Inhibition of Multidrug Efflux Pumps Belonging to the Major Facilitator Superfamily in Bacterial Pathogens
Abstract
1. Bacterial Pathogens
2. Antimicrobial Resistance Mechanisms
3. Antimicrobial Transporter Superfamilies
3.1. Major Facilitator Superfamily (MFS)
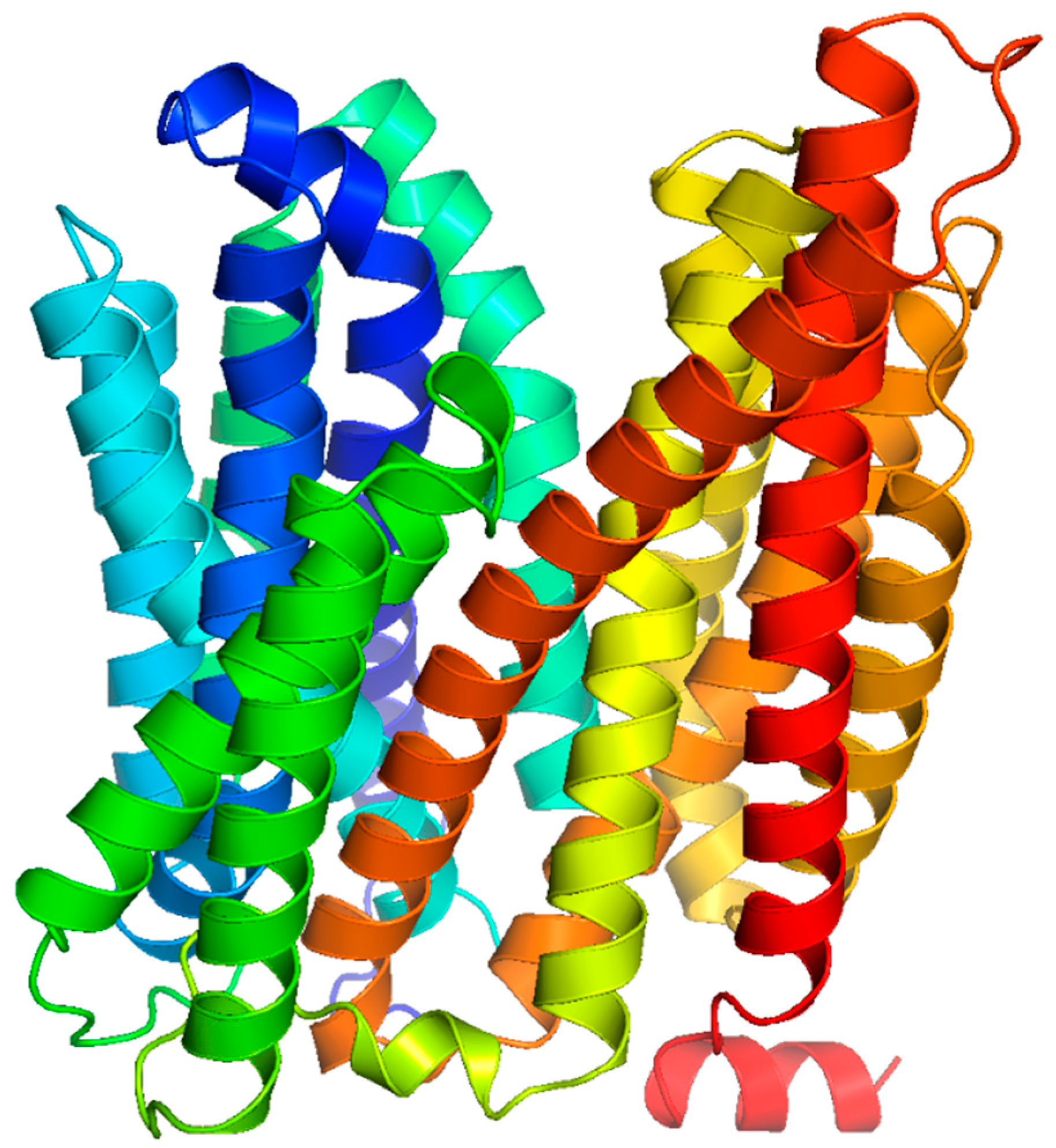
MFS Antimicrobial Efflux Pump Structure and Function
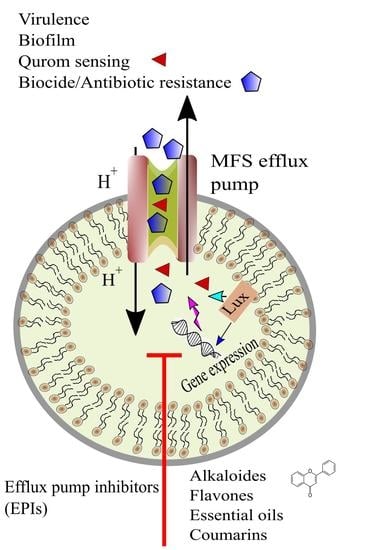
4. Efflux Pump Inhibition
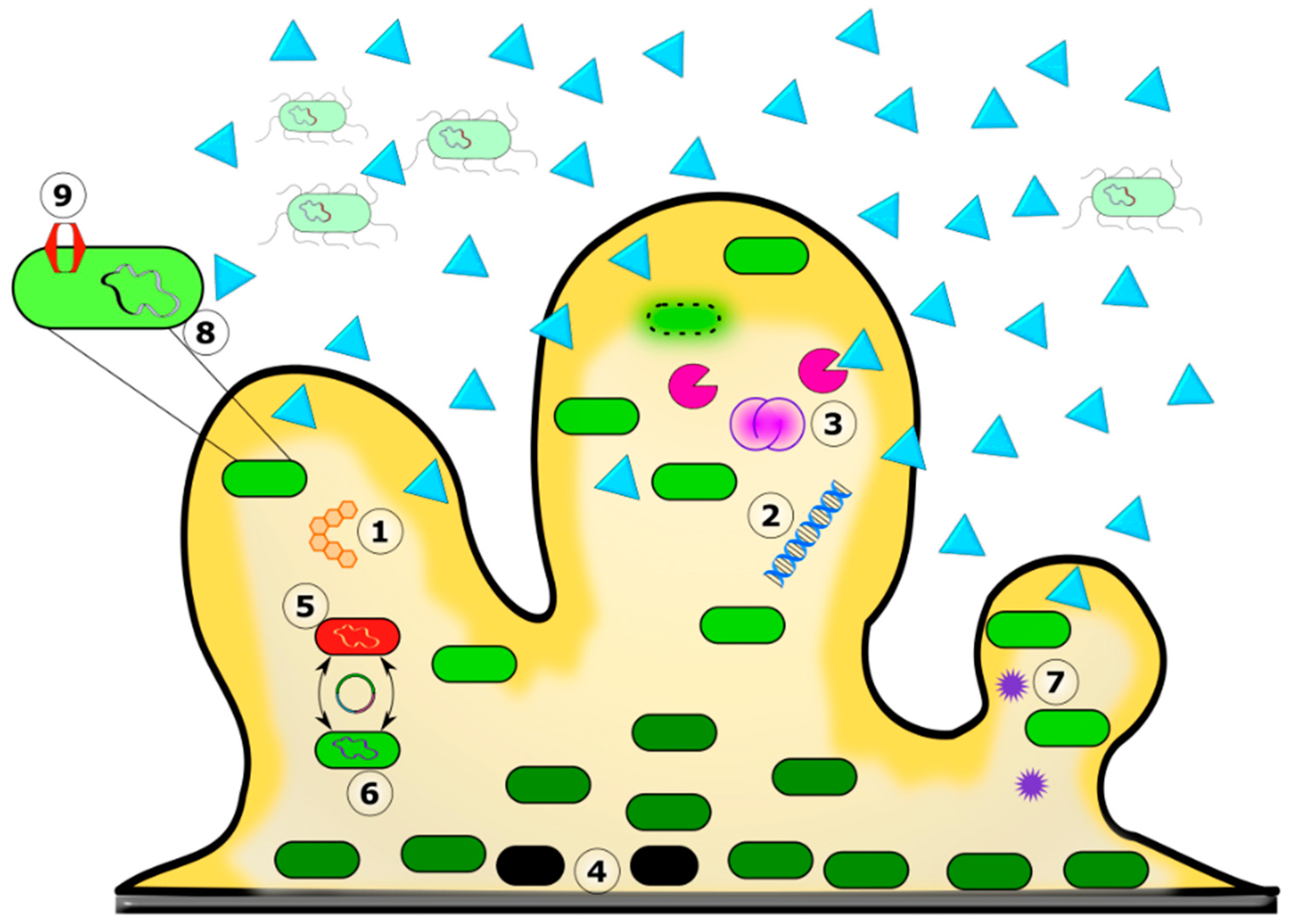
5. Antimicrobial Resistance and Biofilms
Dynamics between Biofilm Formation and Antibiotic Resistance
6. Role of the Biofilm Matrix in Antibiotic Recalcitrance
7. MFS Transporters and Biofilms
8. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stephen, J.; Salam, F.; Lekshmi, M.; Kumar, S.H.; Varela, M.F. The Major Facilitator Superfamily and Antimicrobial Resistance Efflux Pumps of the ESKAPEE Pathogen Staphylococcus aureus. Antibiotics 2023, 12, 343. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Buroni, S.; Swain, S.S.; Bonacorsi, A.; da Fonseca Amorim, E.A.; Kulshrestha, M.; da Silva, L.C.N.; Tiwari, V. Recent Advances to Combat ESKAPE Pathogens with Special Reference to Essential Oils. Front. Microbiol. 2022, 13, 1029098. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and Acquired Resistance Mechanisms in Enterococcus. Virulence 2012, 3, 421. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance in Enterococci. Expert. Rev. Anti Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef]
- Ayobami, O.; Willrich, N.; Reuss, A.; Eckmanns, T.; Markwart, R. The Ongoing Challenge of Vancomycin-Resistant Enterococcus faecium and Enterococcus faecalis in Europe: An Epidemiological Analysis of Bloodstream Infections. Emerg. Microbes Infect. 2020, 9, 1180–1193. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, Research, and Development of New Antibiotics: The Who Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Andersen, J.L.; He, G.-X.; Kakarla, P.; Kc, R.; Kumar, S.; Lakra, W.S.; Mukherjee, M.M.; Ranaweera, I.; Shrestha, U.; Tran, T.; et al. Multidrug Efflux Pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus Bacterial Food Pathogens. Int. J. Environ. Res. Public Health 2015, 12, 1487–1547. [Google Scholar] [CrossRef]
- Lekshmi, M.; Stephen, J.; Ojha, M.; Kumar, S.; Varela, M. Staphylococcus aureus Antimicrobial Efflux Pumps and Their Inhibitors: Recent Developments. AIMS Med. Sci. 2022, 9, 367–393. [Google Scholar] [CrossRef]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, Prevalence, and Management of MRSA Bacteremia across Patient Populations—A Review of Recent Developments in MRSA Management and Treatment. Crit. Care 2017, 21, 211. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Frees, D.; Ingmer, H. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Nandhini, P.; Kumar, P.; Mickymaray, S.; Alothaim, A.S.; Somasundaram, J.; Rajan, M. Recent Developments in Methicillin-Resistant Staphylococcus aureus (MRSA) Treatment: A Review. Antibiotics 2022, 11, 606. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Aqib, A.I.; Muzammil, I.; Majeed, N.; Bhutta, Z.A.; Kulyar, M.F.-A.; Fatima, M.; Zaheer, C.-N.F.; Muneer, A.; Murtaza, M.; et al. MRSA Compendium of Epidemiology, Transmission, Pathophysiology, Treatment, and Prevention within One Health Framework. Front. Microbiol. 2023, 13, 1067284. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical Relevance of the ESKAPE Pathogens. Expert. Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the Labyrinth of Antibacterial Discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Cabot, G.; López-Causapé, C.; Ocampo-Sosa, A.A.; Sommer, L.M.; Domínguez, M.Á.; Zamorano, L.; Juan, C.; Tubau, F.; Rodríguez, C.; Moyà, B.; et al. Deciphering the Resistome of the Widespread Pseudomonas aeruginosa Sequence Type 175 International High-Risk Clone through Whole-Genome Sequencing. Antimicrob. Agents Chemother. 2016, 60, 7415–7423. [Google Scholar] [CrossRef] [PubMed]
- Treepong, P.; Kos, V.N.; Guyeux, C.; Blanc, D.S.; Bertrand, X.; Valot, B.; Hocquet, D. Global Emergence of the Widespread Pseudomonas aeruginosa ST235 Clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef]
- Lobanovska, M.; Pilla, G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future? Yale J. Biol. Med. 2017, 90, 135–145. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and Evolution of Antibiotic Resistance: The Common Mechanisms of Emergence and Spread in Water Bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef]
- Wright, G.D. Molecular mechanisms of antibiotic resistance. Chem. Commun. 2011, 47, 4055–4061. [Google Scholar] [CrossRef]
- Zapun, A.; Contreras-Martel, C.; Vernet, T. Penicillin-Binding Proteins and β-Lactam Resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef]
- Jacoby, G.A. Mechanisms of Resistance to Quinolones. Clin. Infect. Dis. 2005, 41, S120–S126. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Rao, M.; Padyana, S.; Dipin, K.; Kumar, S.; Nayak, B.; Varela, M.F. Antimicrobial Compounds of Plant Origin as Efflux Pump Inhibitors: New Avenues for Controlling Multidrug Resistant Pathogens. J. Antimicrob. Agents 2018, 4, 1–6. [Google Scholar] [CrossRef]
- Henderson, P.J. Studies of Translocation Catalysis. Biosci. Rep. 1991, 11, 477–538; discussion 534–538. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H. Vectorial Metabolism and the Evolution of Transport Systems. J. Bacteriol. 2000, 182, 5029–5035. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent Advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 Update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- Griffith, J.; Sansom, C. The Transporter Factsbook; Elsevier Science: Amsterdam, The Netherlands, 1997; ISBN 978-0-08-054265-2. [Google Scholar]
- Broome-Smith, J.K.; Symposium, S.; Baumberg, S.; Stirling, C.J.; Ward, F.B. Transport of Molecules Across Microbial Membranes; Cambridge University Press: Cambridge, UK, 1999; ISBN 978-0-521-77270-9. [Google Scholar]
- Burata, O.E.; Yeh, T.J.; Macdonald, C.B.; Stockbridge, R.B. Still Rocking in the Structural Era: A Molecular Overview of the Small Multidrug Resistance (SMR) Transporter Family. J. Biol. Chem. 2022, 298, 102482. [Google Scholar] [CrossRef]
- Hvorup, R.N.; Winnen, B.; Chang, A.B.; Jiang, Y.; Zhou, X.F.; Saier, M.H., Jr. The Multidrug/Oligosaccharidyl-Lipid/Polysaccharide (Mop) Exporter Superfamily. Eur. J. Biochem. 2003, 270, 799–813. [Google Scholar] [CrossRef]
- Kuroda, T.; Tsuchiya, T. Multidrug Efflux Transporters in the MATE Family. Biochim. Biophys. Acta 2009, 1794, 763–768. [Google Scholar] [CrossRef]
- Nikaido, H. RND Transporters in the Living World. Res. Microbiol. 2018, 169, 363–371. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.H.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.P.D.; Pokhrel, A. Pacing across the Membrane: The Novel Pace Family of Efflux Pumps Is Widespread in Gram-Negative Pathogens. Res. Microbiol. 2018, 169, 450–454. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Beatty, J.T.; Goffeau, A.; Harley, K.T.; Heijne, W.H.; Huang, S.C.; Jack, D.L.; Jahn, P.S.; Lew, K.; Liu, J.; et al. The Major Facilitator Superfamily. J. Mol. Microbiol. Biotechnol. 1999, 1, 257–279. [Google Scholar]
- Kumar, S.; He, G.; Kakarla, P.; Shrestha, U.; Ranjana, K.C.; Ranaweera, I.; Willmon, T.M.; Barr, S.R.; Hernandez, A.J.; Varela, M.F. Bacterial Multidrug Efflux Pumps of the Major Facilitator Superfamily as Targets for Modulation. Infect. Disord. Drug Targets 2016, 16, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H. Major Facilitator Superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Saidijam, M.; Benedetti, G.; Ren, Q.; Xu, Z.; Hoyle, C.J.; Palmer, S.L.; Ward, A.; Bettaney, K.E.; Szakonyi, G.; Meuller, J.; et al. Microbial Drug Efflux Proteins of the Major Facilitator Superfamily. Curr. Drug Targets 2006, 7, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.K.; Baker, M.E.; Rouch, D.A.; Page, M.G.; Skurray, R.A.; Paulsen, I.T.; Chater, K.F.; Baldwin, S.A.; Henderson, P.J. Membrane Transport Proteins: Implications of Sequence Comparisons. Curr. Opin. Cell. Biol. 1992, 4, 684–695. [Google Scholar] [CrossRef]
- Kumar, S.; Ranjana, K.; Sanford, L.M.; Hernandez, A.J.; Kakarla, P.; Varela, M.F. Structural and Functional Roles of Two Evolutionarily Conserved Amino Acid Sequence Motifs within Solute Transporters of the Major Facilitator Superfamily. Trends Cell. Mol. Biol. 2016, 11, 41–53. [Google Scholar]
- Levy, S.B.; McMurry, L. Plasmid-Determined Tetracycline Resistance Involves New Transport Systems for Tetracycline. Nature 1978, 276, 90–92. [Google Scholar] [CrossRef]
- McMurry, L.M.; Cullinane, J.C.; Petrucci, R.E., Jr.; Levy, S.B. Active Uptake of Tetracycline by Membrane Vesicles from Susceptible Escherichia coli. Antimicrob. Agents Chemother. 1981, 20, 307–313. [Google Scholar] [CrossRef]
- McMurry, L.; Petrucci, R.E., Jr.; Levy, S.B. Active Efflux of Tetracycline Encoded by Four Genetically Different Tetracycline Resistance Determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 1980, 77, 3974–3977. [Google Scholar] [CrossRef]
- Yoshida, H.; Bogaki, M.; Nakamura, S.; Ubukata, K.; Konno, M. Nucleotide Sequence and Characterization of the Staphylococcus aureus norA Gene, Which Confers Resistance to Quinolones. J. Bacteriol. 1990, 172, 6942–6949. [Google Scholar] [CrossRef]
- Edgar, R.; Bibi, E. MdfA, an Escherichia coli Multidrug Resistance Protein with an Extraordinarily Broad Spectrum of Drug Recognition. J. Bacteriol. 1997, 179, 2274–2280. [Google Scholar] [CrossRef] [PubMed]
- Tennent, J.M.; Lyon, B.R.; Midgley, M.; Jones, I.G.; Purewal, A.S.; Skurray, R.A. Physical and Biochemical Characterization of the qacA Gene Encoding Antiseptic and Disinfectant Resistance in Staphylococcus aureus. J. Gen. Microbiol. 1989, 135, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; He, X.; Szewczyk, P.; Nguyen, T.; Chang, G. Structure of the Multidrug Transporter EmrD from Escherichia coli. Science 2006, 312, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhao, Y.; Wang, X.; Fan, J.; Heng, J.; Liu, X.; Feng, W.; Kang, X.; Huang, B.; Liu, J.; et al. Structure of the YajR Transporter Suggests a Transport Mechanism Based on the Conserved Motif A. Proc. Natl. Acad. Sci. USA 2013, 110, 14664–14669. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhao, Y.; Fan, J.; Liu, X.; Wu, Y.; Feng, W.; Zhang, X.C. Atomic Resolution Structure of the E. coli YajR Transporter YAM Domain. Biochem. Biophys. Res. Commun. 2014, 450, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.; Zhao, Y.; Liu, M.; Liu, Y.; Fan, J.; Wang, X.; Zhang, X.C. Substrate-Bound Structure of the E. coli Multidrug Resistance Transporter MdfA. Cell. Res. 2015, 25, 1060–1073. [Google Scholar] [CrossRef]
- Xiao, Q.; Sun, B.; Zhou, Y.; Wang, C.; Guo, L.; He, J.; Deng, D. Visualizing the Nonlinear Changes of a Drug-Proton Antiporter from Inward-Open to Occluded State. Biochem. Biophys. Res. Commun. 2021, 534, 272–278. [Google Scholar] [CrossRef]
- Kumar, S.; Mahendran, I.; Athreya, A.; Ranjan, R.; Penmatsa, A. Isolation and Structural Characterization of a Zn(2+)-Bound Single-Domain Antibody against NorC, a Putative Multidrug Efflux Transporter in Bacteria. J. Biol. Chem. 2020, 295, 55–68. [Google Scholar] [CrossRef]
- Debruycker, V.; Hutchin, A.; Masureel, M.; Ficici, E.; Martens, C.; Legrand, P.; Stein, R.A.; McHaourab, H.S.; Faraldo-Gómez, J.D.; Remaut, H.; et al. An Embedded Lipid in the Multidrug Transporter LmrP Suggests a Mechanism for Polyspecificity. Nat. Struct. Mol. Biol. 2020, 27, 829–835. [Google Scholar] [CrossRef]
- Maloney, P.C. Bacterial Transporters. Curr. Opin. Cell. Biol. 1994, 6, 571–582. [Google Scholar] [CrossRef]
- Huang, Y.; Lemieux, M.J.; Song, J.; Auer, M.; Wang, D.-N. Structure and Mechanism of the Glycerol-3-Phosphate Transporter from Escherichia coli. Science 2003, 301, 616–620. [Google Scholar] [CrossRef]
- Ranaweera, I.; Shrestha, U.; Ranjana, K.C.; Kakarla, P.; Willmon, T.M.; Hernandez, A.J.; Mukherjee, M.M.; Barr, S.R.; Varela, M.F. Structural Comparison of Bacterial Multidrug Efflux Pumps of the Major Facilitator Superfamily. Trends Cell. Mol. Biol. 2015, 10, 131–140. [Google Scholar] [PubMed]
- Fowler, P.W.; Orwick-Rydmark, M.; Radestock, S.; Solcan, N.; Dijkman, P.M.; Lyons, J.A.; Kwok, J.; Caffrey, M.; Watts, A.; Forrest, L.R.; et al. Gating Topology of the Proton-Coupled Oligopeptide Symporters. Structure 2015, 23, 290–301. [Google Scholar] [CrossRef]
- Tanford, C. Mechanism of Free Energy Coupling in Active Transport. Annu. Rev. Biochem. 1983, 52, 379–409. [Google Scholar] [CrossRef]
- Jencks, W.P. From Chemistry to Biochemistry to Catalysis to Movement. Annu. Rev. Biochem. 1997, 66, 1–18. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Skurray, R.A. Topology, Structure and Evolution of Two Families of Proteins Involved in Antibiotic and Antiseptic Resistance in Eukaryotes and Prokaryotes—An Analysis. Gene 1993, 124, 1–11. [Google Scholar] [CrossRef]
- Varela, M.F.; Griffith, J.K. Nucleotide and deduced protein sequences of the class D tetracycline resistance determinant: Relationship to other antimicrobial transport proteins. Antimicrob. Agents Chemother. 1993, 37, 1253–1258. [Google Scholar] [CrossRef]
- Varela, M.F.; Sansom, C.E.; Griffith, J.K. Mutational Analysis and Molecular Modelling of an Amino acid Sequence Motif Conserved in Antiporters but not Symporters in a Transporter Superfamily. Mol. Membr. Biol. 1995, 12, 313–319. [Google Scholar] [CrossRef]
- Rouch, D.A.; Cram, D.S.; DiBerardino, D.; Littlejohn, T.G.; Skurray, R.A. Efflux-Mediated Antiseptic Resistance Gene qacA from Staphylococcus aureus: Common Ancestry with Tetracycline- and Sugar-Transport Proteins. Mol. Microbiol. 1990, 4, 2051–2062. [Google Scholar] [CrossRef]
- Maiden, M.C.; Davis, E.O.; Baldwin, S.A.; Moore, D.C.; Henderson, P.J. Mammalian and Bacterial Sugar Transport Proteins Are Homologous. Nature 1987, 325, 641–643. [Google Scholar] [CrossRef]
- Ginn, S.L.; Brown, M.H.; Skurray, R.A. The TetA (K) Tetracycline/H+ Antiporter from Staphylococcus aureus: Mutagenesis and Functional Analysis of Motif C. J. Bacteriol. 2000, 182, 1492–1498. [Google Scholar] [CrossRef]
- Konishi, S.; Iwaki, S.; Kimura-Someya, T.; Yamaguchi, A. Cysteine-Scanning Mutagenesis around Transmembrane Segment VI of Tn10-Encoded Metal-Tetracycline/H(+) Antiporter. FEBS Lett. 1999, 461, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Krulwich, T.A. Site-Directed Mutagenesis Studies of Selected Motif and Charged Residues and of Cysteines of the Multifunctional Tetracycline Efflux Protein Tet(L). J. Bacteriol. 2002, 184, 1796–1800. [Google Scholar] [CrossRef]
- De Jesus, M.; Jin, J.; Guffanti, A.A.; Krulwich, T.A. Importance of the GP Dipeptide of the Antiporter Motif and Other Membrane-Embedded Proline and Glycine Residues in Tetracycline Efflux Protein Tet(L). Biochemistry 2005, 44, 12896–12904. [Google Scholar] [CrossRef]
- Saraceni-Richards, C.A.; Levy, S.B. Evidence for Interactions between Helices 5 and 8 and a Role for the Interdomain Loop in Tetracycline Resistance Mediated by Hybrid Tet Proteins. J. Biol. Chem. 2000, 275, 6101–6106. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Galea, M.; Wu, J.; Mitchell, B.A.; Skurray, R.A.; Brown, M.H. Functional Effects of Intramembranous Proline Substitutions in the Staphylococcal Multidrug Transporter QacA. FEMS Microbiol. Lett. 2006, 263, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Pasrija, R.; Banerjee, D.; Prasad, R. Structure and Function Analysis of CaMdr1p, a Major Facilitator Superfamily Antifungal Efflux Transporter Protein of Candida albicans: Identification of Amino Acid Residues Critical for Drug/H+ Transport. Eukaryot. Cell. 2007, 6, 443–453. [Google Scholar] [CrossRef]
- Yaffe, D.; Radestock, S.; Shuster, Y.; Forrest, L.R.; Schuldiner, S. Identification of Molecular Hinge Points Mediating Alternating Access in the Vesicular Monoamine Transporter VMAT2. Proc. Natl. Acad. Sci. USA 2013, 110, E1332–E1341. [Google Scholar] [CrossRef]
- Luo, J.; Parsons, S.M. Conformational Propensities of Peptides Mimicking Transmembrane Helix 5 and Motif C in Wild-Type and Mutant Vesicular Acetylcholine Transporters. ACS Chem. Neurosci. 2010, 1, 381–390. [Google Scholar] [CrossRef]
- Lekshmi, M.; Ammini, P.; Adjei, J.; Sanford, L.M.; Shrestha, U.; Kumar, S.; Varela, M.F. Modulation of Antimicrobial Efflux Pumps of the Major Facilitator Superfamily in Staphylococcus aureus. AIMS Microbiol. 2018, 4, 1–18. [Google Scholar] [CrossRef]
- Lekshmi, M.; Parvathi, A.; Kumar, S.; Varela, M.F. Efflux Pump-Mediated Quorum Sensing: New Avenues for Modulation of Antimicrobial Resistance and Bacterial Virulence. In Biotechnological Applications of Quorum Sensing Inhibitors; Kalia, V.C., Ed.; Springer: Singapore, 2018; pp. 127–142. ISBN 978-981-10-9026-4. [Google Scholar]
- Henderson, P.J.F. Proton-Linked Sugar Transport Systems in Bacteria. J. Bioenerg. Biomembr. 1990, 22, 525–569. [Google Scholar] [CrossRef]
- Tamura, N.; Konishi, S.; Yamaguchi, A. Mechanisms of Drug/H+ Antiport: Complete Cysteine-Scanning Mutagenesis and the Protein Engineering Approach. Curr. Opin. Chem. Biol. 2003, 7, 570–579. [Google Scholar] [CrossRef]
- Nelson, M.L.; Levy, S.B. The History of the Tetracyclines. Ann. N. Y. Acad. Sci. 2011, 1241, 17–32. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Ono, N.; Akasaka, T.; Noumi, T.; Sawai, T. Metal-Tetracycline/H+ Antiporter of Escherichia coli Encoded by a Transposon, Tn10. The Role of the Conserved Dipeptide, Ser65-Asp66, in Tetracycline Transport. J. Biol. Chem. 1990, 265, 15525–15530. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Akasaka, T.; Kimura, T.; Sakai, T.; Adachi, Y.; Sawai, T. Role of the Conserved Quartets of Residues Located in the N- and C-Terminal Halves of the Transposon Tn10-Encoded Metal-Tetracycline/H+ Antiporter of Escherichia coli. Biochemistry 1993, 32, 5698–5704. [Google Scholar] [CrossRef]
- Kimura, T.; Shiina, Y.; Sawai, T.; Yamaguchi, A. Cysteine-Scanning Mutagenesis around Transmembrane Segment III of Tn10-Encoded Metal-Tetracycline/H+ Antiporter. J. Biol. Chem. 1998, 273, 5243–5247. [Google Scholar] [CrossRef] [PubMed]
- Bolhuis, H.; Poelarends, G.; van Veen, H.W.; Poolman, B.; Driessen, A.J.; Konings, W.N. The Lactococcal lmrP Gene Encodes a Proton Motive Force-Dependent Drug Transporter. J. Biol. Chem. 1995, 270, 26092–26098. [Google Scholar] [CrossRef]
- Masureel, M.; Martens, C.; Stein, R.A.; Mishra, S.; Ruysschaert, J.-M.; Mchaourab, H.S.; Govaerts, C. Protonation Drives the Conformational Switch in the Multidrug Transporter LmrP. Nat. Chem. Biol. 2014, 10, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.F.; Kumar, S. Strategies for Discovery of New Molecular Targets for Anti-Infective Drugs. Curr. Opin. Pharmacol. 2019, 48, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Stephen, J.; Lekshmi, M.; Ammini, P.; Kumar, S.H.; Varela, M.F. Membrane Efflux Pumps of Pathogenic Vibrio Species: Role in Antimicrobial Resistance and Virulence. Microorganisms 2022, 10, 382. [Google Scholar] [CrossRef]
- Mohanty, H.; Pachpute, S.; Yadav, R.P. Mechanism of Drug Resistance in Bacteria: Efflux Pump Modulation for Designing of New Antibiotic Enhancers. Folia Microbiol. (Praha) 2021, 66, 727–739. [Google Scholar] [CrossRef]
- Bruns, M.M.; Kakarla, P.; Floyd, J.T.; Mukherjee, M.M.; Ponce, R.C.; Garcia, J.A.; Ranaweera, I.; Sanford, L.M.; Hernandez, A.J.; Willmon, T.M.; et al. Modulation of the Multidrug Efflux Pump EmrD-3 from Vibrio cholerae by Allium sativum Extract and the Bioactive Agent Allyl Sulfide plus Synergistic Enhancement of Antimicrobial Susceptibility by A. sativum Extract. Arch. Microbiol. 2017, 199, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Kumar, S.; Varela, M.F. Identification, Cloning, and Functional Characterization of EmrD-3, a Putative Multidrug Efflux Pump of the Major Facilitator Superfamily from Vibrio cholerae O395. Arch. Microbiol. 2009, 191, 903–911. [Google Scholar] [CrossRef]
- Floyd, J.L.; Smith, K.P.; Kumar, S.H.; Floyd, J.T.; Varela, M.F. LmrS Is a Multidrug Efflux Pump of the Major Facilitator Superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5406–5412. [Google Scholar] [CrossRef]
- Kumar, G.; Kiran Tudu, A. Tackling Multidrug-Resistant Staphylococcus aureus by Natural Products and Their Analogues Acting as NorA Efflux Pump Inhibitors. Bioorg. Med. Chem. 2023, 80, 117187. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.E.; de Freitas, T.S.; Xavier, J.C.; Pereira, R.L.S.; Pereira Junior, F.N.; Nogueira, C.E.S.; Marinho, M.M.; Bandeira, P.N.; Rodrigues, L.G.; Marinho, E.S.; et al. ADMET Study, Spectroscopic Characterization and Effect of Synthetic Nitro Chalcone in Combination with Norfloxacin, Ciprofloxacin, and Ethidium Bromide against Staphylococcus aureus Efflux Pumps. Fundam. Clin. Pharmacol. 2023, 37, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Santos Araujo, N.J.; Pereira da Silva, A.R.; Costa, M.D.S.; Pereira Silva, C.A.; Sampaio de Freitas, T.; Oliveira de Sousa, E.; Barbosa Filho, J.M.; Lobo Soares de Matos, Y.M.; Melo Coutinho, H.D.; Andrade-Pinheiro, J.C. Evaluation of the Antibacterial Activity of Hecogenin Acetate and Its Inhibitory Potential of NorA and MepA Efflux Pumps from Staphylococcus aureus. Microb. Pathog. 2023, 174, 105925. [Google Scholar] [CrossRef]
- Oliveira-Tintino, C.D.d.M.; Tintino, S.R.; Justino de Araújo, A.C.; dos Santos Barbosa, C.R.; Ramos Freitas, P.; Araújo Neto, J.B.d.; Begnini, I.M.; Rebelo, R.A.; Silva, L.E.d.; Mireski, S.L.; et al. Efflux Pump (QacA, QacB, and QacC) and β-Lactamase Inhibitors? An Evaluation of 1,8-Naphthyridines against Staphylococcus aureus Strains. Molecules 2023, 28, 1819. [Google Scholar] [CrossRef]
- Tong, Y.; Zhang, J.; Sun, N.; Wang, X.M.; Wei, Q.; Zhang, Y.; Huang, R.; Pu, Y.; Dai, H.; Ren, B.; et al. Berberine Reverses Multidrug Resistance in Candida albicans by Hijacking the Drug Efflux Pump Mdr1p. Sci. Bull. Beijing 2021, 66, 1895–1905. [Google Scholar] [CrossRef]
- Li, Y.; Ge, X. Role of Berberine as a Potential Efflux Pump Inhibitor against MdfA from Escherichia coli: In Vitro and In Silico Studies. Microbiol. Spectr. 2023, 11, e03324-22. [Google Scholar] [CrossRef]
- Gil, F.; Laiolo, J.; Bayona-Pacheco, B.; Cannon, R.D.; Ferreira-Pereira, A.; Carpinella, M.C. Extracts from Argentinian Native Plants Reverse Fluconazole Resistance in Candida Species by Inhibiting the Efflux Transporters Mdr1 and Cdr1. BMC Complement. Med. Ther. 2022, 22, 264. [Google Scholar] [CrossRef]
- Khan, I.A.; Mirza, Z.M.; Kumar, A.; Verma, V.; Qazi, G.N. Piperine, a Phytochemical Potentiator of Ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 810–812. [Google Scholar] [CrossRef]
- Nargotra, A.; Sharma, S.; Koul, J.L.; Sangwan, P.L.; Khan, I.A.; Kumar, A.; Taneja, S.C.; Koul, S. Quantitative Structure Activity Relationship (QSAR) of Piperine Analogsfor Bacterial NorA Efflux Pump Inhibitors. Eur. J. Med. Chem. 2009, 44, 4128–4135. [Google Scholar] [CrossRef]
- Seukep, A.J.; Kuete, V.; Nahar, L.; Sarker, S.D.; Guo, M. Plant-Derived Secondary Metabolites as the Main Source of Efflux Pump Inhibitors and Methods for Identification. J. Pharm. Anal. 2020, 10, 277–290. [Google Scholar] [CrossRef]
- Bame, J.R.; Graf, T.N.; Junio, H.A.; Iii, R.O.B.; Jarmusch, S.A.; El-Elimat, T.; Iii, J.O.F.; Oberlies, N.H.; Cech, R.A.; Cech, N.B. Sarothrin from Alkanna orientalis Is an Antimicrobial Agent and Efflux Pump Inhibitor. Planta Med. 2013, 79, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Sun, Z.-L.; Liu, T.; Gibbons, S.; Zhang, W.-J.; Qing, M. Flavonoids from Sophora moorcroftiana and Their Synergistic Antibacterial Effects on MRSA. Phytother. Res. PTR 2014, 28, 1071–1076. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Scriven, L.N.; Tegos, G.; Lewis, K. Two Flavonols from Artemisa annua which Potentiate the Activity of Berberine and Norfloxacin Against a Resistant Strain of Staphylococcus aureus. Planta Med. 2002, 68, 1140–1141. [Google Scholar] [CrossRef]
- Smith, E.C.J.; Williamson, E.M.; Wareham, N.; Kaatz, G.W.; Gibbons, S. Antibacterials and Modulators of Bacterial Resistance from the Immature Cones of Chamaecyparis lawsoniana. Phytochemistry 2007, 68, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.C.J.; Kaatz, G.W.; Seo, S.M.; Wareham, N.; Williamson, E.M.; Gibbons, S. The Phenolic Diterpene Totarol Inhibits Multidrug Efflux Pump Activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 4480–4483. [Google Scholar] [CrossRef]
- Morel, C.; Stermitz, F.R.; Tegos, G.; Lewis, K. Isoflavones As Potentiators of Antibacterial Activity. J. Agric. Food Chem. 2003, 51, 5677–5679. [Google Scholar] [CrossRef]
- Roy, S.K.; Kumari, N.; Pahwa, S.; Agrahari, U.C.; Bhutani, K.K.; Jachak, S.M.; Nandanwar, H. NorA Efflux Pump Inhibitory Activity of Coumarins from Mesua ferrea. Fitoterapia 2013, 90, 140–150. [Google Scholar] [CrossRef]
- Holler, J.G.; Christensen, S.B.; Slotved, H.-C.; Rasmussen, H.B.; Gúzman, A.; Olsen, C.-E.; Petersen, B.; Mølgaard, P. Novel Inhibitory Activity of the Staphylococcus aureus NorA Efflux Pump by a Kaempferol Rhamnoside Isolated from Persea lingue Nees. J. Antimicrob. Chemother. 2012, 67, 1138–1144. [Google Scholar] [CrossRef]
- Marquez, B.; Neuville, L.; Moreau, N.J.; Genet, J.-P.; dos Santos, A.F.; Caño de Andrade, M.C.; Goulart Sant’Ana, A.E. Multidrug Resistance Reversal Agent from Jatropha elliptica. Phytochemistry 2005, 66, 1804–1811. [Google Scholar] [CrossRef]
- Ponnusamy, K.; Ramasamy, M.; Savarimuthu, I.; Paulraj, M.G. Indirubin Potentiates Ciprofloxacin Activity in the NorA Efflux Pump of Staphylococcus aureus. Scand. J. Infect. Dis. 2010, 42, 500–505. [Google Scholar] [CrossRef]
- Holler, J.G.; Slotved, H.-C.; Mølgaard, P.; Olsen, C.E.; Christensen, S.B. Chalcone Inhibitors of the NorA Efflux Pump in Staphylococcus aureus Whole Cells and Enriched Everted Membrane Vesicles. Bioorg. Med. Chem. 2012, 20, 4514–4521. [Google Scholar] [CrossRef]
- Hellewell, L.; Bhakta, S. Chalcones, Stilbenes and Ketones Have Anti-Infective Properties via Inhibition of Bacterial Drug-Efflux and Consequential Synergism with Antimicrobial Agents. Access. Microbiol. 2020, 2, acmi000105. [Google Scholar] [CrossRef]
- Pereda-Miranda, R.; Kaatz, G.W.; Gibbons, S. Polyacylated Oligosaccharides from Medicinal Mexican Morning Glory Species as Antibacterials and Inhibitors of Multidrug Resistance in Staphylococcus aureus. J. Nat. Prod. 2006, 69, 406–409. [Google Scholar] [CrossRef]
- Sabatini, S.; Gosetto, F.; Iraci, N.; Barreca, M.L.; Massari, S.; Sancineto, L.; Manfroni, G.; Tabarrini, O.; Dimovska, M.; Kaatz, G.W.; et al. Re-Evolution of the 2-Phenylquinolines: Ligand-Based Design, Synthesis, and Biological Evaluation of a Potent New Class of Staphylococcus aureus NorA Efflux Pump Inhibitors to Combat Antimicrobial Resistance. J. Med. Chem. 2013, 56, 4975–4989. [Google Scholar] [CrossRef]
- Felicetti, T.; Mangiaterra, G.; Cannalire, R.; Cedraro, N.; Pietrella, D.; Astolfi, A.; Massari, S.; Tabarrini, O.; Manfroni, G.; Barreca, M.L.; et al. C-2 Phenyl Replacements to Obtain Potent Quinoline-Based Staphylococcus aureus NorA Inhibitors. J. Enzym. Inhib. Med. Chem. 2020, 35, 584–597. [Google Scholar] [CrossRef]
- Felicetti, T.; Cannalire, R.; Nizi, M.G.; Tabarrini, O.; Massari, S.; Barreca, M.L.; Manfroni, G.; Schindler, B.D.; Cecchetti, V.; Kaatz, G.W.; et al. Studies on 2-Phenylquinoline Staphylococcus aureus NorA Efflux Pump Inhibitors: New Insights on the C-6 Position. Eur. J. Med. Chem. 2018, 155, 428–433. [Google Scholar] [CrossRef]
- Gibbons, S.; Oluwatuyi, M.; Kaatz, G.W. A Novel Inhibitor of Multidrug Efflux Pumps in Staphylococcus aureus. J. Antimicrob. Chemother. 2003, 51, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Chovanová, R.; Mezovská, J.; Vaverková, Š.; Mikulášová, M. The Inhibition the Tet(K) Efflux Pump of Tetracycline Resistant Staphylococcus epidermidis by Essential Oils from Three Salvia Species. Lett. Appl. Microbiol. 2015, 61, 58–62. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Tawara-Matsuda, J.; Lorenz, P.; Mueller, P.; Zenewicz, L.; Lewis, K. 5′-Methoxyhydnocarpin-D and Pheophorbide A: Berberis Species Components That Potentiate Berberine Growth Inhibition of Resistant Staphylococcus aureus. J. Nat. Prod. 2000, 63, 1146–1149. [Google Scholar] [CrossRef]
- Kakarla, P.; Floyd, J.; Mukherjee, M.; Devireddy, A.R.; Inupakutika, M.A.; Ranweera, I.; Kc, R.; Shrestha, U.; Cheeti, U.R.; Willmon, T.M.; et al. Inhibition of the Multidrug Efflux Pump LmrS from Staphylococcus aureus by Cumin Spice Cuminum cyminum. Arch. Microbiol. 2017, 199, 465–474. [Google Scholar] [CrossRef]
- Samosorn, S.; Tanwirat, B.; Muhamad, N.; Casadei, G.; Tomkiewicz, D.; Lewis, K.; Suksamrarn, A.; Prammananan, T.; Gornall, K.C.; Beck, J.L.; et al. Antibacterial Activity of Berberine-NorA Pump Inhibitor Hybrids with a Methylene Ether Linking Group. Bioorg. Med. Chem. 2009, 17, 3866–3872. [Google Scholar] [CrossRef]
- Waditzer, M.; Bucar, F. Flavonoids as Inhibitors of Bacterial Efflux Pumps. Molecules 2021, 26, 6904. [Google Scholar] [CrossRef]
- Shiu, W.K.P.; Malkinson, J.P.; Rahman, M.M.; Curry, J.; Stapleton, P.; Gunaratnam, M.; Neidle, S.; Mushtaq, S.; Warner, M.; Livermore, D.M.; et al. A New Plant-Derived Antibacterial Is an Inhibitor of Efflux Pumps in Staphylococcus aureus. Int. J. Antimicrob. Agents 2013, 42, 513–518. [Google Scholar] [CrossRef]
- Chérigo, L.; Pereda-Miranda, R.; Fragoso-Serrano, M.; Jacobo-Herrera, N.; Kaatz, G.W.; Gibbons, S. Inhibitors of Bacterial Multidrug Efflux Pumps from the Resin Glycosides of Ipomoea murucoides. J. Nat. Prod. 2008, 71, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Tiwari, N.; Gupta, P.; Verma, S.; Pal, A.; Srivastava, S.K.; Darokar, M.P. A Clerodane Diterpene from Polyalthia longifolia as a Modifying Agent of the Resistance of Methicillin Resistant Staphylococcus aureus. Phytomedicine 2016, 23, 654–661. [Google Scholar] [CrossRef]
- Kalia, N.P.; Mahajan, P.; Mehra, R.; Nargotra, A.; Sharma, J.P.; Koul, S.; Khan, I.A. Capsaicin, a Novel Inhibitor of the NorA Efflux Pump, Reduces the Intracellular Invasion of Staphylococcus aureus. J. Antimicrob. Chemother. 2012, 67, 2401–2408. [Google Scholar] [CrossRef]
- Singh, S.; Kalia, N.P.; Joshi, P.; Kumar, A.; Sharma, P.R.; Kumar, A.; Bharate, S.B.; Khan, I.A. Boeravinone B, A Novel Dual Inhibitor of NorA Bacterial Efflux Pump of Staphylococcus aureus and Human P-Glycoprotein, Reduces the Biofilm Formation and Intracellular Invasion of Bacteria. Front. Microbiol. 2017, 8, 1868. [Google Scholar] [CrossRef]
- Tintino, S.R.; Oliveira-Tintino, C.D.; Campina, F.F.; Silva, R.L.; Costa Mdo, S.; Menezes, I.R.; Calixto-Junior, J.T.; Siqueira-Junior, J.P.; Coutinho, H.D.; Leal-Balbino, T.C.; et al. Evaluation of the Tannic Acid Inhibitory Effect against the NorA Efflux Pump of Staphylococcus aureus. Microb. Pathog. 2016, 97, 9–13. [Google Scholar] [CrossRef]
- Ojeda-Sana, A.M.; Repetto, V.; Moreno, S. Carnosic Acid Is an Efflux Pumps Modulator by Dissipation of the Membrane Potential in Enterococcus faecalis and Staphylococcus aureus. World J. Microbiol. Biotechnol. 2013, 29, 137–144. [Google Scholar] [CrossRef]
- Vázquez, N.M.; Fiorilli, G.; Cáceres Guido, P.A.; Moreno, S. Carnosic Acid Acts Synergistically with Gentamicin in Killing Methicillin-Resistant Staphylococcus aureus Clinical Isolates. Phytomedicine 2016, 23, 1337–1343. [Google Scholar] [CrossRef]
- Chan, B.C.L.; Han, X.Q.; Lui, S.L.; Wong, C.W.; Wang, T.B.Y.; Cheung, D.W.S.; Cheng, S.W.; Ip, M.; Han, S.Q.B.; Yang, X.-S.; et al. Combating against Methicillin-Resistant Staphylococcus aureus-Two Fatty Acids from Purslane (Portulaca Oleracea L.) Exhibit Synergistic Effects with Erythromycin. J. Pharm. Pharmacol. 2015, 67, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.; Moser, E.; Kaatz, G.W. Catechin Gallates Inhibit Multidrug Resistance (MDR) in Staphylococcus aureus. Planta Med. 2004, 70, 1240–1242. [Google Scholar] [CrossRef]
- Joshi, P.; Singh, S.; Wani, A.; Sharma, S.; Jain, S.K.; Singh, B.; Gupta, B.D.; Satti, N.K.; Koul, S.; Khan, I.A.; et al. Osthol and Curcumin as Inhibitors of Human Pgp and Multidrug Efflux Pumps of Staphylococcus aureus: Reversing the Resistance against Frontline Antibacterial Drugs. Med. Chem. Comm. 2014, 5, 1540–1547. [Google Scholar] [CrossRef]
- Sobisch, L.-Y.; Rogowski, K.M.; Fuchs, J.; Schmieder, W.; Vaishampayan, A.; Oles, P.; Novikova, N.; Grohmann, E. Biofilm Forming Antibiotic Resistant Gram-Positive Pathogens Isolated From Surfaces on the International Space Station. Front. Microbiol. 2019, 10, 543. [Google Scholar] [CrossRef]
- Davies, D. Understanding Biofilm Resistance to Antibacterial Agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Bridier, A.; Sanchez-Vizuete, P.; Guilbaud, M.; Piard, J.-C.; Naitali, M.; Briandet, R. Biofilm-Associated Persistence of Food-Borne Pathogens. Food Microbiol. 2015, 45, 167–178. [Google Scholar] [CrossRef]
- Fernández, L.; Escobedo, S.; Gutiérrez, D.; Portilla, S.; Martínez, B.; García, P.; Rodríguez, A. Bacteriophages in the Dairy Environment: From Enemies to Allies. Antibiotics 2017, 6, 27. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-P.; Yang, Y.; Lu, D.-P.; Niu, Z.-S.; Feng, J.-N.; Chen, Y.-R.; Tou, F.-Y.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a Sink for Antibiotic Resistance Genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- Ratajczak, M.; Kamińska, D.; Nowak-Malczewska, D.M.; Schneider, A.; Dlugaszewska, J. Relationship between Antibiotic Resistance, Biofilm Formation, Genes Coding Virulence Factors and Source of Origin of Pseudomonas aeruginosa Clinical Strains. Ann. Agric. Environ. Med. AAEM 2021, 28, 306–313. [Google Scholar] [CrossRef]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside Antibiotics Induce Bacterial Biofilm Formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Høiby, N.; Henneberg, K.-Å.; Wang, H.; Stavnsbjerg, C.; Bjarnsholt, T.; Ciofu, O.; Johansen, U.R.; Sams, T. Formation of Pseudomonas aeruginosa Inhibition Zone during Tobramycin Disk Diffusion Is Due to Transition from Planktonic to Biofilm Mode of Growth. Int. J. Antimicrob. Agents 2019, 53, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular Mechanisms of Biofilm-Based Antibiotic Resistance and Tolerance in Pathogenic Bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Pletzer, D.; Mansour, S.C.; Hancock, R.E.W. Synergy between Conventional Antibiotics and Anti-Biofilm Peptides in a Murine, Sub-Cutaneous Abscess Model Caused by Recalcitrant ESKAPE Pathogens. PLoS Pathog. 2018, 14, e1007084. [Google Scholar] [CrossRef] [PubMed]
- Dastgheyb, S.; Parvizi, J.; Shapiro, I.M.; Hickok, N.J.; Otto, M. Effect of Biofilms on Recalcitrance of Staphylococcal Joint Infection to Antibiotic Treatment. J. Infect. Dis. 2015, 211, 641–650. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. MMBR 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin Resistance and the Biofilm Phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased Antibiotic Resistance of Escherichia coli in Mature Biofilms. Appl. Environ. Microbiol. 2009, 75, 4093–4100. [Google Scholar] [CrossRef]
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of Efflux Pumps Abolishes Bacterial Biofilm Formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382. [Google Scholar] [CrossRef] [PubMed]
- Christena, L.R.; Subramaniam, S.; Vidhyalakshmi, M.; Mahadevan, V.; Sivasubramanian, A.; Nagarajan, S. Dual Role of Pinostrobin-a Flavonoid Nutraceutical as an Efflux Pump Inhibitor and Antibiofilm Agent to Mitigate Food Borne Pathogens. RSC Adv. 2015, 5, 61881–61887. [Google Scholar] [CrossRef]
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.V.; Webber, M.A. Loss of or Inhibition of All Multidrug Resistance Efflux Pumps of Salmonella enterica Serovar Typhimurium Results in Impaired Ability to Form a Biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417. [Google Scholar] [CrossRef]
- Rezaie, P.; Pourhajibagher, M.; Chiniforush, N.; Hosseini, N.; Bahador, A. The Effect of Quorum-Sensing and Efflux Pumps Interactions in Pseudomonas aeruginosa Against Photooxidative Stress. J. Lasers Med. Sci. 2018, 9, 161–167. [Google Scholar] [CrossRef]
- Kaur, B.; Gupta, J.; Sharma, S.; Sharma, D.; Sharma, S. Focused Review on Dual Inhibition of Quorum Sensing and Efflux Pumps: A Potential Way to Combat Multi Drug Resistant Staphylococcus aureus Infections. Int. J. Biol. Macromol. 2021, 190, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Rahmati, S.; Yang, S.; Davidson, A.L.; Zechiedrich, E.L. Control of the AcrAB Multidrug Efflux Pump by Quorum-Sensing Regulator SdiA. Mol. Microbiol. 2002, 43, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Schembri, M.A.; Kjaergaard, K.; Klemm, P. Global Gene Expression in Escherichia coli Biofilms. Mol. Microbiol. 2003, 48, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Waite, R.D.; Papakonstantinopoulou, A.; Littler, E.; Curtis, M.A. Transcriptome Analysis of Pseudomonas aeruginosa Growth: Comparison of Gene Expression in Planktonic Cultures and Developing and Mature Biofilms. J. Bacteriol. 2005, 187, 6571–6576. [Google Scholar] [CrossRef] [PubMed]
- Short, F.L.; Liu, Q.; Shah, B.; Clift, H.E.; Naidu, V.; Li, L.; Prity, F.T.; Mabbutt, B.C.; Hassan, K.A.; Paulsen, I.T. The Acinetobacter baumannii Disinfectant Resistance Protein, AmvA, Is a Spermidine and Spermine Efflux Pump. Commun. Biol. 2021, 4, 1114. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, J.; Guo, L.; Zhao, W.; Hu, X.; Wei, X. Inactivation of a Putative Efflux Pump (LmrB) in Streptococcus Mutans Results in Altered Biofilm Structure and Increased Exopolysaccharide Synthesis: Implications for Biofilm Resistance. Biofouling 2017, 33, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef]
- Lamut, A.; Peterlin Mašič, L.; Kikelj, D.; Tomašič, T. Efflux Pump Inhibitors of Clinically Relevant Multidrug Resistant Bacteria. Med. Res. Rev. 2019, 39, 2460–2504. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef]
- Singh, S.; Datta, S.; Narayanan, K.B.; Rajnish, K.N. Bacterial Exo-Polysaccharides in Biofilms: Role in Antimicrobial Resistance and Treatments. J. Genet. Eng. Biotechnol. 2021, 19, 140. [Google Scholar] [CrossRef]
- Tomaś, N.; Myszka, K.; Wolko, Ł.; Nuc, K.; Szwengiel, A.; Grygier, A.; Majcher, M. Effect of Black Pepper Essential Oil on Quorum Sensing and Efflux Pump Systems in the Fish-Borne Spoiler Pseudomonas psychrophila KM02 Identified by RNA-Seq, RT-QPCR and Molecular Docking Analyses. Food Control 2021, 130, 108284. [Google Scholar] [CrossRef]
- Aggarwal, S.; Singh, D.V. Efflux Pumps and Biofilm Formation by Both Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus Strains. Indian. J. Exp. Biol. IJEB 2020, 58, 527–538. [Google Scholar] [CrossRef]
- Christena, L.R.; Mangalagowri, V.; Pradheeba, P.; Ahmed, K.B.A.; Shalini, B.I.S.; Vidyalakshmi, M.; Anbazhagan, V.; Subramanian, N.S. Copper Nanoparticles as an Efflux Pump Inhibitor to Tackle Drug Resistant Bacteria. RSC Adv. 2015, 5, 12899–12909. [Google Scholar] [CrossRef]
- Yu, Y.; Zhao, Y.; He, Y.; Pang, J.; Yang, Z.; Zheng, M.; Yin, R. Inhibition of Efflux Pump Encoding Genes and Biofilm Formation by Sub-Lethal Photodynamic Therapy in Methicillin Susceptible and Resistant Staphylococcus aureus. Photodiagnosis Photodyn. Ther. 2022, 39, 102900. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.; Klinger-Strobel, M.; Bohnert, J.A.; Wendler, S.; Rödel, J.; Pletz, M.W.; Löffler, B.; Tuchscherr, L. Clinically Approved Drugs Inhibit the Staphylococcus aureus Multidrug NorA Efflux Pump and Reduce Biofilm Formation. Front. Microbiol. 2019, 10, 2762. [Google Scholar] [CrossRef]
- Abd El-Baky, R.M.; Sandle, T.; John, J.; Abuo-Rahma, G.E.-D.A.; Hetta, H.F. A Novel Mechanism of Action of Ketoconazole: Inhibition of the NorA Efflux Pump System and Biofilm Formation in Multidrug-Resistant Staphylococcus aureus. Infect. Drug Resist. 2019, 12, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, K.; Furukawa, S.; Ogihara, H.; Morinaga, Y. Roles of Multidrug Efflux Pumps on the Biofilm Formation of Escherichia coli K-12. Biocontrol Sci. 2011, 16, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Pasqua, M.; Grossi, M.; Scinicariello, S.; Aussel, L.; Barras, F.; Colonna, B.; Prosseda, G. The MFS Efflux Pump EmrKY Contributes to the Survival of Shigella within Macrophages. Sci. Rep. 2019, 9, 2906. [Google Scholar] [CrossRef]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a Therapeutic Tool to Combat Microbial Resistance. Adv. Drug Deliv. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef]
- Nejabatdoust, A.; Zamani, H.; Salehzadeh, A. Functionalization of ZnO Nanoparticles by Glutamic Acid and Conjugation with Thiosemicarbazide Alters Expression of Efflux Pump Genes in Multiple Drug-Resistant Staphylococcus aureus Strains. Microb. Drug Resist. 2019, 25, 966–974. [Google Scholar] [CrossRef]
- Seena, S.; Rai, A. Nanoengineering Approaches to Fight Multidrug-Resistant Bacteria. In Non-Traditional Approaches to Combat. Antimicrobial Drug Resistance; Wani, M.Y., Ahmad, A., Eds.; Springer Nature: Singapore, 2023; pp. 221–248. ISBN 978-981-19916-7-7. [Google Scholar]
- Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms 2017, 5, 11. [Google Scholar] [CrossRef]
- Lowrence, R.C.; Subramaniapillai, S.G.; Ulaganathan, V.; Nagarajan, S. Tackling Drug Resistance with Efflux Pump Inhibitors: From Bacteria to Cancerous Cells. Crit. Rev. Microbiol. 2019, 45, 334–353. [Google Scholar] [CrossRef]

| Inhibitor | Efflux Pump | Reference |
|---|---|---|
| Plant-derived alkaloid compounds (reserpine, piperines, and piperine analogs) | NorA, Bmr, MdeA, LmrA, PmrA | [105,106,107] |
| Flavonoids (genistein, sarothrin) | NorA | [108,109] |
| Flavones (chrysosplenol-D and chrysoplenetin) | NorA | [110] |
| Diterpenes (ferruginol) | NorA | [111,112] |
| Isolflavones | NorA | [113] |
| Coumarins | NorA | [114] |
| Kaempferol rhamnoside | NorA | [115] |
| 2,6-dimethyl-4-phenyl-pyridine-3,5-dicarboxylic acid diethyl ester | NorA, MsrA | [116] |
| Indirubin | NorA | [117] |
| Chalcones (4-phenoxy-4′-dimethylamino ethoxy chalcone) | NorA | [118,119] |
| Oligosaccharides (orizabin) | NorA | [120] |
| Derivatives of 2-phenylquinoline | NorA | [121,122,123] |
| Abietane diterpenes | Tet(K), Msr(A) | [124] |
| Essential oils | TetK | [125] |
| 5′-Methoxyhydnocarpin-D and Pheophorbide A | NorA | [126] |
| Cumin seed oil, cumin aldehyde | LmrS | [127] |
| Plant-derived alkaloid compounds (berberine and palmatine) | NorA, MdfA | [103,128,129] |
| Sarothrin | NorA | [108] |
| Olympicin | NorA | [130] |
| Murucoidins | NorA | [131] |
| Clerodane diterpene 16α- hydroxycleroda-3,13 (14)-Z-dien-15,16-olid 6 | NorB, NorC | [132] |
| Verapamil, capsaicin, boeravinone B | NorA, QacA | [133,134] |
| Cholecalciferol and alpha-tocopherol | TetK, MsrA | [101,135] |
| Carnosic acid | MsrA, TetK, and NorA | [136,137] |
| Linoleic and oleic acids | MsrA | [138] |
| Epigallocatechin gallate, Epicatechin gallate | TetK | [139] |
| Osthtol | NorA, MdeA, TetK, MsrA | [140] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varela, M.F.; Stephen, J.; Bharti, D.; Lekshmi, M.; Kumar, S. Inhibition of Multidrug Efflux Pumps Belonging to the Major Facilitator Superfamily in Bacterial Pathogens. Biomedicines 2023, 11, 1448. https://doi.org/10.3390/biomedicines11051448
Varela MF, Stephen J, Bharti D, Lekshmi M, Kumar S. Inhibition of Multidrug Efflux Pumps Belonging to the Major Facilitator Superfamily in Bacterial Pathogens. Biomedicines. 2023; 11(5):1448. https://doi.org/10.3390/biomedicines11051448
Chicago/Turabian StyleVarela, Manuel F., Jerusha Stephen, Deeksha Bharti, Manjusha Lekshmi, and Sanath Kumar. 2023. "Inhibition of Multidrug Efflux Pumps Belonging to the Major Facilitator Superfamily in Bacterial Pathogens" Biomedicines 11, no. 5: 1448. https://doi.org/10.3390/biomedicines11051448
APA StyleVarela, M. F., Stephen, J., Bharti, D., Lekshmi, M., & Kumar, S. (2023). Inhibition of Multidrug Efflux Pumps Belonging to the Major Facilitator Superfamily in Bacterial Pathogens. Biomedicines, 11(5), 1448. https://doi.org/10.3390/biomedicines11051448