Adult IDH Wild-Type Glioblastoma Ultrastructural Investigation Suggests a Possible Correlation between Morphological Biomarkers and Ki-67 Index
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients Characteristics
2.2. Histopathological Procedures and Molecular Characterization of Glioblastoma Tumors
2.3. Light and Transmission Electron Microscopy Protocols
2.4. Semiquantitative Evaluation of Ultrastructural Characteristics
3. Results
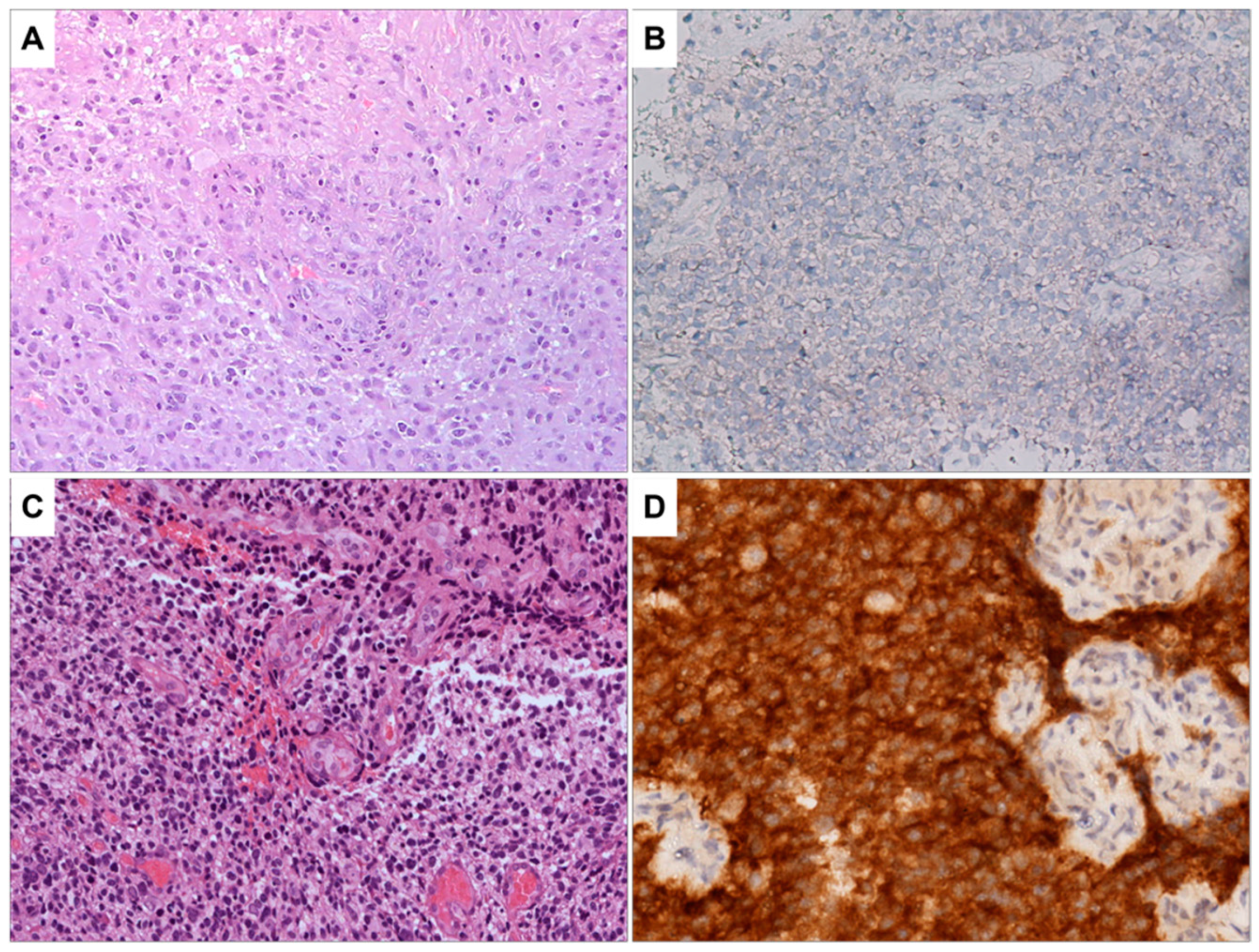
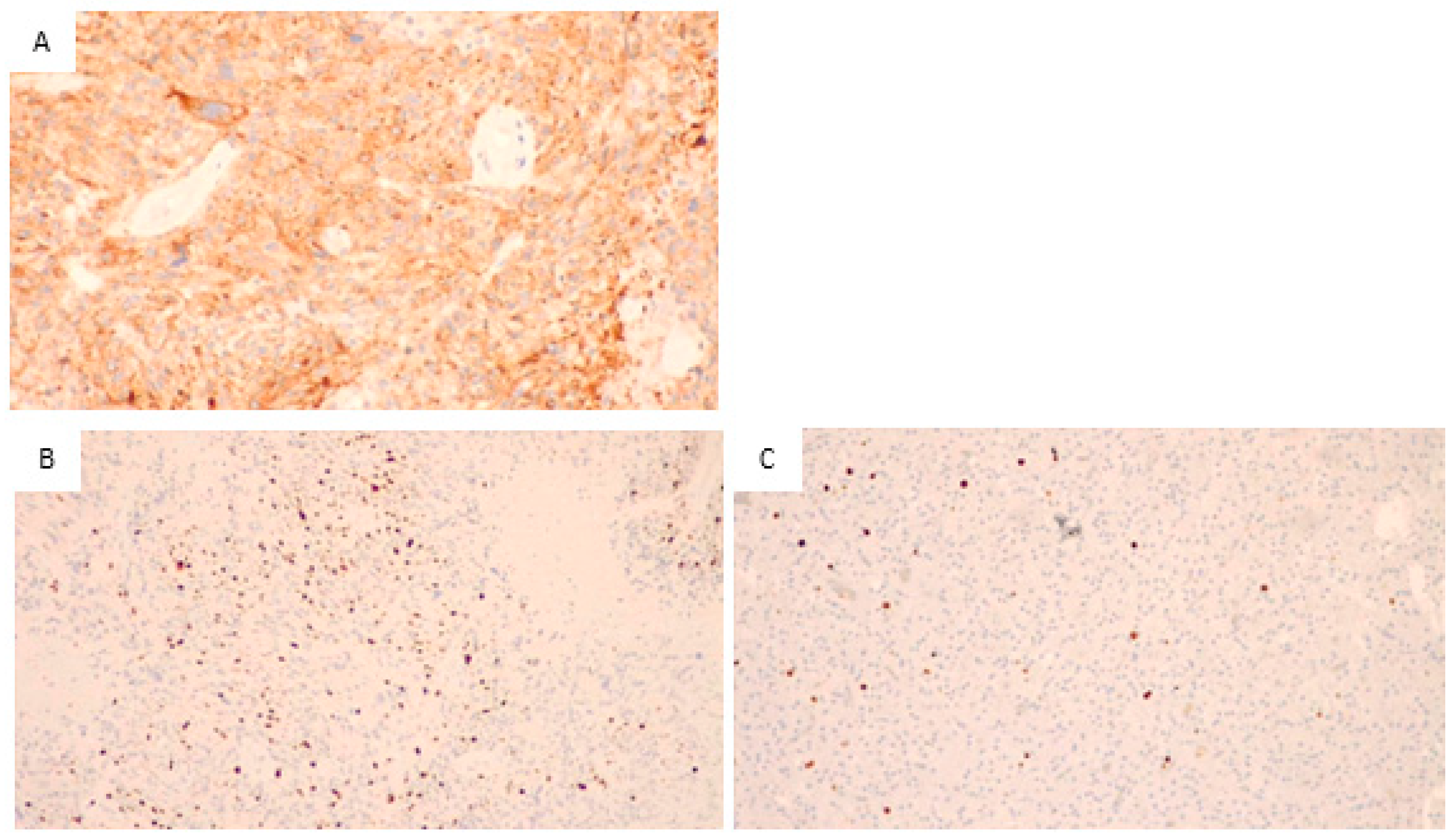
3.1. Histopathological and Immunohistochemical Analysis
3.2. Light Microscopy Analysis of Glioblastoma Tissue
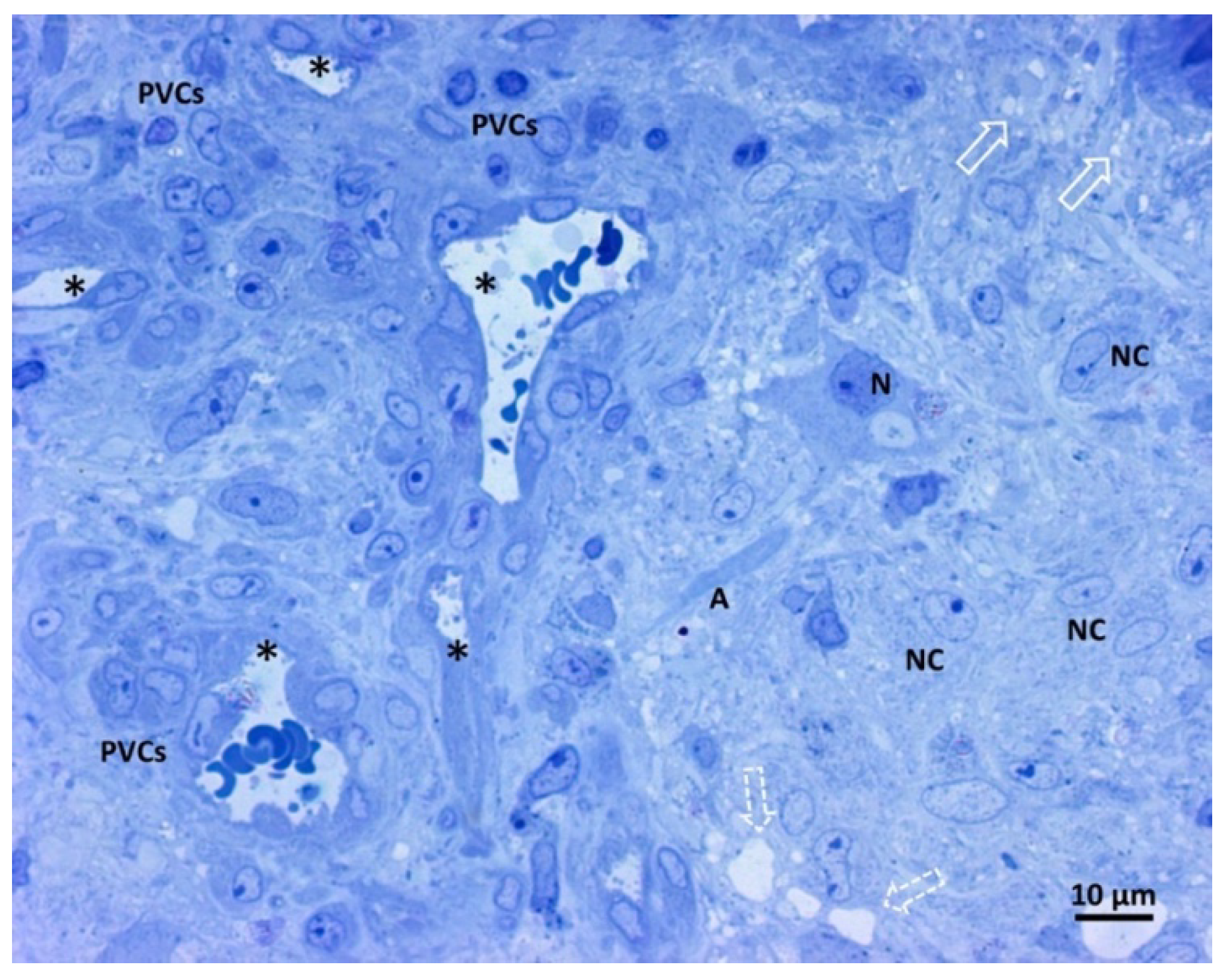
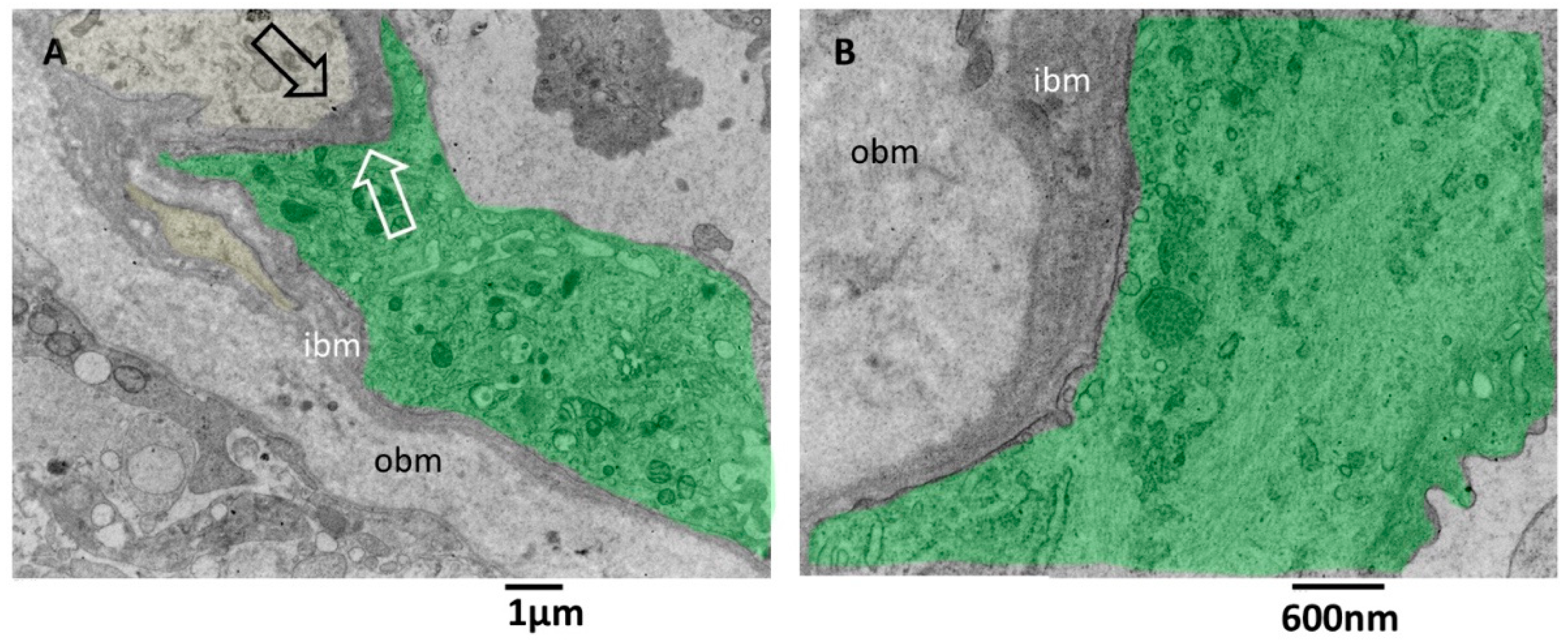
3.3. Transmission Electron Microscopy Analysis of Glioblastoma Tissue
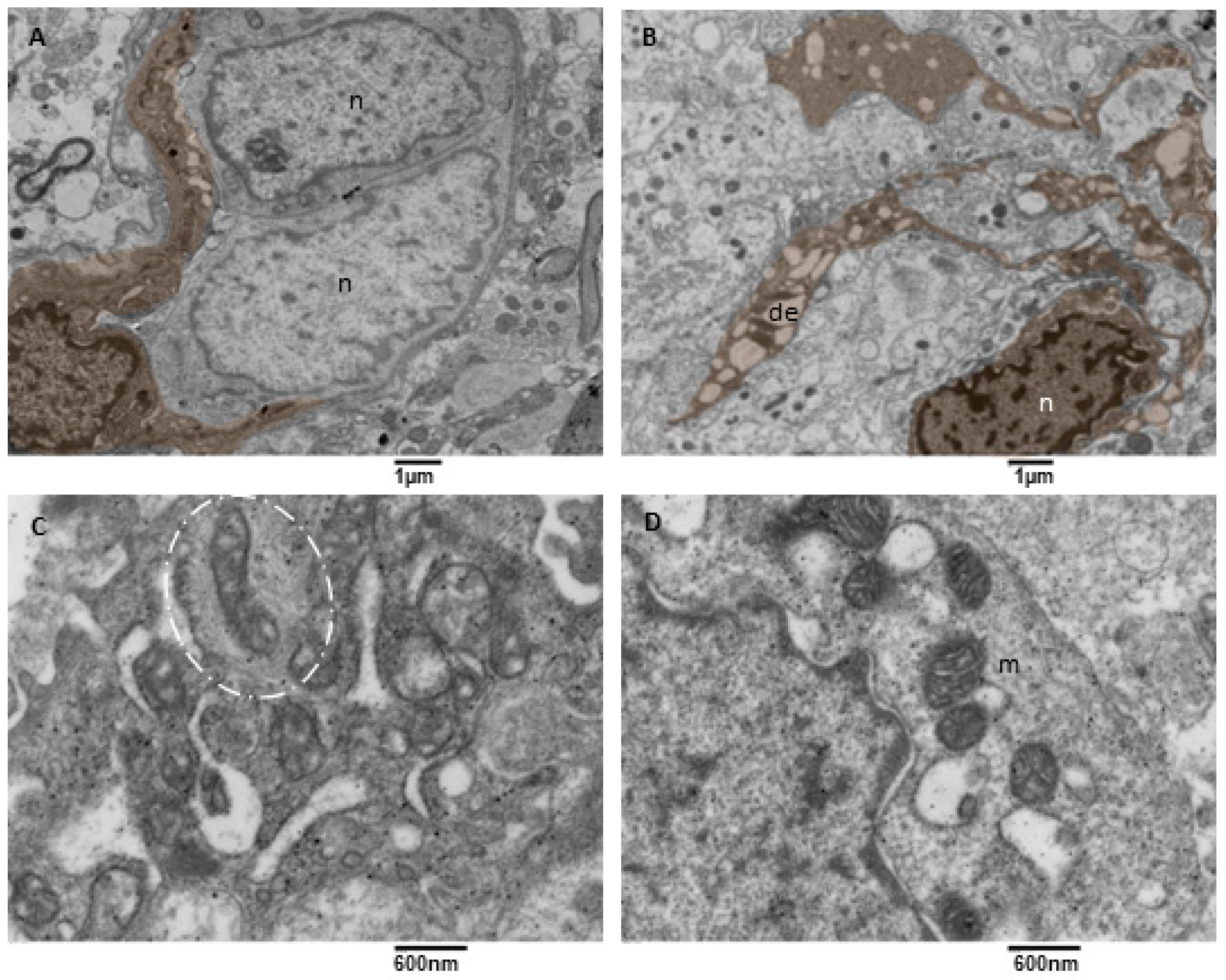
3.3.1. Small Vessels
3.3.2. Neoplastic Cells
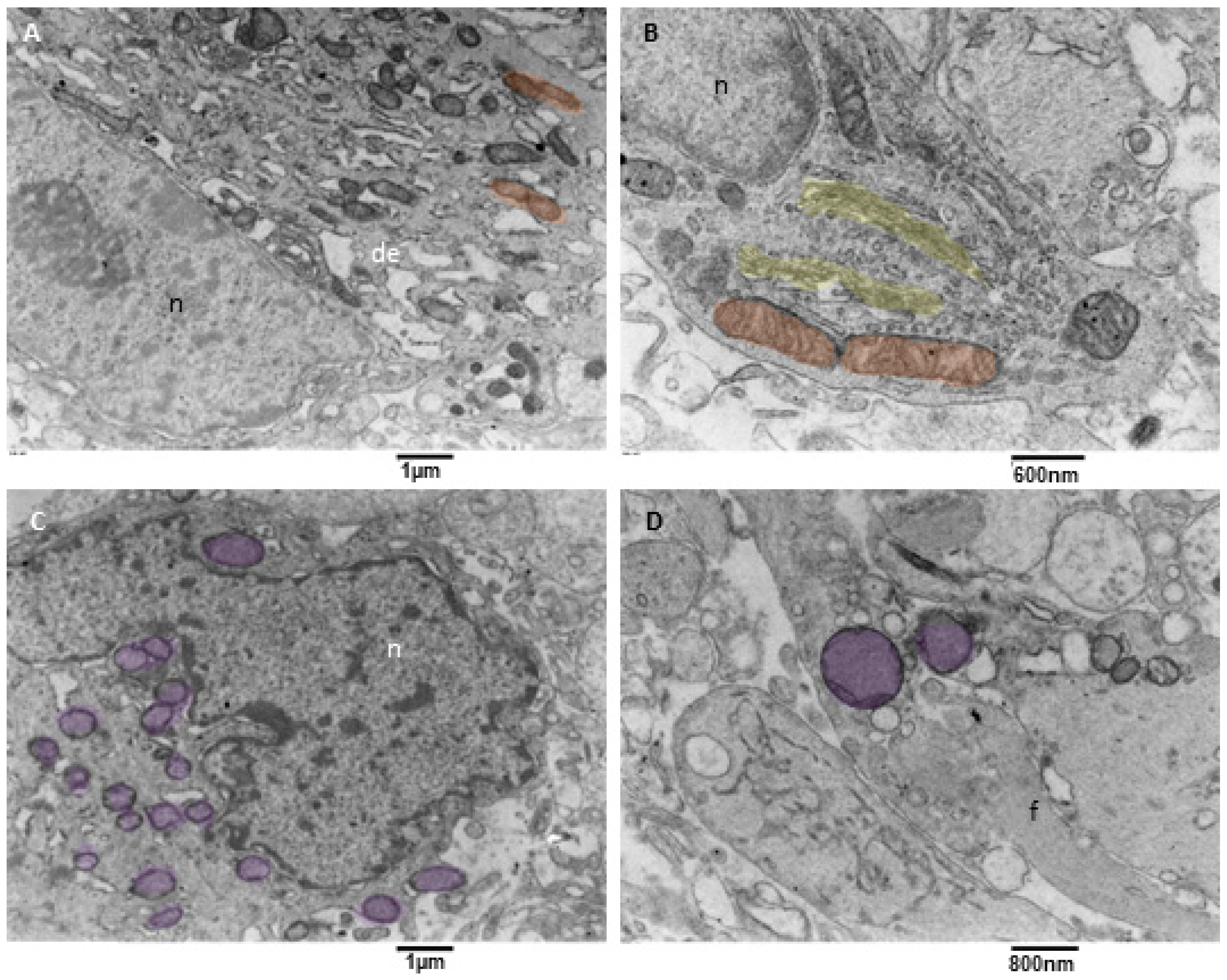
3.3.3. Neoplastic Cells Mitophagy (NCsM)
3.3.4. Microglia Cells
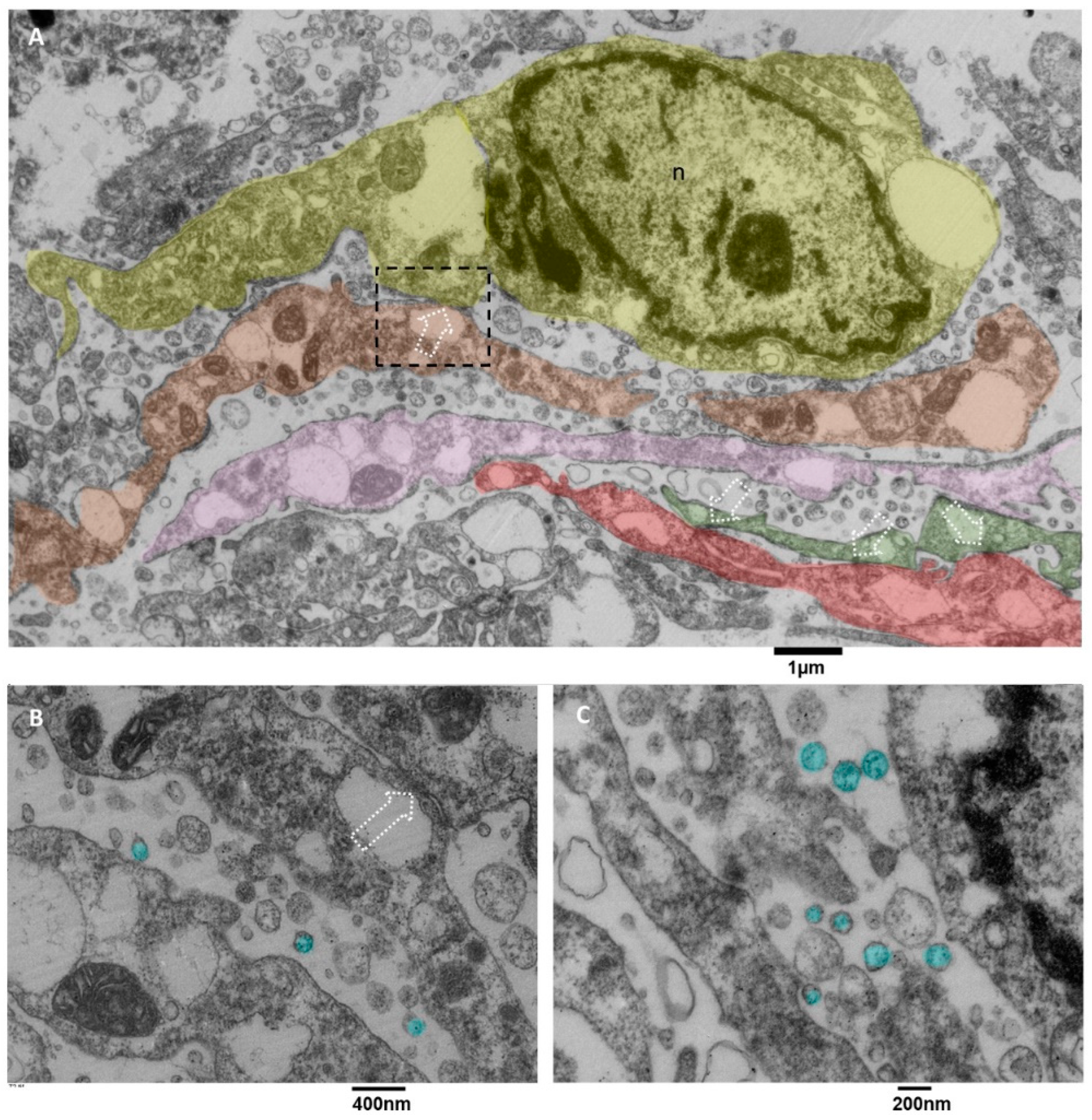
3.3.5. Microglia Cells Secreting sEVs (MsEVs)
3.3.6. Microglia Cells Storing Lipid Vesicles (MsLVs)
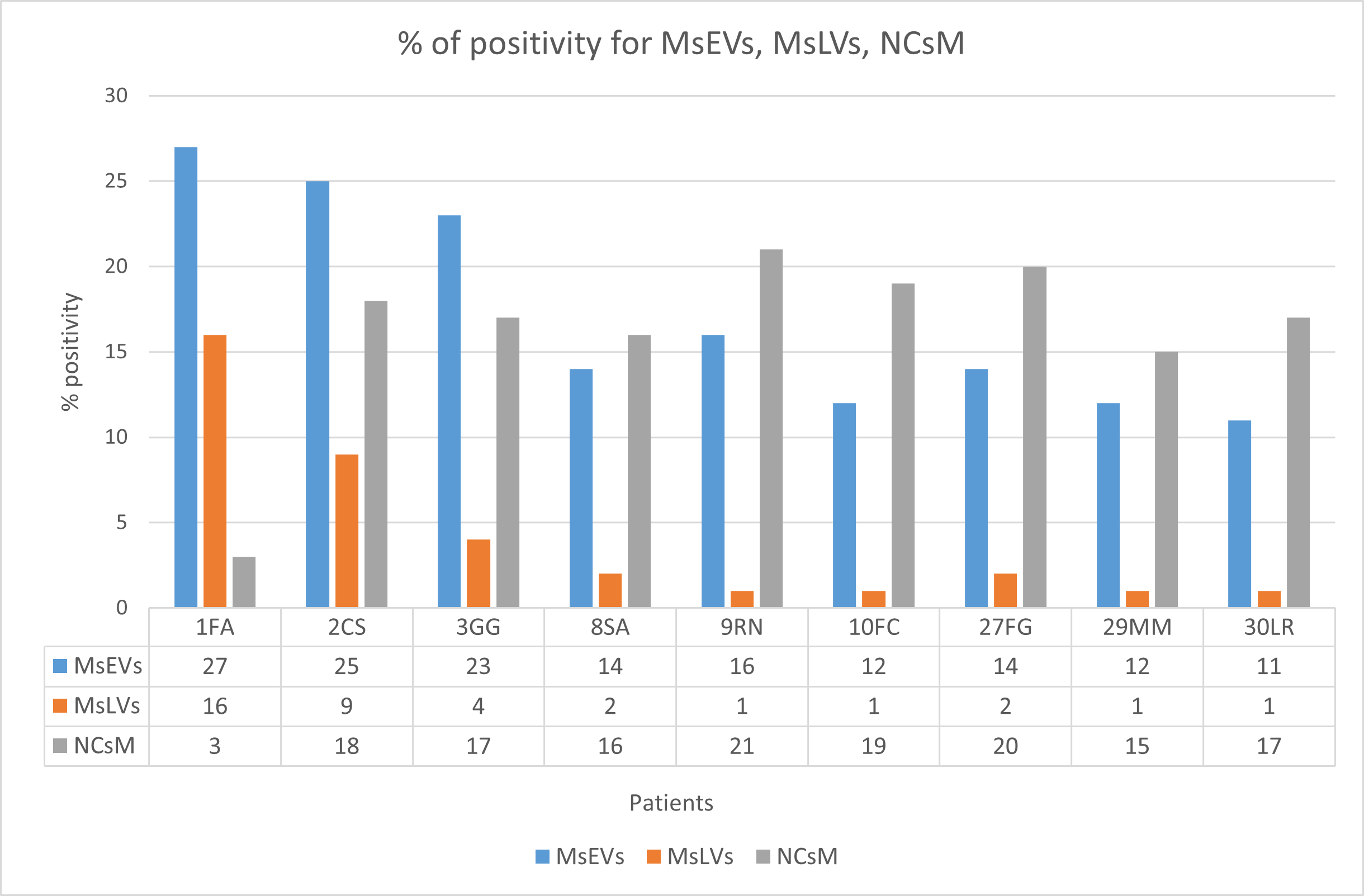
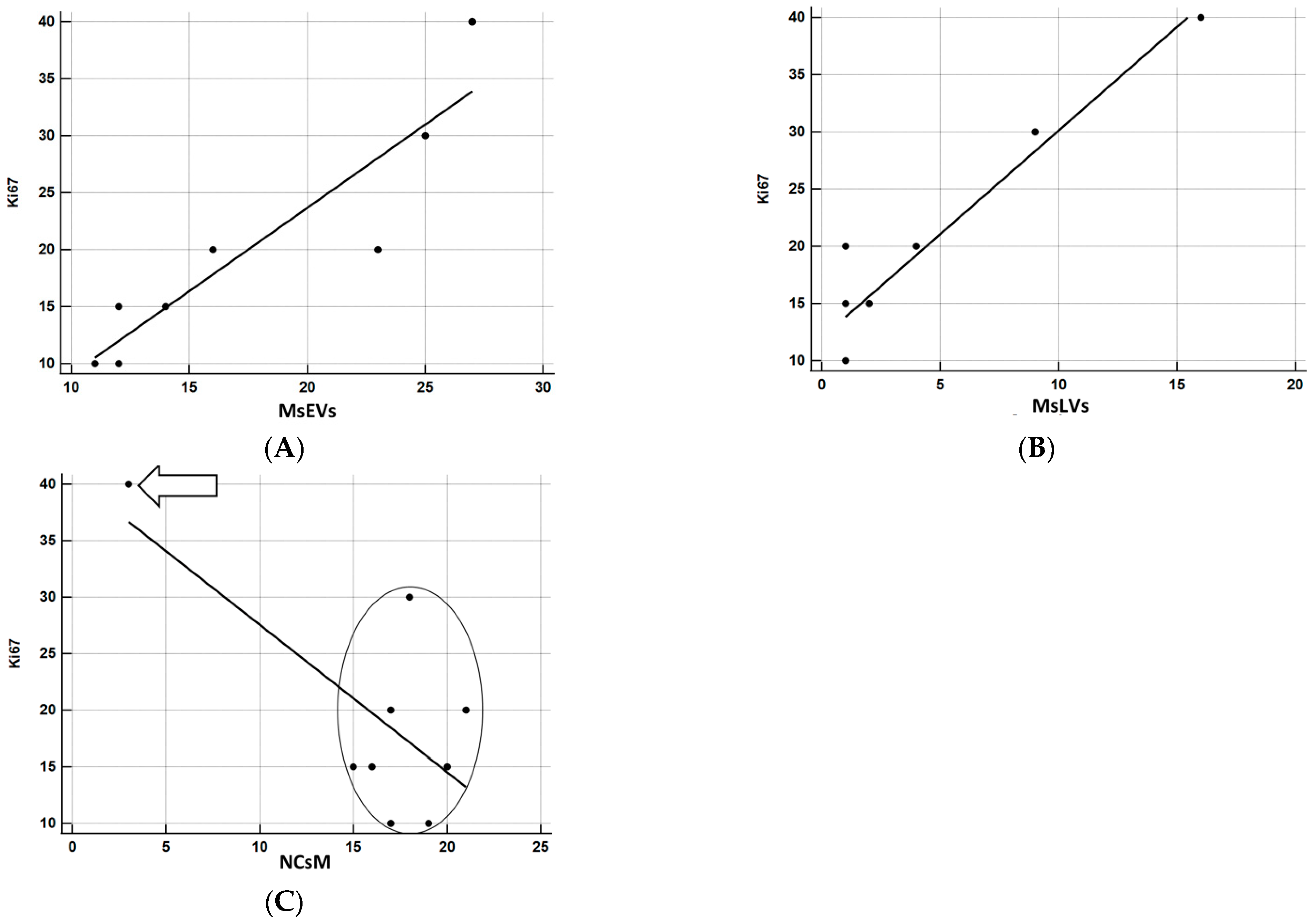
3.3.7. Statistical Analysis
4. Discussion
- Visualization of the anatomical basis of astrocyte-endothelial cells’ crosstalk
- 2.
- Ultrastructural in situ imaging of clusters of hyperactivated microglia cells (MsEVs)
- 3.
- Ultrastructural in situ imaging of microglia cells storing lipid vesicles (MsLVs)
- 4.
- Ultrastructural in situ imaging of neoplastic cells mitophagy (NCsM)
- 5.
- Statistical analysis of ultrastructural data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme-Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Gerber, N.K.; Goenka, A.; Turcan, S.; Reyngold, M.; Makarov, V.; Kannan, K.; Beal, K.; Omuro, A.; Yamada, Y.; Gutin, P.; et al. Transcriptional diversity of long-term glioblastoma survivors. Neuro Oncol. 2014, 16, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Qin, T.; Mullan, B.; Ravindran, R.; Messinger, D.; Siada, R.; Cummings, J.R.; Harris, M.; Muruganand, A.; Pyaram, K.; Miklja, Z.; et al. ATRX loss in glioma results in dysregulation of cell-cycle phase transition and ATM inhibitor radio-sensitization. Cell Rep. 2022, 38, 110216. [Google Scholar] [CrossRef]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef]
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, A.B.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Nagahashi, M.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012, 3, 709–722. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Zhang, Y.; Dube, C.; Gibert, M., Jr.; Cruickshanks, N.; Wang, B.; Coughlan, M.; Yang, Y.; Setiady, I.; Deveau, C.; Saoud, K.; et al. The p53 Pathway in Glioblastoma. Cancers 2018, 10, 297. [Google Scholar] [CrossRef]
- Uceda-Castro, R.; van Asperen, J.V.; Vennin, C.; Sluijs, J.A.; van Bodegraven, E.J.; Margarido, A.S.; Robe, P.A.J.; van Rheenen, J.; Hol, E.M. GFAP splice variants fine-tune glioma cell invasion and tumour dynamics by modulating migration persistence. Sci. Rep. 2022, 12, 424. [Google Scholar] [CrossRef]
- Xu, H.; Zong, H.; Ma, C.; Ming, X.; Shang, M.; Li, K.; He, X.; Du, H.; Cao, L. Epidermal growth factor receptor in glioblastoma. Oncol. Lett. 2017, 14, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, J.; Schwab, U.; Lemke, H.; Stein, H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int. J. Cancer 1983, 31, 13–20. [Google Scholar] [CrossRef]
- Chen, W.J.; He, D.S.; Tang, R.X.; Ren, F.H.; Chen, G. Ki-67 is a valuable prognostic factor in gliomas: Evidence from a systematic review and meta-analysis. Asian Pac. J. Cancer Prev. 2015, 16, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Arshad, H.; Ahmad, Z.; Hasan, S.H. Gliomas: Correlation of histologic grade, Ki67 and p53 expression with patient survival. Asian Pac. J. Cancer Prev. 2010, 11, 1637–1640. [Google Scholar]
- Dahlrot, R.H.; Bangsø, J.A.; Petersen, J.K.; Rosager, A.M.; Sørensen, M.D.; Reifenberger, G.; Hansen, S.; Kristensen, B.W. Prognostic role of Ki-67 in glioblastomas excluding contribution from non-neoplastic cells. Sci. Rep. 2021, 11, 17918. [Google Scholar] [CrossRef] [PubMed]
- Du, N.; Shu, W.; Li, K.; Deng, Y.; Xu, X.; Ye, Y.; Tang, F.; Mao, R.; Lin, G.; Li, S.; et al. An initial study on the predictive value using multiple MRI characteristics for Ki-67 labeling index in glioma. J. Transl. Med. 2023, 21, 119. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Bergers, G. Glioblastoma: Defining Tumor Niches. Trends Cancer 2015, 1, 252–265. [Google Scholar] [CrossRef]
- Geribaldi-Doldán, N.; Fernández-Ponce, C.; Quiroz, R.N.; Sánchez-Gomar, I.; Escorcia, L.G.; Velásquez, E.P.; Quiroz, E.N. The Role of Microglia in Glioblastoma. Front. Oncol. 2021, 10, 603495. [Google Scholar] [CrossRef]
- Buonfiglioli, A.; Hambardzumyan, D. Macrophages and microglia: The cerberus of glioblastoma. Acta Neuropathol. Commun. 2021, 9, 54. [Google Scholar] [CrossRef]
- Rajendran, L.; Bali, J.; Barr, M.M.; Court, F.A.; Krämer-Albers, E.M.; Picou, F.; Raposo, G.; van der Vos, K.E.; van Niel, G.; Wang, J.; et al. Emerging roles of extracellular vesicles in the nervous system. J. Neurosci. 2014, 34, 15482–15489. [Google Scholar] [CrossRef]
- Ciregia, F.; Urbani, A.; Palmisano, G. Extracellular Vesicles in Brain Tumors and Neurodegenerative Diseases. Front. Mol. Neurosci. 2017, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, C.A.; Kaye, A.H.; Drummond, K.J.; Widodo, S.S.; Mantamadiotis, T.; Vella, L.J.; Stylli, S.S. Extracellular vesicles and their role in glioblastoma. Crit. Rev. Clin. Lab. Sci. 2019, 57, 227–252. [Google Scholar] [CrossRef] [PubMed]
- Gassama, Y.; Favereaux, A. Emerging Roles of Extracellular Vesicles in the Central Nervous System: Physiology, Pathology, and Therapeutic Perspectives. Front. Cell. Neurosci. 2021, 15, 626043. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, A.; Roy, S.; Fleming, T.; Leong, H.S.; Schuurmans, C. The Emerging Role of Extracellular Vesicles in the Glioma Microenvironment: Biogenesis and Clinical Relevance. Cancers 2020, 12, 1964. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, S.; Tripathi, A.; Bhan, A.; Acharya, M.M.; Pillai, P. Emerging Concepts on the Role of Extracellular Vesicles and Its Cargo Contents in Glioblastoma-Microglial Crosstalk. Mol. Neurobiol. 2022, 59, 2822–2837. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, C.; Cai, J.; Chen, G.; Zhang, D.; Zhang, Z.; Dong, Z. PINK1/Parkin-mediated mitophagy is activated in cisplatin nephrotoxicity to protect against kidney injury. Cell Death Dis. 2018, 4, 1113. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, X.; Huang, J.; Zhuo, Z.; Chen, H.; Zeng, R.; Wu, H.; Guo, K.; Yang, Q.; Ye, H.; et al. Development and validation of a novel mitophagy-related gene prognostic signature for glioblastoma multiforme. BMC Cancer 2022, 22, 644. [Google Scholar] [CrossRef]
- Petan, T.; Jarc, E.; Jusović, M. Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23, 1941. [Google Scholar] [CrossRef]
- Leblond, M.M.; Gerault, A.N.; Corroyer-Dulmont, A.; MacKenzie, E.T.; Petit, E.; Bernaudin, M.; Valable, S. Hypoxia induces macrophage polarization and re-education toward an angiogenic phenotype in U87 and U251 glioblastoma models. Oncoimmunology 2018, 7, e1438106. [Google Scholar]
- Cruz, A.L.; Barreto, E.D.A.; Fazolini, N.P.; Viola, J.P.; Bozza, P.T. Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020, 11, 105. [Google Scholar] [CrossRef]
- Cottignoli, V.; Relucenti, M.; Agrosì, G.; Cavarretta, E.; Familiari, G.; Salvador, L.; Maras, A. Biological Niches within Human Calcified Aortic Valves: Towards Understanding of the Pathological Biomineralization Process. BioMed Res. Int. 2015, 2015, 542687. [Google Scholar] [CrossRef] [PubMed]
- Nardoni, M.; Della Valle, E.; Liberti, M.; Relucenti, M.; Casadei, M.A.; Paolicelli, P.; Apollonio, F.; Petralito, S. Can Pulsed Electromagnetic Fields Trigger On-Demand Drug Release from High-Tm Magnetoliposomes? Nanomaterials 2018, 8, 196. [Google Scholar] [CrossRef]
- Relucenti, M.; Francescangeli, F.; De Angelis, M.L.; D’Andrea, V.; Miglietta, S.; Pilozzi, E.; Li, X.; Boe, A.; Chen, R.; Zeuner, A.; et al. The Ultrastructural Analysis of Human Colorectal Cancer Stem Cell-Derived Spheroids and Their Mouse Xenograft Shows That the Same Cells Types Have Different Ratios. Biology 2021, 10, 929. [Google Scholar] [CrossRef] [PubMed]
- Relucenti, M.; Francescangeli, F.; De Angelis, M.L.; D’Andrea, V.; Miglietta, S.; Donfrancesco, O.; Li, X.; Chen, R.; Zeuner, A.; Familiari, G. A Different Exosome Secretion Pattern Characterizes Patient-Derived Colorectal Cancer Multicellular Spheroids and Their Mouse Xenografts. Biology 2022, 11, 1427. [Google Scholar] [CrossRef] [PubMed]
- Petralito, S.; Paolicelli, P.; Nardoni, M.; Trilli, J.; Di Muzio, L.; Cesa, S.; Relucenti, M.; Matassa, R.; Vitalone, A.; Adrover, A.; et al. Gelation of the internal core of liposomes as a strategy for stabilization and modified drug delivery I. Physico-chemistry study. Int. J. Pharm. 2020, 585, 119467. [Google Scholar] [CrossRef] [PubMed]
- Relucenti, M.; Miglietta, S.; Bove, G.; Donfrancesco, O.; Battaglione, E.; Familiari, P.; Barbaranelli, C.; Covelli, E.; Barbara, M.; Familiari, G. SEM BSE 3D Image Analysis of Human Incus Bone Affected by Cholesteatoma Ascribes to Osteoclasts the Bone Erosion and VpSEM dEDX Analysis Reveals New Bone Formation. Scanning 2020, 2020, 9371516. [Google Scholar] [CrossRef]
- Lo Torto, F.; Relucenti, M.; Familiari, G.; Vaia, N.; Casella, D.; Matassa, R.; Miglietta, S.; Marinozzi, F.; Bini, F.; Fratoddi, I.; et al. The Effect of Postmastectomy Radiation Therapy on Breast Implants: Material Analysis on Silicone and Polyurethane Prosthesis. Ann. Plast. Surg. 2018, 81, 228–234. [Google Scholar] [CrossRef]
- Adjei-Sowah, E.A.; O’Connor, S.A.; Veldhuizen, J.; Lo Cascio, C.; Plaisier, C.; Mehta, S.; Nikkhah, M. Investigating the Interactions of Glioma Stem Cells in the Perivascular Niche at Single-Cell Resolution using a Microfluidic Tumor Microenvironment Model. Adv. Sci. 2022, 9, e2201436. [Google Scholar] [CrossRef]
- McCoy, M.G.; Nyanyo, D.; Hung, C.K.; Goerger, J.P.; Zipfel, R.W.; Williams, R.M.; Nishimura, N.; Fischbach, C. Endothelial cells promote 3D invasion of GLIOBLASTOMA by IL-8-dependent induction of cancer stem cell properties. Sci. Rep. 2019, 9, 9069. [Google Scholar] [CrossRef]
- Akindona, F.A.; Frederico, S.C.; Hancock, J.C.; Gilbert, M.R. Exploring the origin of the cancer stem cell niche and its role in anti-angiogenic treatment for glioblastoma. Front. Oncol. 2022, 12, 947634. [Google Scholar] [CrossRef]
- Dinda, A.K.; Sarkar, C.; Roy, S.; Kharbanda, K.; Mathur, M.; Khosla, A.K.; Banerji, A.K. A transmission and scanning electron microscopic study of tumoral and peritumoral microblood vessels in human gliomas. J. Neuro Oncol. 1993, 16, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Cribaro, G.P.; Saavedra-López, E.; Romarate, L.; Mitxitorena, I.; Díaz, L.R.; Casanova, P.V.; Roig-Martínez, M.; Gallego, J.M.; Perez-Vallés, A.; Barcia, C. Three-dimensional vascular microenvironment landscape in human glioblastoma. Acta Neuropathol. Commun. 2021, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.C.; Herman, M.M.; Rubinstein, L.J. Electron microscopic observations on human glioblastomas and astrocytomas maintained in organ culture systems. Am. J. Pathol. 1973, 73, 589–606. [Google Scholar] [PubMed]
- Ferese, R.; Lenzi, P.; Fulceri, F.; Biagioni, F.; Fabrizi, C.; Gambardella, S.; Familiari, P.; Frati, A.; Limanaqi, F.; Fornai, F. Quantitative Ultrastructural Morphometry and Gene Expression of mTOR-Related Mitochondriogenesis within Glioblastoma Cells. Int. J. Mol. Sci. 2020, 21, 4570. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, P.; Ferese, R.; Biagioni, F.; Fulceri, F.; Busceti, C.L.; Falleni, A.; Gambardella, S.; Frati, A.; Fornai, F. Rapamycin Ameliorates Defects in Mitochondrial Fission and Mitophagy in Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 5379. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; D’Alessandro, G.; Lepore, F.; Corazzari, M.; Caldarola, S.; Valacca, C.; Faienza, F.; Esposito, V.; Limatola, C.; Cecconi, F.; et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol. 2015, 9, 1612–1625. [Google Scholar] [CrossRef]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nature reviews. Immunology 2018, 18, 225–242. [Google Scholar]
- Wolf, S.A.; Boddeke, H.W.; Kettenmann, H. Microglia in Physiology and Disease. Annu. Rev. Physiol. 2017, 79, 619–643. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef]
- Graeber, M.B.; Li, W.; Rodriguez, M.L. Role of microglia in CNS inflammation. FEBS Lett. 2011, 585, 3798–3805. [Google Scholar] [CrossRef]
- Augusto-Oliveira, M.; Arrifano, G.P.; Delage, C.I.; Tremblay, M.È.; Crespo-Lopez, M.E.; Verkhratsky, A. Plasticity of microglia. Biol. Rev. Camb. Philos. Soc. 2022, 97, 217–250. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, N.; Leube, R.E. Intermediate Filaments as Organizers of Cellular Space: How They Affect Mitochondrial Structure and Function. Cells 2016, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, M.-K.; Carrier, M.; Ibáñez, F.G.; Šimončičová, E.; Wallman, M.-J.; Vallières, L.; Parent, M.; Tremblay, M.-È. Ultrastructural characterization of dark microglia during aging in a mouse model of Alzheimer’s disease pathology and in human post-mortem brain samples. J. Neuroinflamm. 2022, 19, 235. [Google Scholar] [CrossRef] [PubMed]
- Bisht, K.; Sharma, K.P.; Lecours, C.; Sánchez, M.G.; El Hajj, H.; Milior, G.; Olmos-Alonso, A.; Gómez-Nicola, D.; Luheshi, G.; Vallières, L.; et al. Dark microglia: A new phenotype predominantly associated with pathological states. Glia 2016, 64, 826–839. [Google Scholar] [CrossRef]
- Yoshimura, T. The production of monocyte chemoattractant protein-1 (MCP−1)/CCL2 in tumor microenvironments. Cytokine 2017, 98, 71–78. [Google Scholar] [CrossRef]
- Rong, Y.; Ji, C.; Wang, Z.; Ge, X.; Wang, J.; Ye, W.; Tang, P.; Jiang, D.; Fan, J.; Yin, G.; et al. Small extracellular vesicles encapsulating CCL2 from activated astrocytes induce microglial activation and neuronal apoptosis after traumatic spinal cord injury. J. Neuroinflamm. 2021, 18, 196. [Google Scholar] [CrossRef]
- Grimaldi, A.; Serpe, C.; Chece, G.; Nigro, V.; Sarra, A.; Ruzicka, B.; Relucenti, M.; Familiari, G.; Ruocco, G.; Pascucci, G.R.; et al. Microglia-Derived Microvesicles Affect Microglia Phenotype in Glioma. Front. Cell. Neurosci. 2019, 13, 41. [Google Scholar] [CrossRef]
- Serpe, C.; Monaco, L.; Relucenti, M.; Iovino, L.; Familiari, P.; Scavizzi, F.; Raspa, M.; Familiari, G.; Civiero, L.; D’Agnano, I.; et al. Microglia-Derived Small Extracellular Vesicles Reduce Glioma Growth by Modifying Tumor Cell Metabolism and Enhancing Glutamate Clearance through miR-124. Cells 2021, 10, 2066. [Google Scholar] [CrossRef]
- Fitzner, D.; Bader, J.M.; Penkert, H.; Bergner, C.G.; Su, M.; Weil, M.T.; Surma, M.A.; Mann, M.; Klose, C.; Simons, M. Cell-Type- and Brain-Region-Resolved Mouse Brain Lipidome. Cell Rep. 2020, 32, 108132. [Google Scholar] [CrossRef]
- Geng, F.; Cheng, X.; Wu, X.; Yoo, J.Y.; Cheng, C.; Guo, J.Y.; Mo, X.; Ru, P.; Hurwitz, B.; Kim, S.H.; et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin. Cancer Res. 2016, 22, 5337–5348. [Google Scholar] [CrossRef]
- Hayes, A.F. Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression-Based Approach; The Guilford Press: New York, NY, USA, 2022. [Google Scholar]
- Reiter-Brennan, C.; Semmler, L.; Klein, A. The effects of 2-hydroxyglutarate on the tumorigenesis of gliomas. Contemp. Oncol. 2018, 22, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zheng, S.; Zheng, Y.; Huang, R.; An, N.; Liang, A.; Hu, C. Glioma-derived isocitrate dehydrogenase-2 mutations induced up-regulation of HIF-1α and β-catenin signaling: Possible impact on glioma cell metastasis and chemo-resistance. Int. J. Biochem. Cell Biol. 2012, 44, 770–775. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, E.; Chong, K.; Ryu, S.W.; Kim, C.; Choi, K.; Kim, J.H.; Choi, C. Atypical induction of HIF-1α expression by pericellular Notch1 signaling suffices for the malignancy of glioblastoma multiforme cells. Cell. Mol. Life Sci. 2022, 79, 537. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, S.; Abdellatef, S.; Haddad, M.A.; Fakhoury, I.; El-Sibai, M. Hypoxia and EGF Stimulation Regulate VEGF Expression in Human Glioblastoma Multiforme (GLIOBLASTOMA) Cells by Differential Regulation of the PI3K/Rho-GTPase and MAPK Pathways. Cells 2019, 8, 1397. [Google Scholar] [CrossRef]
- Domènech, M.; Hernández, A.; Plaja, A.; Martínez-Balibrea, E.; Balañà, C. Hypoxia: The Cornerstone of Glioblastoma. Int. J. Mol. Sci. 2021, 22, 12608. [Google Scholar] [CrossRef]
- He, J.; Mansouri, A.; Das, S. Alpha Thalassemia/Mental Retardation Syndrome X-Linked, the Alternative Lengthening of Telomere Phenotype, and Gliomagenesis: Current Understandings and Future Potential. Front. Oncol. 2018, 7, 322. [Google Scholar] [CrossRef]
- Danussi, C.; Bose, P.; Parthasarathy, P.T.; Silberman, P.C.; Van Arnam, J.S.; Vitucci, M.; Tang, O.Y.; Heguy, A.; Wang, Y.; Chan, T.A.; et al. Atrx inactivation drives disease-defining phenotypes in glioma cells of origin through global epigenomic remodeling. Nat. Commun. 2018, 9, 1057. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- Mao, H.; Lebrun, D.G.; Yang, J.; Zhu, V.F.; Li, M. Deregulated signaling pathways in glioblastoma multiforme: Molecular mechanisms and therapeutic targets. Cancer Investig. 2012, 30, 48–56. [Google Scholar] [CrossRef]
- Pearson, J.R.D.; Regad, T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct. Target. Ther. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Restrepo, A.; Smith, C.A.; Agnihotri, S.; Shekarforoush, M.; Kongkham, P.N.; Seol, H.J.; Northcott, P.; Rutka, J.T. Epigenetic regulation of glial fibrillary acidic protein by DNA methylation in human malignant gliomas. Neuro Oncol. 2011, 13, 42–50. [Google Scholar] [CrossRef]
- Bignami, A.; Eng, L.F.; Dahl, D.; Uyeda, C.T. Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Res. 1972, 43, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Oprita, A.; Baloi, S.C.; Staicu, G.A.; Alexandru, O.; Tache, D.E.; Danoiu, S.; Micu, E.S.; Sevastre, A.S. Updated Insights on EGFR Signaling Pathways in Glioma. Int. J. Mol. Sci. 2021, 22, 587. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, K.; Clavreul, A.; Lagarce, F. Toward an effective strategy in glioblastoma treatment. Part I: Resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov. Today 2015, 20, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, H.; Kleihues, P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009, 100, 2235–2241. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Takano, S.; Ishikawa, E.; Sakamoto, N.; Matsuda, M.; Akutsu, H.; Noguchi, M.; Kato, Y.; Yamamoto, T.; Matsumura, A. Immunohistochemistry on IDH 1/2, ATRX, p53 and Ki-67 substitute molecular genetic testing and predict patient prognosis in grade III adult diffuse gliomas. Brain Tumor Pathol. 2016, 33, 107–116. [Google Scholar] [CrossRef]
- Armocida, D.; Frati, A.; Salvati, M.; Santoro, A.; Pesce, A. Is Ki-67 index overexpression in IDH wild type glioblastoma a predictor of shorter Progression Free survival? A clinical and Molecular analytic investigation. Clin. Neurol. Neurosurg. 2020, 198, 106126. [Google Scholar] [CrossRef]
- Nakod, P.S.; Kim, Y.; Rao, S.S. The Impact of Astrocytes and Endothelial Cells on Glioblastoma Stemness Marker Expression in Multicellular Spheroids. Cell. Mol. Bioeng. 2021, 14, 639–651. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, H.J.; Kim, M.S.; Choi, H.J.; He, J.; Wu, Q.; Aldape, K.; Weinberg, J.S.; Yung, W.K.; Conrad, C.A.; et al. Macitentan, a Dual Endothelin Receptor Antagonist, in Combination with Temozolomide Leads to Glioblastoma Regression and Long-term Survival in Mice. Clin. Cancer Res. 2015, 21, 4630–4641. [Google Scholar] [CrossRef]
- Horng, S.; Therattil, A.; Moyon, S.; Gordon, A.; Kim, K.; Argaw, A.T.; Hara, Y.; Mariani, J.N.; Sawai, S.; Flodby, P.; et al. Astrocytic tight junctions control inflammatory CNS lesion pathogenesis. J. Clin. Investig. 2017, 127, 3136–3151. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.; Kim, E.Y.; Badr, C.E.; Weissleder, R.; Mempel, T.R.; Tannous, B.A. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015, 6, 7029. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Kerbel, R.S.; Allison, A.C.; Rak, J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. USA 2009, 106, 3794–3799. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Mukherjee, S.; Ali, S.; Bhardwaj, U.; Choudhary, R.K.; Balakrishnan, S.; Naseem, A.; Mir, S.A.; Banawas, S.; Alaidarous, M.; et al. Pioneer Role of Extracellular Vesicles as Modulators of Cancer Initiation in Progression, Drug Therapy, and Vaccine Prospects. Cells 2022, 11, 490. [Google Scholar] [CrossRef]
- Murgoci, A.N.; Duhamel, M.; Raffo-Romero, A.; Mallah, K.; Aboulouard, S.; Lefebvre, C.; Salzet, M. Location of neonatal microglia drives small extracellular vesicles content and biological functions in vitro. J. Extracell. Vesicles 2020, 9, 1727637. [Google Scholar] [CrossRef]
- Muñoz Herrera, O.M.; Zivkovic, A.M. Microglia and Cholesterol Handling: Implications for Alzheimer’s Disease. Biomedicines 2022, 10, 3105. [Google Scholar] [CrossRef]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-Droplet-Accumulating Microglia Represent a Dysfunctional and Proinflammatory State in the Aging Brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef]
- Colombo, A.; Dinkel, L.; Müller, S.A.; Sebastian Monasor, L.; Schifferer, M.; Cantuti-Castelvetri, L.; König, J.; Vidatic, L.; Bremova-Ertl, T.; Lieberman, A.P.; et al. Loss of NPC1 Enhances Phagocytic Uptake and Impairs Lipid Trafficking in Microglia. Nat. Commun. 2021, 12, 1158. [Google Scholar] [CrossRef]
- Hanisch, U.K.; Kettenmann, H. Microglia: Active Sensor and Versatile Effector Cells in the Normal and Pathologic Brain. Nat. Neurosci. 2007, 10, 1387–1394. [Google Scholar] [CrossRef]
- Bernardini, J.P.; Lazarou, M.; Dewson, G. Parkin and mitophagy in cancer. Oncogene 2017, 36, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Maiti, P.; Scott, J.; Sengupta, D.; Al-Gharaibeh, A.; Dunbar, G.L. Curcumin and solid lipid curcumin particles induce autophagy, but inhibit mitophagy and the PI3K-Akt/mTOR pathway in cultured glioblastoma cells. Int. J. Mol. Sci. 2019, 20, 399. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Peng, Z.; Li, P.; Fu, H.; Feng, J.; Zhang, Y.; Liu, T.; Liu, Y.; Liu, Q.; Liu, Q.; et al. lncRNA RMST suppressed glioblastoma cell mitophagy through enhancing FUS SUMOylation. Mol. Ther. Nucleic Acids 2020, 19, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Xu, T.; Wang, Y.; Zhou, Y.; Yu, D.; Wang, Z.; He, L.; Chen, Z.; Zhang, Y.; Davidson, D.; et al. Cannabidiol inhibits human glioma by induction of lethal mitophagy through activating TRPV4. Autophagy 2021, 17, 3592–3606. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Lu, S.; Wang, X.Z.; Wang, C.C.; Wang, L.; Liang, S.P.; Luo, T.-F.; Wang, Z.-C.; Piao, M.-H.; Chi, G.-F.; et al. FOXO3a protects glioma cells against temozolomide-induced DNA double strand breaks via promotion of BNIP3-mediated mitophagy. Acta Pharmacol. Sin. 2021, 2, 1324–1337. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, L.; Xue, Y.; Ma, Y.; Liu, X.; Li, Z.; Li, Z.; Liu, Y. Endothelial monocyte-activating polypeptide-II induces BNIP3-mediated mitophagy to enhance temozolomide cytotoxicity of glioma stem cells via down-regulating MiR-24-3p. Front. Mol. Neurosci. 2018, 11, 92. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Familiari, P.; Relucenti, M.; Lapolla, P.; Palmieri, M.; Antonelli, M.; Cristiano, L.; Barbaranelli, C.; Catalano, M.; D’Angelo, L.; Familiari, G.; et al. Adult IDH Wild-Type Glioblastoma Ultrastructural Investigation Suggests a Possible Correlation between Morphological Biomarkers and Ki-67 Index. Biomedicines 2023, 11, 1968. https://doi.org/10.3390/biomedicines11071968
Familiari P, Relucenti M, Lapolla P, Palmieri M, Antonelli M, Cristiano L, Barbaranelli C, Catalano M, D’Angelo L, Familiari G, et al. Adult IDH Wild-Type Glioblastoma Ultrastructural Investigation Suggests a Possible Correlation between Morphological Biomarkers and Ki-67 Index. Biomedicines. 2023; 11(7):1968. https://doi.org/10.3390/biomedicines11071968
Chicago/Turabian StyleFamiliari, Pietro, Michela Relucenti, Pierfrancesco Lapolla, Mauro Palmieri, Manila Antonelli, Loredana Cristiano, Claudio Barbaranelli, Myriam Catalano, Luca D’Angelo, Giuseppe Familiari, and et al. 2023. "Adult IDH Wild-Type Glioblastoma Ultrastructural Investigation Suggests a Possible Correlation between Morphological Biomarkers and Ki-67 Index" Biomedicines 11, no. 7: 1968. https://doi.org/10.3390/biomedicines11071968
APA StyleFamiliari, P., Relucenti, M., Lapolla, P., Palmieri, M., Antonelli, M., Cristiano, L., Barbaranelli, C., Catalano, M., D’Angelo, L., Familiari, G., Santoro, A., Frati, A., & Bruzzaniti, P. (2023). Adult IDH Wild-Type Glioblastoma Ultrastructural Investigation Suggests a Possible Correlation between Morphological Biomarkers and Ki-67 Index. Biomedicines, 11(7), 1968. https://doi.org/10.3390/biomedicines11071968









