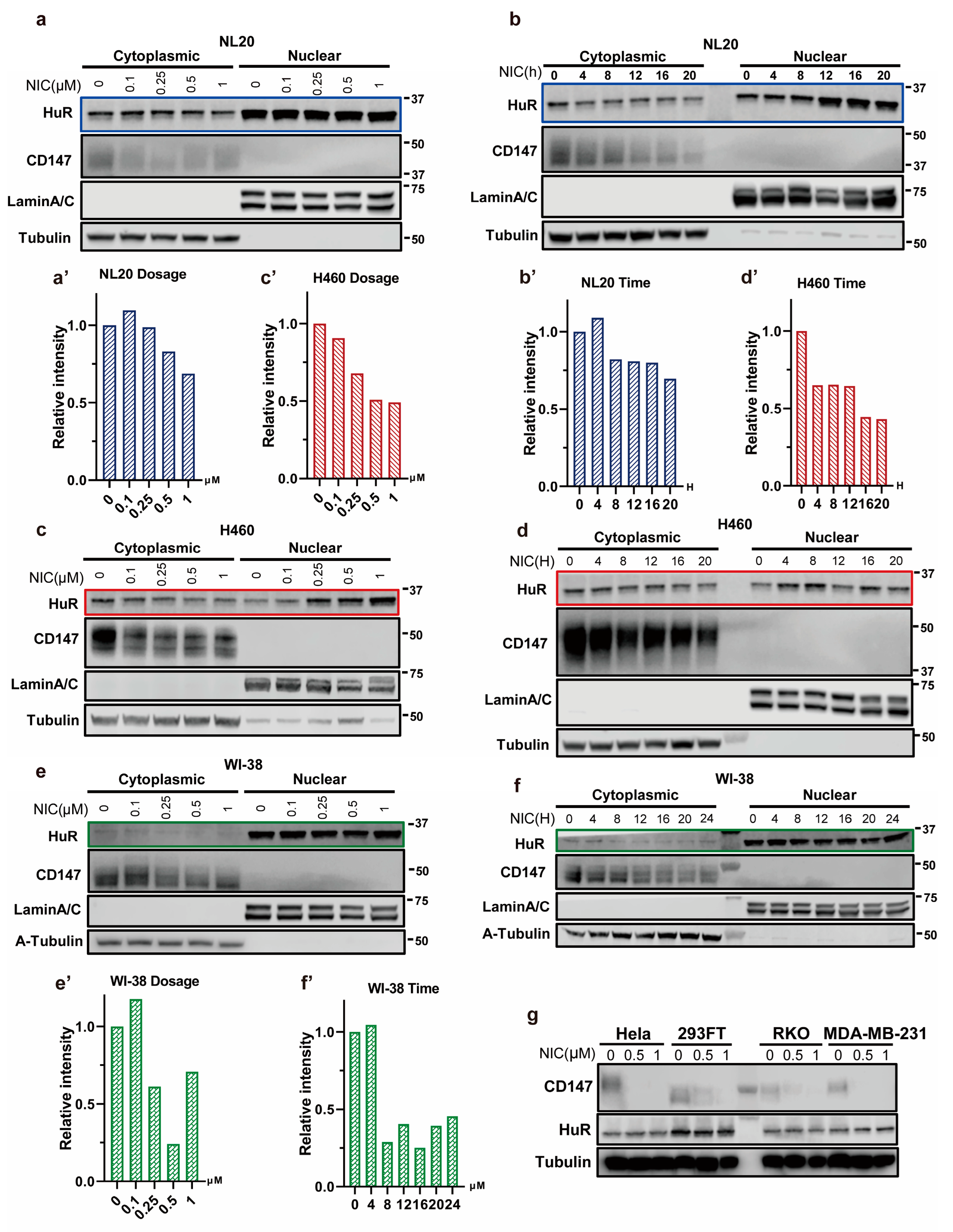
3.2. Niclosamide Inhibits HuR Nucleocytoplasmic Translocation
3.3. Niclosamide Significantly Reduces Overall CD147 Protein Levels
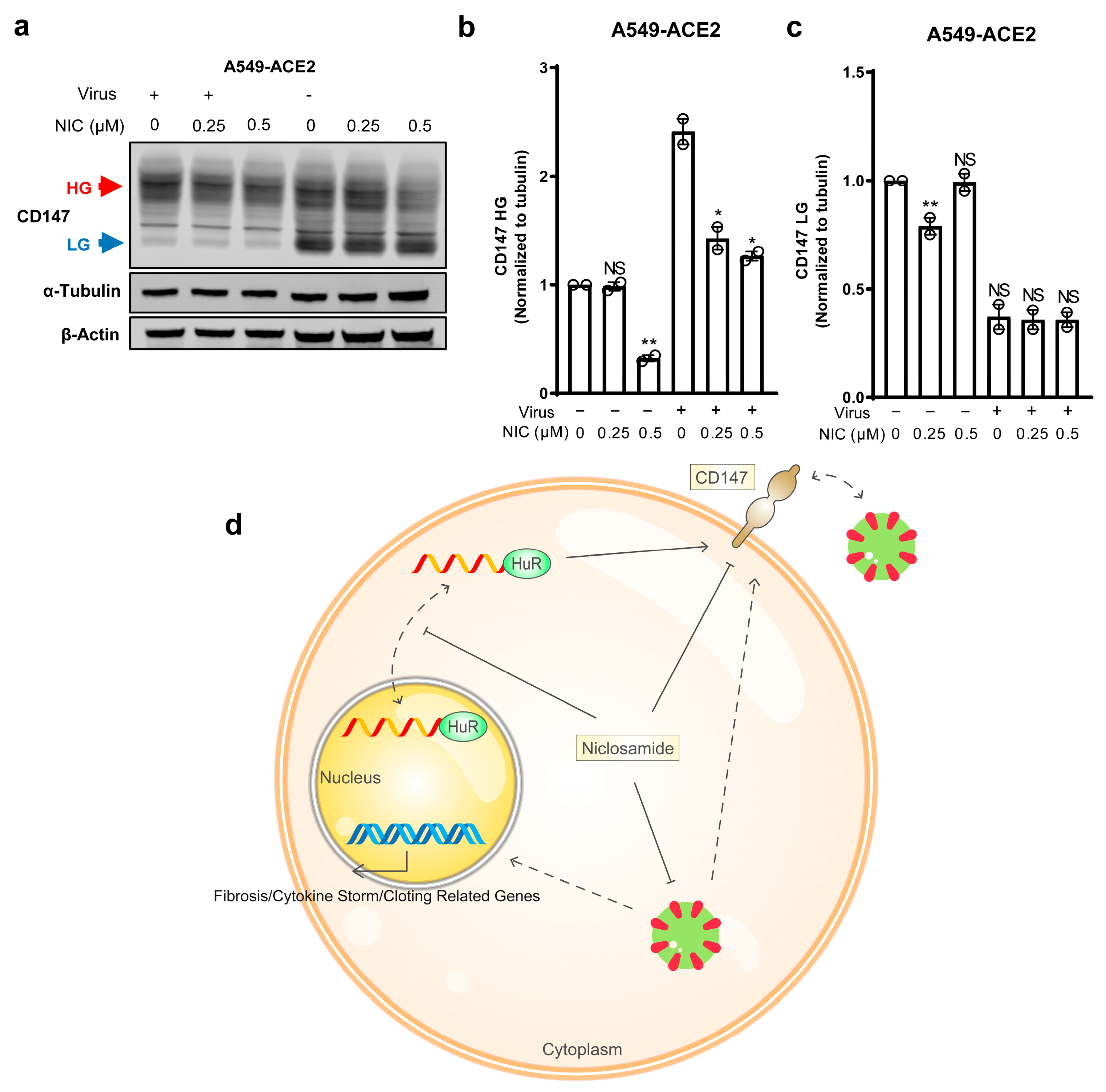
3.4. Niclosamide Effectively Inhibits SARS-CoV-2-Induced CD147
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| COVID-19 | Coronavirus disease 2019 |
| HuR | Human-antigen R |
| 3′-UTR | 3′-untranslated region |
| FDA | Food and Drug Administration |
| BSG | Basigin |
| ACE2 | Angiotensin-converting enzyme 2 |
| HEK-293FT | Human embryonal kidney 293-FT |
| LL/2 | Lewis lung-2 |
| ATCC | American Type Culture Collection |
| FBS | Fetal bovine serum |
| RNP-IP | Ribonucleoprotein immunoprecipitation |
| RNA-IP | RNA immunoprecipitation |
| MOI | Multiplicity of infection |
| ICC | Immunocytochemical |
| IHC | Immunohistochemical |
| HG | High glycosylated |
| LG | Low glycosylated |
| A549-ACE2 | ACE2-expressing A549 cells |
References
- WHO. WHO COVID-19 Dashboard; World Health Organization: Geneva, Switzerland, 2020; Available online: https://covid19.who.int/ (accessed on 11 June 2023).
- Grubaugh, N.D.; Cobey, S. Of variants and vaccines. Cell 2021, 184, 6222–6223. [Google Scholar] [CrossRef]
- Shukri, A.M.A.; Wang, S.M.; Chia, S.L.; Nawi, S. The SARS-CoV-2 Variants and their Impacts. J. Pure Appl. Microbiol. 2022, 16, 1409–1424. [Google Scholar] [CrossRef]
- Marzi, M.; Vakil, M.K.; Bahmanyar, M.; Zarenezhad, E. Paxlovid: Mechanism of Action, Synthesis, and In Silico Study. BioMed Res. Int. 2022, 2022, 7341493. [Google Scholar] [CrossRef]
- Kumar, G.N.; Rodrigues, A.D.; Buko, A.M.; Denissen, J.F. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J. Pharmacol. Exp. Ther. 1996, 277, 423–431. [Google Scholar]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19-Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Adamsick, M.L.; Gandhi, R.G.; Bidell, M.R.; Elshaboury, R.H.; Bhattacharyya, R.P.; Kim, A.Y.; Nigwekar, S.; Rhee, E.P.; Sise, M.E. Remdesivir in Patients with Acute or Chronic Kidney Disease and COVID-19. J. Am. Soc. Nephrol. 2020, 31, 1384. [Google Scholar] [CrossRef]
- Gevers, S.; Welink, J.; van Nieuwkoop, C. Remdesivir in COVID-19 Patients with Impaired Renal Function. J. Am. Soc. Nephrol. 2021, 32, 518. [Google Scholar] [CrossRef]
- Santi Laurini, G.; Montanaro, N.; Motola, D. Safety Profile of Molnupiravir in the Treatment of COVID-19: A Descriptive Study Based on FAERS Data. J. Clin. Med. 2023, 12, 34. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Updated Healthcare Infection Prevention and Control Recommendations in Response to COVID-19 Vaccination. 2021. Available online: https://www.cdc.gov/ (accessed on 11 June 2023).
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Zhou, Y.Q.; Wang, K.; Wang, X.Y.; Cui, H.Y.; Zhao, Y.; Zhu, P.; Chen, Z.N. SARS-CoV-2 pseudovirus enters the host cells through spike protein-CD147 in an Arf6-dependent manner. Emerg. Microbes Infect. 2022, 11, 1135–1144. [Google Scholar] [CrossRef]
- Kong, L.-M.; Liao, C.-G.; Zhang, Y.; Xu, J.; Li, Y.; Huang, W.; Zhang, Y.; Bian, H.; Chen, Z.-N. A Regulatory Loop Involving miR-22, Sp1, and c-Myc Modulates CD147 Expression in Breast Cancer Invasion and Metastasis. Cancer Res. 2014, 74, 3764–3778. [Google Scholar] [CrossRef]
- Lu, M.; Wu, J.; Hao, Z.-W.; Shang, Y.-K.; Xu, J.; Nan, G.; Li, X.; Chen, Z.-N.; Bian, H. Basolateral CD147 induces hepatocyte polarity loss by E-cadherin ubiquitination and degradation in hepatocellular carcinoma progress. Hepatology 2018, 68, 317–332. [Google Scholar] [CrossRef]
- Li, L.; Tang, W.; Wu, X.; Karnak, D.; Meng, X.; Thompson, R.; Hao, X.; Li, Y.; Qiao, X.T.; Lin, J.; et al. HAb18G/CD147 Promotes pSTAT3-Mediated Pancreatic Cancer Development via CD44s. Clin. Cancer Res. 2013, 19, 6703–6715. [Google Scholar] [CrossRef]
- Pushkarsky, T.; Zybarth, G.; Dubrovsky, L.; Yurchenko, V.; Tang, H.; Guo, H.; Toole, B.; Sherry, B.; Bukrinsky, M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 2001, 98, 6360–6365. [Google Scholar] [CrossRef]
- Badeti, S.; Jiang, Q.; Naghizadeh, A.; Tseng, H.-c.; Bushkin, Y.; Marras, S.A.E.; Nisa, A.; Tyagi, S.; Chen, F.; Romanienko, P.; et al. Development of a novel human CD147 knock-in NSG mouse model to test SARS-CoV-2 viral infection. Cell Biosci. 2022, 12, 88. [Google Scholar] [CrossRef]
- Kalejaiye, T.D.; Bhattacharya, R.; Burt, M.A.; Travieso, T.; Okafor, A.E.; Mou, X.; Blasi, M.; Musah, S. SARS-CoV-2 Employ BSG/CD147 and ACE2 Receptors to Directly Infect Human Induced Pluripotent Stem Cell-Derived Kidney Podocytes. Front. Cell Dev. Biol. 2022, 10, 855340. [Google Scholar] [CrossRef]
- Trugilho, M.R.O.; Azevedo-Quintanilha, I.G.; Gesto, J.S.M.; Moraes, E.C.S.; Mandacaru, S.C.; Campos, M.M.; Oliveira, D.M.; Dias, S.S.G.; Bastos, V.A.; Santos, M.D.M.; et al. Platelet proteome reveals features of cell death, antiviral response and viral replication in COVID-19. Cell Death Discov. 2022, 8, 324. [Google Scholar] [CrossRef]
- Barrett, T.J.; Bilaloglu, S.; Cornwell, M.; Burgess, H.M.; Virginio, V.W.; Drenkova, K.; Ibrahim, H.; Yuriditsky, E.; Aphinyanaphongs, Y.; Lifshitz, M.; et al. Platelets contribute to disease severity in COVID-19. J. Thromb. Haemost. 2021, 19, 3139–3153. [Google Scholar] [CrossRef]
- Maugeri, N.; De Lorenzo, R.; Clementi, N.; Antonia Diotti, R.; Criscuolo, E.; Godino, C.; Tresoldi, C.; Bonini, C.; Clementi, M.; Ciceri, F.; et al. Unconventional CD147-dependent platelet activation elicited by SARS-CoV-2 in COVID-19. J. Thromb. Haemost. 2021, 20, 434–448. [Google Scholar] [CrossRef]
- Avolio, E.; Carrabba, M.; Milligan, R.; Williamson, M.K.; Beltrami, A.P.; Gupta, K.; Elvers, K.T.; Gamez, M.; Foster, R.R.; Gillespie, K.; et al. The SARS-CoV-2 Spike protein disrupts human cardiac pericytes function through CD147 receptor-mediated signalling: A potential non-infective mechanism of COVID-19 microvascular disease. Clin. Sci. 2021, 135, 2667–2689. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Qin, C.; Huo, F.; Liang, X.; Yang, X.; Zhang, K.; Lin, P.; Liu, J.; Feng, Z.; et al. CD147 contributes to SARS-CoV-2-induced pulmonary fibrosis. Signal Transduct. Target. Ther. 2022, 7, 382. [Google Scholar] [CrossRef]
- Bian, H.; Zheng, Z.-H.; Wei, D.; Wen, A.; Zhang, Z.; Lian, J.-Q.; Kang, W.-Z.; Hao, C.-Q.; Wang, J.; Xie, R.-H.; et al. Safety and efficacy of meplazumab in healthy volunteers and COVID-19 patients: A randomized phase 1 and an exploratory phase 2 trial. Signal Transduct. Target. Ther. 2021, 6, 194. [Google Scholar] [CrossRef]
- Geng, J.J.; Chen, L.; Yuan, Y.F.; Wang, K.; Wang, Y.C.; Qin, C.A.; Wu, G.Z.; Chen, R.; Zhang, Z.; Wei, D.; et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target. Ther. 2021, 6, 347. [Google Scholar] [CrossRef]
- Wu, C.-J.; Jan, J.-T.; Chen, C.-M.; Hsieh, H.-P.; Hwang, D.-R.; Liu, H.-W.; Liu, C.-Y.; Huang, H.-W.; Chen, S.-C.; Hong, C.-F.; et al. Inhibition of Severe Acute Respiratory Syndrome Coronavirus Replication by Niclosamide. Antimicrob. Agents Chemother. 2004, 48, 2693. [Google Scholar] [CrossRef]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 5770. [Google Scholar] [CrossRef]
- Jeon, S.; Ko, M.; Lee, J.; Choi, I.; Byun, S.Y.; Park, S.; Shum, D.; Kim, S. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 2020, 64, e00819-20. [Google Scholar] [CrossRef]
- Volle, R.; Murer, L.; Petkidis, A.; Andriasyan, V.; Savi, A.; Bircher, C.; Meili, N.; Fischer, L.; Sequeira, D.P.; Mark, D.K.; et al. Methylene blue, Mycophenolic acid, Posaconazole, and Niclosamide inhibit SARS-CoV-2 Omicron variant BA.1 infection of human airway epithelial organoids. Curr. Res. Microb. Sci. 2022, 3, 100158. [Google Scholar] [CrossRef]
- Weiss, A.; Touret, F.; Baronti, C.; Gilles, M.; Hoen, B.; Nougairède, A.; de Lamballerie, X.; Sommer, M.O.A. Niclosamide shows strong antiviral activity in a human airway model of SARS-CoV-2 infection and a conserved potency against the Alpha (B.1.1.7), Beta (B.1.351) and Delta variant (B.1.617.2). PLoS ONE 2021, 16, e0260958. [Google Scholar] [CrossRef]
- Gyamfi, J.; Lee, Y.-H.; Min, B.S.; Choi, J. Niclosamide reverses adipocyte induced epithelial-mesenchymal transition in breast cancer cells via suppression of the interleukin-6/STAT3 signalling axis. Sci. Rep. 2019, 9, 11336. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.L.; Carpenter, B.L.; West, D.S.; Knifley, T.; Liu, L.; Wang, C.; Weiss, H.L.; Gal, T.S.; Durbin, E.B.; Arnold, S.M.; et al. S100A4 drives non-small cell lung cancer invasion, associates with poor prognosis, and is effectively targeted by the FDA-approved anti-helminthic agent niclosamide. Oncotarget 2016, 7, 34630–34642. [Google Scholar] [CrossRef] [PubMed]
- Braga, L.; Ali, H.; Secco, I.; Chiavacci, E.; Neves, G.; Goldhill, D.; Penn, R.; Jimenez-Guardeño, J.M.; Ortega-Prieto, A.M.; Bussani, R.; et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature 2021, 594, 88–93. [Google Scholar] [CrossRef]
- Miner, K.; Labitzke, K.; Liu, B.; Wang, P.; Henckels, K.; Gaida, K.; Elliott, R.; Chen, J.J.; Liu, L.; Leith, A.; et al. Drug Repurposing: The Anthelmintics Niclosamide and Nitazoxanide Are Potent TMEM16A Antagonists That Fully Bronchodilate Airways. Front. Pharmacol. 2019, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Figarola, J.L.; Singhal, J.; Singhal, S.; Kusari, J.; Riggs, A. Bioenergetic modulation with the mitochondria uncouplers SR4 and niclosamide prevents proliferation and growth of treatment-naïve and vemurafenib-resistant melanomas. Oncotarget 2018, 9, 36945–36965. [Google Scholar]
- de Almeida, L.; da Silva, A.L.N.; Rodrigues, T.S.; Oliveira, S.; Ishimoto, A.Y.; Seribelli, A.A.; Becerra, A.; Andrade, W.A.; Ataide, M.A.; Caetano, C.C.S.; et al. Identification of immunomodulatory drugs that inhibit multiple inflammasomes and impair SARS-CoV-2 infection. Sci. Adv. 2022, 8, eabo5400. [Google Scholar] [CrossRef]
- Chang, X.; Zhen, X.; Liu, J.; Ren, X.; Hu, Z.; Zhou, Z.; Zhu, F.; Ding, K.; Nie, J. The antihelmenthic phosphate niclosamide impedes renal fibrosis by inhibiting homeodomain-interacting protein kinase 2 expression. Kidney Int. 2017, 92, 612–624. [Google Scholar] [CrossRef]
- Boyapally, R.; Pulivendala, G.; Bale, S.; Godugu, C. Niclosamide alleviates pulmonary fibrosis in vitro and in vivo by attenuation of epithelial-to-mesenchymal transition, matrix proteins & Wnt/β-catenin signaling: A drug repurposing study. Life Sci. 2019, 220, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, K.; Ousingsawat, J.; Cabrita, I.; Doušová, T.; Bähr, A.; Janda, M.; Schreiber, R.; Benedetto, R. TMEM16A in Cystic Fibrosis: Activating or Inhibiting? Front. Pharmacol. 2019, 10, 3. [Google Scholar] [CrossRef]
- Spagnolo, P.; Balestro, E.; Aliberti, S.; Cocconcelli, E.; Biondini, D.; Casa, G.D.; Sverzellati, N.; Maher, T.M. Pulmonary fibrosis secondary to COVID-19: A call to arms? Lancet Respir. Med. 2020, 8, 750–752. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef] [PubMed]
- George, P.M.; Wells, A.U.; Jenkins, R.G. Pulmonary fibrosis and COVID-19: The potential role for antifibrotic therapy. Lancet Respir. Med. 2020, 8, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Pillat, M.M. CD147 as a Target for COVID-19 Treatment: Suggested Effects of Azithromycin and Stem Cell Engagement. Stem Cell Rev. Rep. 2020, 16, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, P.; Kandel, S.; Mamani, U.F.; Mustafa, B.; Hao, S.; Qiu, J.; Fetse, J.; Liu, Y.; Ibrahim, N.M.; Li, Y.; et al. Discovery of Small Anti-ACE2 Peptides to Inhibit SARS-CoV-2 Infectivity. Adv. Ther. 2021, 4, 2100087. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Ning, K.; Kuz, C.A.; Vorhies, K.; Yan, Z.; Qiu, J. Long-Term Modeling of SARS-CoV-2 Infection of In Vitro Cultured Polarized Human Airway Epithelium. mBio 2020, 11, 6. [Google Scholar] [CrossRef]
- Zou, W.; Xiong, M.; Hao, S.; Zhang, E.Y.; Baumlin, N.; Kim, M.D.; Salathe, M.; Yan, Z.; Qiu, J. The SARS-CoV-2 Transcriptome and the Dynamics of the S Gene Furin Cleavage Site in Primary Human Airway Epithelia. mBio 2021, 12, 3. [Google Scholar] [CrossRef]
- McClain, M.T.; Constantine, F.J.; Henao, R.; Liu, Y.; Tsalik, E.L.; Burke, T.W.; Steinbrink, J.M.; Petzold, E.; Nicholson, B.P.; Rolfe, R.; et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat. Commun. 2021, 12, 1079. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]
- Katsanou, V.; Papadaki, O.; Milatos, S.; Blackshear, P.J.; Anderson, P.; Kollias, G.; Kontoyiannis, D.L. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 2005, 19, 777–789. [Google Scholar] [CrossRef]
- Trivlidis, J.; Aloufi, N.; Al-Habeeb, F.; Nair, P.; Azuelos, I.; Eidelman, D.H.; Baglole, C.J. HuR drives lung fibroblast differentiation but not metabolic reprogramming in response to TGF-beta and hypoxia. Respir. Res. 2021, 22, 323. [Google Scholar] [CrossRef]
- Woodhoo, A.; Iruarrizaga-Lejarreta, M.; Beraza, N.; Garcia-Rodriguez, J.L.; Embade, N.; Fernandez-Ramos, D.; Martinez-Lopez, N.; Gutierrez-De Juan, V.; Arteta, B.; Caballeria, J.; et al. Human antigen R contributes to hepatic stellate cell activation and liver fibrosis. Hepatology 2012, 56, 1870–1882. [Google Scholar] [CrossRef]
- Shang, J.; Zhao, Z.Z. Emerging role of HuR in inflammatory response in kidney diseases. Acta Biochim. Et Biophys. Sin. 2017, 49, 753–763. [Google Scholar] [CrossRef]
- Wu, X.Q.; Gardashova, G.; Lan, L.; Han, S.; Zhong, C.C.; Marquez, R.T.; Wei, L.J.; Wood, S.; Roy, S.; Gowthaman, R.; et al. Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis. Commun. Biol. 2020, 3, 193. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.N.; Lou, H. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 2008, 65, 3168. [Google Scholar] [CrossRef]
- Hu, N.; Fu, Y.; Li, W.-F.; Yang, X.-R.; Cao, M.; Li, F.-F.; Chen, J.-H.; Chen, X.-Y.; Zhao, H.; Sun, Z.-J.; et al. Chemical mitochondrial uncouplers share common inhibitory effect on NLRP3 inflammasome activation through inhibiting NFκB nuclear translocation. Toxicol. Appl. Pharmacol. 2021, 414, 115426. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Chang, S.B.; Hemler, M.E. Links between CD147 Function, Glycosylation, and Caveolin-1. Mol. Biol. Cell 2004, 15, 4043–4050. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.-Y.; Zhao, Y.-C.; Zhao, C.-X.; Gu, Z.-C.; Lu, X.-Y.; Jiang, W.-L.; Gao, L.-C.; Li, W.-L.; Qin, Z.-H.; Ge, H.; et al. The Role of CD147 in Pathological Cardiac Hypertrophy Is Regulated by Glycosylation. Oxid. Med. Cell. Longev. 2022, 2022, 6603296. [Google Scholar] [CrossRef]
- Shilts, J.; Crozier, T.W.M.; Greenwood, E.J.D.; Lehner, P.J.; Wright, G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021, 11, 413. [Google Scholar] [CrossRef]
- Geng, J.J.; Zhang, K.; Chen, L.N.; Miao, J.L.; Yao, M.; Ren, Y.; Fu, Z.G.; Chen, Z.N.; Zhu, P. Enhancement of CD147 on M1 macrophages induces differentiation of Th17 cells in the lung interstitial fibrosis. Biochim. Biophys. Acta-Mol. Basis Dis. 2014, 1842, 1770–1782. [Google Scholar] [CrossRef]
- Shi, W.P.; Ju, D.; Li, H.; Yuan, L.; Cui, J.; Luo, D.; Chen, Z.N.; Bian, H.J. CD147 Promotes CXCL1 Expression and Modulates Liver Fibrogenesis. Int. J. Mol. Sci. 2018, 19, 1145. [Google Scholar] [CrossRef]
- Yan, Z.Y.; Qu, K.; Zhang, J.; Huang, Q.C.; Qu, P.; Xu, X.S.; Yuan, P.; Huang, X.J.; Shao, Y.P.; Liu, C.; et al. CD147 promotes liver fibrosis progression via VEGF-A/VEGFR2 signalling-mediated cross-talk between hepatocytes and sinusoidal endothelial cells. Clin. Sci. 2015, 129, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Sanda, M.; Morrison, L.; Goldman, R. N- and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 2021, 93, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-term cardiovascular outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Zhang, Q.; Wu, X.; Hao, S.; Hao, X.; Jones, E.; Zhang, Y.; Qiu, J.; Xu, L. Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147. Biomedicines 2023, 11, 2019. https://doi.org/10.3390/biomedicines11072019
Yang Z, Zhang Q, Wu X, Hao S, Hao X, Jones E, Zhang Y, Qiu J, Xu L. Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147. Biomedicines. 2023; 11(7):2019. https://doi.org/10.3390/biomedicines11072019
Chicago/Turabian StyleYang, Zhe, Qi Zhang, Xiaoqing Wu, Siyuan Hao, Xinbao Hao, Elizabeth Jones, Yuxia Zhang, Jianming Qiu, and Liang Xu. 2023. "Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147" Biomedicines 11, no. 7: 2019. https://doi.org/10.3390/biomedicines11072019
APA StyleYang, Z., Zhang, Q., Wu, X., Hao, S., Hao, X., Jones, E., Zhang, Y., Qiu, J., & Xu, L. (2023). Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147. Biomedicines, 11(7), 2019. https://doi.org/10.3390/biomedicines11072019






