Abstinence from Escalation of Cocaine Intake Changes the microRNA Landscape in the Cortico-Accumbal Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Rat Brain Samples
2.2. Rat Brain Dissection and Small RNA Sequencing (sRNA-Seq)
2.3. Statistical Analysis
2.4. Bioinformatics Analysis
3. Results
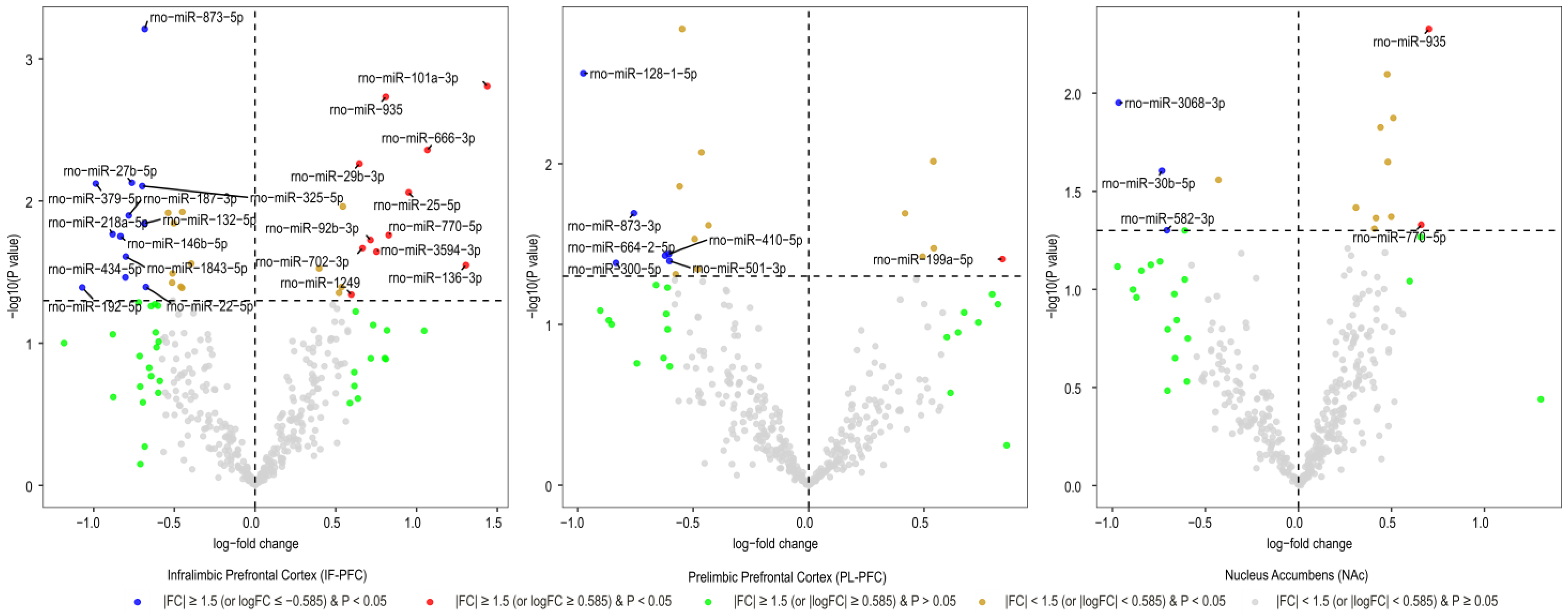
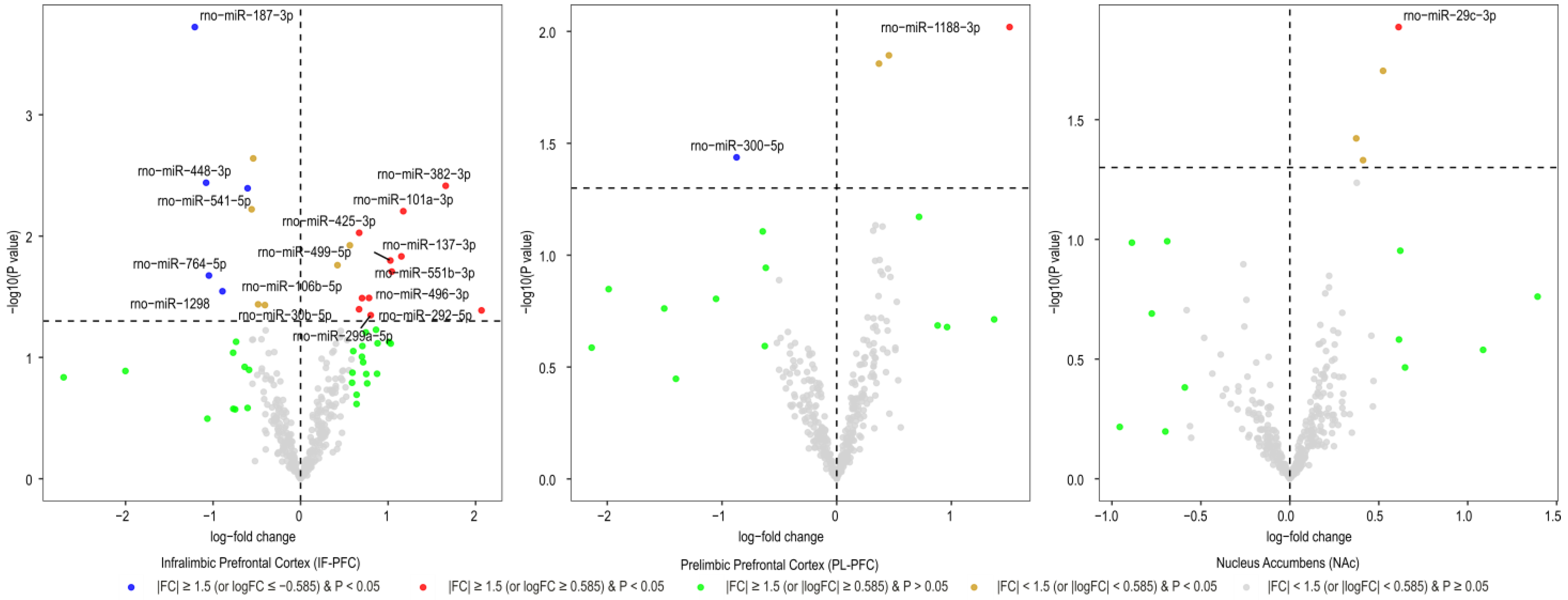
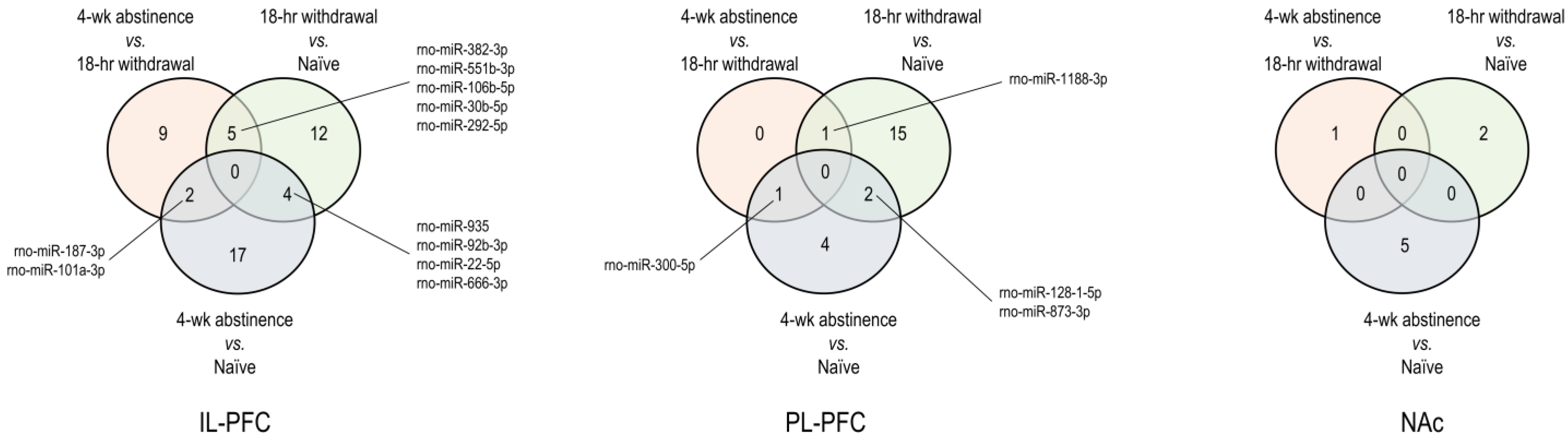
3.1. Brain Region-Specific miRNA Expression Changes Due to Acute Withdrawal or Protracted Abstinence following Cocaine Self-Administration (SA)
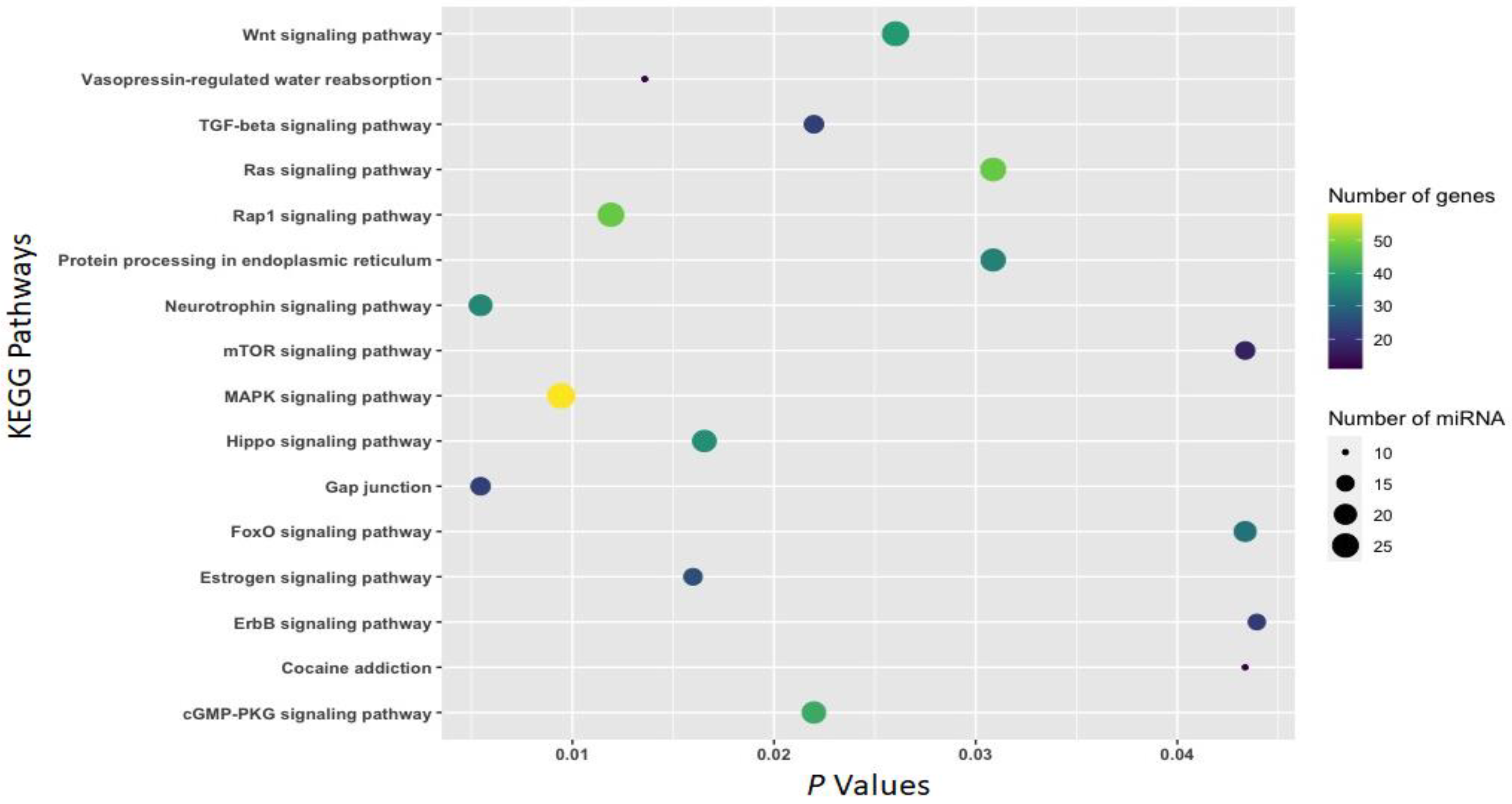
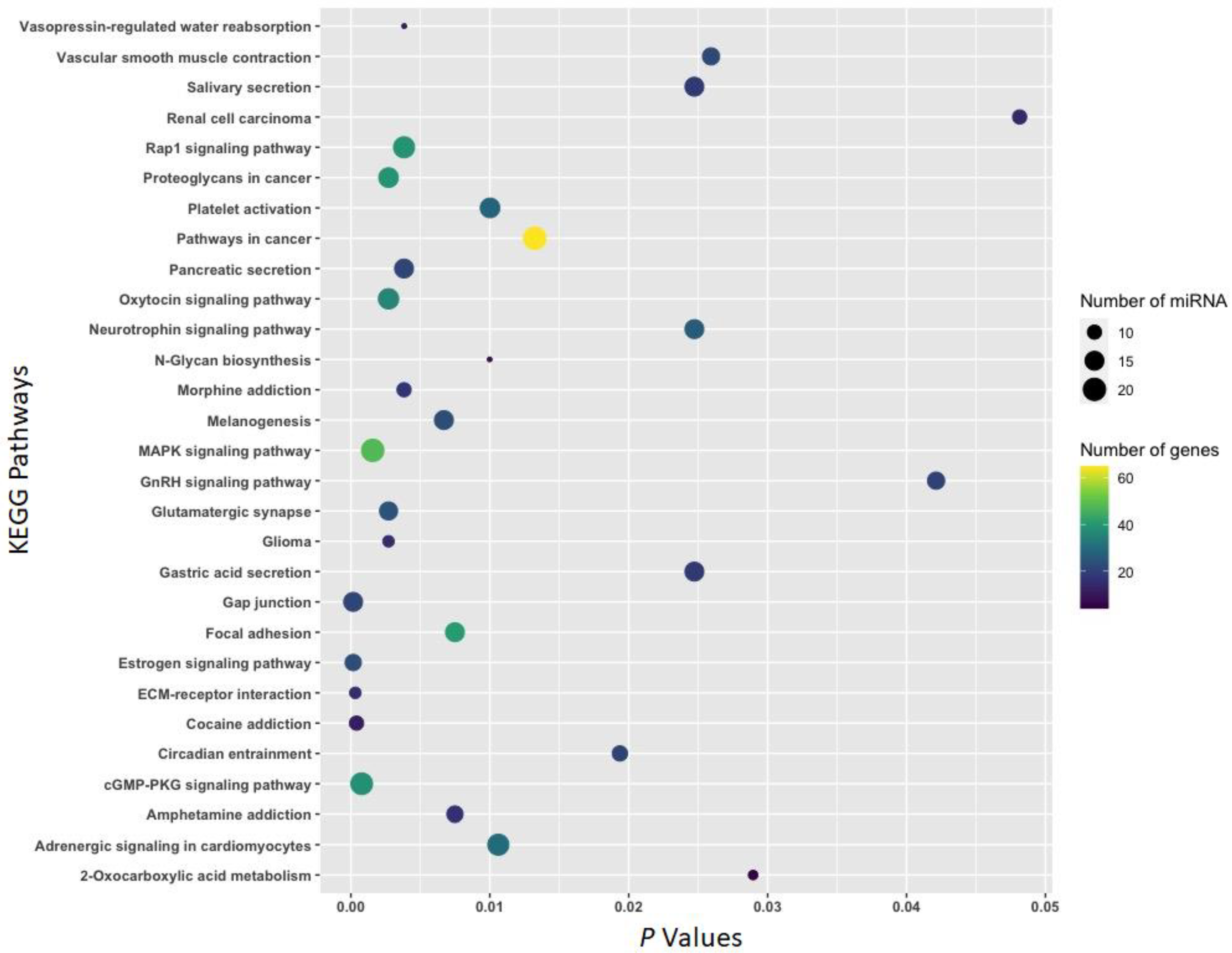
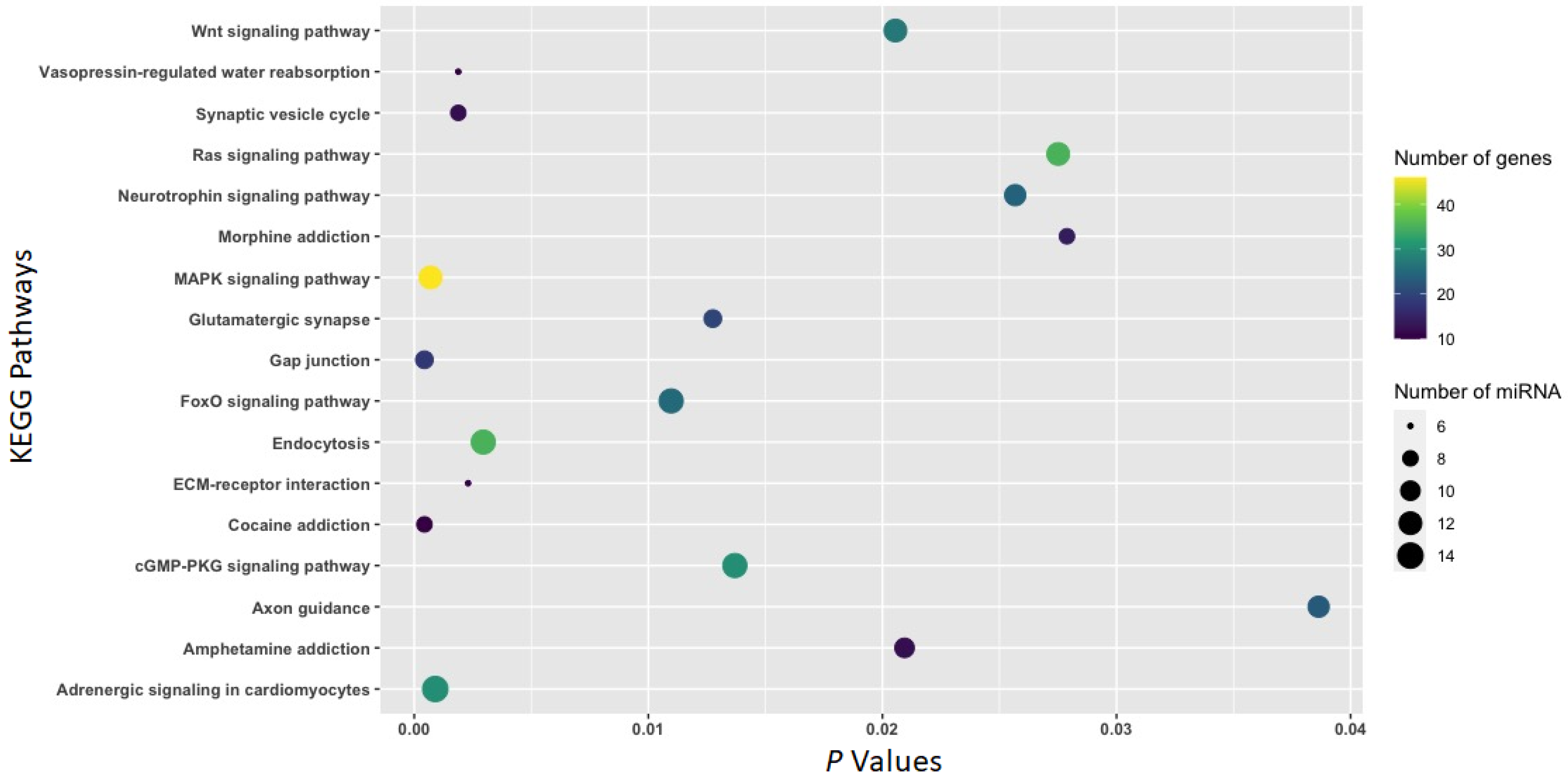
3.2. Function of Withdrawal-Associated Brain miRNAs Predicted by DIANA-mirPath
3.3. Cocaine Withdrawal or Abstinence-Associated miRNA−mRNA-Pathway Network Predicted by IPA
3.4. Addiction-like Behaviors and Their Correlation with Expression Levels of Differentially Expressed miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- John, W.S.; Wu, L.T. Trends and correlates of cocaine use and cocaine use disorder in the United States from 2011 to 2015. Drug Alcohol Depend. 2017, 180, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Skolnick, P.; Volkow, N.D. Addiction therapeutics: Obstacles and opportunities. Biol. Psychiatry 2012, 72, 890–891. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.; Koob, G.F. Escalation of drug self-administration as a hallmark of persistent addiction liability. Behav. Pharm. 2013, 24, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Kariisa, M.; Scholl, L.; Wilson, N.; Seth, P.; Hoots, B. Drug Overdose Deaths Involving Cocaine and Psychostimulants with Abuse Potential—United States, 2003–2017. MMWR Morb. Mortal Wkly. Rep. 2019, 68, 388–395. [Google Scholar] [CrossRef]
- Mattson, M.E.; Lynch, S. Medication Prescribing and Behavioral Treatment for Substance Use Disorders in Physician Office Settings. In The CBHSQ Report; Substance Abuse and Mental Health Services Administration (US): Rockville, MD, USA, 2013; pp. 1–6. [Google Scholar]
- Deak, J.D.; Johnson, E.C. Genetics of substance use disorders: A review. Psychol. Med. 2021, 51, 2189–2200. [Google Scholar] [CrossRef]
- Cabana-Dominguez, J.; Shivalikanjli, A.; Fernandez-Castillo, N.; Cormand, B. Genome-wide association meta-analysis of cocaine dependence: Shared genetics with comorbid conditions. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 94, 109667. [Google Scholar] [CrossRef]
- Huggett, S.B.; Stallings, M.C. Cocaine’omics: Genome-wide and transcriptome-wide analyses provide biological insight into cocaine use and dependence. Addict. Biol. 2020, 25, e12719. [Google Scholar] [CrossRef]
- Kenny, P.J. Epigenetics, microRNA, and addiction. Dialogues Clin. Neurosci. 2014, 16, 335–344. [Google Scholar] [CrossRef]
- Hamilton, P.J.; Nestler, E.J. Epigenetics and addiction. Curr. Opin. Neurobiol. 2019, 59, 128–136. [Google Scholar] [CrossRef]
- Kocerha, J.; Kauppinen, S.; Wahlestedt, C. microRNAs in CNS disorders. Neuromol. Med. 2009, 11, 162–172. [Google Scholar] [CrossRef]
- Bali, P.; Kenny, P.J. MicroRNAs and Drug Addiction. Front. Genet. 2013, 4, 43. [Google Scholar] [CrossRef]
- Blount, G.S.; Coursey, L.; Kocerha, J. MicroRNA Networks in Cognition and Dementia. Cells 2022, 11, 1882. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.C.W.; Kenny, P.J. MicroRNAs regulate synaptic plasticity underlying drug addiction. Genes Brain Behav. 2018, 17, e12424. [Google Scholar] [CrossRef]
- Cuesta, S.; Restrepo-Lozano, J.M.; Silvestrin, S.; Nouel, D.; Torres-Berrio, A.; Reynolds, L.M.; Arvanitogiannis, A.; Flores, C. Non-Contingent Exposure to Amphetamine in Adolescence Recruits miR-218 to Regulate Dcc Expression in the VTA. Neuropsychopharmacology 2018, 43, 900–911. [Google Scholar] [CrossRef]
- Torres-Berrio, A.; Cuesta, S.; Lopez-Guzman, S.; Nava-Mesa, M.O. Interaction Between Stress and Addiction: Contributions From Latin-American Neuroscience. Front. Psychol. 2018, 9, 2639. [Google Scholar] [CrossRef] [PubMed]
- Fregeac, J.; Moriceau, S.; Poli, A.; Nguyen, L.S.; Oury, F.; Colleaux, L. Loss of the neurodevelopmental disease-associated gene miR-146a impairs neural progenitor differentiation and causes learning and memory deficits. Mol. Autism. 2020, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Gorini, G.; Nunez, Y.O.; Mayfield, R.D. Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PLoS ONE 2013, 8, e82565. [Google Scholar] [CrossRef]
- Rosato, A.J.; Chen, X.; Tanaka, Y.; Farrer, L.A.; Kranzler, H.R.; Nunez, Y.Z.; Henderson, D.C.; Gelernter, J.; Zhang, H. Salivary microRNAs identified by small RNA sequencing and machine learning as potential biomarkers of alcohol dependence. Epigenomics 2019, 11, 739–749. [Google Scholar] [CrossRef]
- Viola, T.W.; Heberle, B.A.; Zaparte, A.; Sanvicente-Vieira, B.; Wainer, L.M.; Fries, G.R.; Walss-Bass, C.; Grassi-Oliveira, R. Peripheral blood microRNA levels in females with cocaine use disorder. J. Psychiatr. Res. 2019, 114, 48–54. [Google Scholar] [CrossRef]
- Grimm, S.L.; Mendez, E.F.; Stertz, L.; Meyer, T.D.; Fries, G.R.; Gandhi, T.; Kanchi, R.; Selvaraj, S.; Teixeira, A.L.; Kosten, T.R.; et al. MicroRNA-mRNA networks are dysregulated in opioid use disorder postmortem brain: Further evidence for opioid-induced neurovascular alterations. Front. Psychiatry 2022, 13, 1025346. [Google Scholar] [CrossRef]
- Epstein, D.H.; Preston, K.L.; Stewart, J.; Shaham, Y. Toward a model of drug relapse: An assessment of the validity of the reinstatement procedure. Psychopharmacology 2006, 189, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Marchant, N.J.; Li, X.; Shaham, Y. Recent developments in animal models of drug relapse. Curr. Opin. Neurobiol. 2013, 23, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.; Spuhler, K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol. Clin. Exp. Res. 1984, 8, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Palmer, A.A. Behavioral Genetic Studies in Rats. Methods Mol. Biol. 2019, 2018, 319–326. [Google Scholar] [CrossRef]
- Carrette, L.L.G.; de Guglielmo, G.; Kallupi, M.; Maturin, L.; Brennan, M.; Boomhower, B.; Conlisk, D.; Sedighim, S.; Tieu, L.; Fannon, M.J.; et al. The Cocaine and Oxycodone Biobanks, Two Repositories from Genetically Diverse and Behaviorally Characterized Rats for the Study of Addiction. eNeuro 2021, 8, 1–12. [Google Scholar] [CrossRef]
- Li, M.; Xu, P.; Xu, Y.; Teng, H.; Tian, W.; Du, Q.; Zhao, M. Dynamic Expression Changes in the Transcriptome of the Prefrontal Cortex after Repeated Exposure to Cocaine in Mice. Front. Pharm. 2017, 8, 142. [Google Scholar] [CrossRef]
- Eipper-Mains, J.E.; Kiraly, D.D.; Duff, M.O.; Horowitz, M.J.; McManus, C.J.; Eipper, B.A.; Graveley, B.R.; Mains, R.E. Effects of cocaine and withdrawal on the mouse nucleus accumbens transcriptome. Genes Brain Behav. 2013, 12, 21–33. [Google Scholar] [CrossRef]
- Walker, D.M.; Cates, H.M.; Loh, Y.E.; Purushothaman, I.; Ramakrishnan, A.; Cahill, K.M.; Lardner, C.K.; Godino, A.; Kronman, H.G.; Rabkin, J.; et al. Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biol. Psychiatry 2018, 84, 867–880. [Google Scholar] [CrossRef]
- Lim, Y.; Beane-Ebel, J.E.; Tanaka, Y.; Ning, B.; Husted, C.R.; Henderson, D.C.; Xiang, Y.; Park, I.H.; Farrer, L.A.; Zhang, H. Exploration of alcohol use disorder-associated brain miRNA-mRNA regulatory networks. Transl. Psychiatry 2021, 11, 504. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Evans, J.; Bhagwate, A.; Middha, S.; Bockol, M.; Yan, H.; Kocher, J.P. CAP-miRSeq: A comprehensive analysis pipeline for microRNA sequencing data. BMC Genom. 2014, 15, 423. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Michaud, J.; Simpson, K.M.; Escher, R.; Buchet-Poyau, K.; Beissbarth, T.; Carmichael, C.; Ritchie, M.E.; Schutz, F.; Cannon, P.; Liu, M.; et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genom. 2008, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Churchill, G.A. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003, 4, 210. [Google Scholar] [CrossRef]
- Baba, K.; Shibata, R.; Sibuya, M. Partial correlation and conditional correlation as measures of conditional independence. Aust. N. Z. J Stat 2004, 46, 657–664. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Kalivas, P.W.; Volkow, N.D. The neural basis of addiction: A pathology of motivation and choice. Am. J. Psychiatry 2005, 162, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Deroche-Gamonet, V.; Belin, D.; Piazza, P.V. Evidence for addiction-like behavior in the rat. Science 2004, 305, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Piazza, P.V.; Deroche-Gamonet, V. A multistep general theory of transition to addiction. Psychopharmacology 2013, 229, 387–413. [Google Scholar] [CrossRef] [PubMed]
- Fouyssac, M.; Puaud, M.; Ducret, E.; Marti-Prats, L.; Vanhille, N.; Ansquer, S.; Zhang, X.; Belin-Rauscent, A.; Giuliano, C.; Houeto, J.L.; et al. Environment-dependent behavioral traits and experiential factors shape addiction vulnerability. Eur. J. Neurosci. 2021, 53, 1794–1808. [Google Scholar] [CrossRef]
- Florez-Salamanca, L.; Secades-Villa, R.; Hasin, D.S.; Cottler, L.; Wang, S.; Grant, B.F.; Blanco, C. Probability and predictors of transition from abuse to dependence on alcohol, cannabis, and cocaine: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Am. J. Drug Alcohol Abus. 2013, 39, 168–179. [Google Scholar] [CrossRef] [PubMed]
- George, O.; Koob, G.F. Individual differences in prefrontal cortex function and the transition from drug use to drug dependence. Neurosci. Biobehav. Rev. 2010, 35, 232–247. [Google Scholar] [CrossRef]
- George, O.; Koob, G.F. Individual differences in the neuropsychopathology of addiction. Dialogues Clin. Neurosci. 2017, 19, 217–229. [Google Scholar] [CrossRef]
- Luscher, C.; Malenka, R.C. Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron 2011, 69, 650–663. [Google Scholar] [CrossRef]
- Wolf, M.E. Synaptic mechanisms underlying persistent cocaine craving. Nat. Rev. Neurosci. 2016, 17, 351–365. [Google Scholar] [CrossRef]
- Duttke, S.H.; Montilla-Perez, P.; Chang, M.W.; Li, H.; Chen, H.; Carrette, L.L.G.; de Guglielmo, G.; George, O.; Palmer, A.A.; Benner, C.; et al. Glucocorticoid Receptor-Regulated Enhancers Play a Central Role in the Gene Regulatory Networks Underlying Drug Addiction. Front. Neurosci. 2022, 16, 858427. [Google Scholar] [CrossRef]
- Peters, J.; Kalivas, P.W.; Quirk, G.J. Extinction circuits for fear and addiction overlap in prefrontal cortex. Learn Mem. 2009, 16, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Everitt, B.J. Neural and psychological mechanisms underlying compulsive drug seeking habits and drug memories—Indications for novel treatments of addiction. Eur. J. Neurosci. 2014, 40, 2163–2182. [Google Scholar] [CrossRef] [PubMed]
- Grimm, J.W.; Hope, B.T.; Wise, R.A.; Shaham, Y. Neuroadaptation. Incubation of cocaine craving after withdrawal. Nature 2001, 412, 141–142. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Grimm, J.W.; Hope, B.T.; Shaham, Y. Incubation of cocaine craving after withdrawal: A review of preclinical data. Neuropharmacology 2004, 47 (Suppl. 1), 214–226. [Google Scholar] [CrossRef] [PubMed]
- Pickens, C.L.; Airavaara, M.; Theberge, F.; Fanous, S.; Hope, B.T.; Shaham, Y. Neurobiology of the incubation of drug craving. Trends Neurosci. 2011, 34, 411–420. [Google Scholar] [CrossRef]
- Parvaz, M.A.; Moeller, S.J.; Goldstein, R.Z. Incubation of Cue-Induced Craving in Adults Addicted to Cocaine Measured by Electroencephalography. JAMA Psychiatry 2016, 73, 1127–1134. [Google Scholar] [CrossRef]
- Koob, G.F.; Le Moal, M. Plasticity of reward neurocircuitry and the ‘dark side’ of drug addiction. Nat. Neurosci. 2005, 8, 1442–1444. [Google Scholar] [CrossRef]
- Kampman, K.M.; Dackis, C.; Lynch, K.G.; Pettinati, H.; Tirado, C.; Gariti, P.; Sparkman, T.; Atzram, M.; O’Brien, C.P. A double-blind, placebo-controlled trial of amantadine, propranolol, and their combination for the treatment of cocaine dependence in patients with severe cocaine withdrawal symptoms. Drug Alcohol. Depend. 2006, 85, 129–137. [Google Scholar] [CrossRef]
- Koob, G.F.; Buck, C.L.; Cohen, A.; Edwards, S.; Park, P.E.; Schlosburg, J.E.; Schmeichel, B.; Vendruscolo, L.F.; Wade, C.L.; Whitfield, T.W., Jr.; et al. Addiction as a stress surfeit disorder. Neuropharmacology 2014, 76 Pt B, 370–382. [Google Scholar] [CrossRef]
- Koob, G.F.; Volkow, N.D. Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry 2016, 3, 760–773. [Google Scholar] [CrossRef]
- Bastle, R.M.; Oliver, R.J.; Gardiner, A.S.; Pentkowski, N.S.; Bolognani, F.; Allan, A.M.; Chaudhury, T.; St Peter, M.; Galles, N.; Smith, C.; et al. In silico identification and in vivo validation of miR-495 as a novel regulator of motivation for cocaine that targets multiple addiction-related networks in the nucleus accumbens. Mol. Psychiatry 2018, 23, 434–443. [Google Scholar] [CrossRef]
- Domingo-Rodriguez, L.; Cabana-Dominguez, J.; Fernandez-Castillo, N.; Cormand, B.; Martin-Garcia, E.; Maldonado, R. Differential expression of miR-1249-3p and miR-34b-5p between vulnerable and resilient phenotypes of cocaine addiction. Addict. Biol. 2022, 27, e13201. [Google Scholar] [CrossRef]
- Mavrikaki, M.; Anastasiadou, E.; Ozdemir, R.A.; Potter, D.; Helmholz, C.; Slack, F.J.; Chartoff, E.H. Overexpression of miR-9 in the Nucleus Accumbens Increases Oxycodone Self-Administration. Int. J. Neuropsychopharmacol./Off. Sci. J. Coll. Int. Neuropsychopharmacol. (CINP) 2019, 22, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Pierce, R.C.; Pierce-Bancroft, A.F.; Prasad, B.M. Neurotrophin-3 contributes to the initiation of behavioral sensitization to cocaine by activating the Ras/Mitogen-activated protein kinase signal transduction cascade. J. Neurosci. 1999, 19, 8685–8695. [Google Scholar] [CrossRef]
- McCracken, C.B.; Hamby, S.M.; Patel, K.M.; Morgan, D.; Vrana, K.E.; Roberts, D.C. Extended cocaine self-administration and deprivation produces region-specific and time-dependent changes in connexin36 expression in rat brain. Synapse 2005, 58, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Leri, F.; Cummins, E.; Hoeschele, M.; Kreek, M.J. Involvement of arginine vasopressin and V1b receptor in heroin withdrawal and heroin seeking precipitated by stress and by heroin. Neuropsychopharmacology 2008, 33, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Sun, W.L.; Young, A.B.; Lee, K.; McGinty, J.F.; See, R.E. Oxytocin reduces cocaine seeking and reverses chronic cocaine-induced changes in glutamate receptor function. Int. J. Neuropsychopharmacol./Off. Sci. J. Coll. Int. Neuropsychopharmacol. (CINP) 2014, 18, pyu009. [Google Scholar] [CrossRef]
- Koob, G.F. Drug Addiction: Hyperkatifeia/Negative Reinforcement as a Framework for Medications Development. Pharmacol. Rev. 2021, 73, 163–201. [Google Scholar] [CrossRef]
- Ferguson, D.; Shao, N.; Heller, E.; Feng, J.; Neve, R.; Kim, H.D.; Call, T.; Magazu, S.; Shen, L.; Nestler, E.J. SIRT1-FOXO3a regulate cocaine actions in the nucleus accumbens. J. Neurosci. 2015, 35, 3100–3111. [Google Scholar] [CrossRef] [PubMed]
- Bali, P.; Kenny, P.J. Transcriptional mechanisms of drug addiction. Dialogues Clin. Neurosci. 2019, 21, 379–387. [Google Scholar] [CrossRef]
- Seney, M.L.; Glausier, J.; Sibille, E. Large-Scale Transcriptomics Studies Provide Insight Into Sex Differences in Depression. Biol. Psychiatry 2022, 91, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Teague, C.D.; Nestler, E.J. Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens. Mol. Psychiatry 2022, 27, 687–709. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Lee, B.R.; Wang, X.; Guo, C.; Liu, L.; Cui, R.; Lan, Y.; Balcita-Pedicino, J.J.; Wolf, M.E.; Sesack, S.R.; et al. Bidirectional modulation of incubation of cocaine craving by silent synapse-based remodeling of prefrontal cortex to accumbens projections. Neuron 2014, 83, 1453–1467. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Holt, L.M.; Huang, H.H.; Sesack, S.R.; Nestler, E.J.; Dong, Y. Astrocytes in cocaine addiction and beyond. Mol. Psychiatry 2022, 27, 652–668. [Google Scholar] [CrossRef] [PubMed]
- Luis, C.; Cannella, N.; Spanagel, R.; Kohr, G. Persistent strengthening of the prefrontal cortex—Nucleus accumbens pathway during incubation of cocaine-seeking behavior. Neurobiol. Learn. Mem. 2017, 138, 281–290. [Google Scholar] [CrossRef]
- Di Chiara, G. Nucleus accumbens shell and core dopamine: Differential role in behavior and addiction. Behav. Brain Res. 2002, 137, 75–114. [Google Scholar] [CrossRef]
- Fattore, L.; Melis, M. Sex differences in impulsive and compulsive behaviors: A focus on drug addiction. Addict. Biol. 2016, 21, 1043–1051. [Google Scholar] [CrossRef] [PubMed]

| Infralimbic Prefrontal Cortex miRNA | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
|---|---|---|---|---|---|---|
| rno-miR-582-3p | −1.30 | 2.98 | −3.73 | −1.30 | 0.001 | 0.292 |
| rno-miR-497-5p | −0.62 | 4.23 | −2.93 | −2.42 | 0.006 | 0.512 |
| rno-miR-485-3p | 0.59 | 10.51 | 2.77 | −2.60 | 0.009 | 0.512 |
| rno-miR-30b-5p | −0.85 | 3.43 | −2.72 | −2.84 | 0.011 | 0.512 |
| rno-miR-935 | 0.61 | 6.30 | 2.63 | −2.87 | 0.013 | 0.512 |
| rno-miR-496-5p | −0.91 | 0.94 | −2.61 | −3.27 | 0.014 | 0.512 |
| rno-miR-92b-3p | 0.73 | 12.03 | 2.58 | −2.93 | 0.015 | 0.512 |
| rno-miR-292-5p | −2.51 | 1.20 | −2.57 | −3.17 | 0.015 | 0.512 |
| rno-miR-379-3p | −0.77 | 5.63 | −2.54 | −3.02 | 0.016 | 0.512 |
| rno-miR-551b-3p | −1.08 | 1.07 | −2.52 | −3.35 | 0.017 | 0.512 |
| rno-miR-22-5p | −0.76 | 6.18 | −2.45 | −3.17 | 0.020 | 0.512 |
| rno-miR-382-3p ** | −1.29 | 2.54 | −2.41 | −3.34 | 0.022 | 0.512 |
| rno-miR-194-5p | −0.80 | 4.86 | −2.38 | −3.30 | 0.024 | 0.512 |
| rno-miR-152-3p | −0.62 | 5.86 | −2.30 | −3.42 | 0.028 | 0.512 |
| rno-miR-666-3p | 0.78 | 0.82 | 2.29 | −3.63 | 0.029 | 0.512 |
| rno-miR-106b-5p | −0.71 | 2.09 | −2.25 | −3.59 | 0.032 | 0.512 |
| rno-miR-24-2-5p | −0.70 | 6.03 | −2.22 | −3.54 | 0.034 | 0.512 |
| rno-miR-195-5p | −0.75 | 3.92 | −2.22 | −3.55 | 0.034 | 0.512 |
| rno-miR-203a-3p | −0.91 | 2.37 | −2.20 | −3.63 | 0.036 | 0.512 |
| rno-miR-409a-3p | 0.63 | 12.53 | 2.11 | −3.73 | 0.043 | 0.512 |
| rno-miR-666-5p | 0.63 | 5.35 | 2.08 | −3.76 | 0.046 | 0.512 |
| Prelimbic Prefrontal Cortex miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
| rno-miR-381-3p | 1.22 | 1.56 | 3.32 | −4.34 | 0.002 | 0.472 |
| rno-miR-193-5p | −0.79 | 2.45 | −2.94 | −4.35 | 0.006 | 0.472 |
| rno-miR-1188-3p | −1.51 | 0.97 | −2.93 | −4.42 | 0.006 | 0.472 |
| rno-miR-150-5p | −0.62 | 9.10 | −2.89 | −4.09 | 0.007 | 0.472 |
| rno-miR-128-1-5p | −0.84 | 4.04 | −2.87 | −4.25 | 0.007 | 0.472 |
| rno-miR-150-3p | −0.68 | 4.41 | −2.86 | −4.23 | 0.008 | 0.472 |
| rno-miR-194-3p | −0.90 | 1.50 | −2.50 | −4.46 | 0.018 | 0.520 |
| rno-miR-877 | −0.82 | 4.76 | −2.42 | −4.32 | 0.022 | 0.520 |
| rno-miR-505-5p | −0.67 | 4.92 | −2.41 | −4.32 | 0.022 | 0.520 |
| rno-miR-667-3p | −0.77 | 6.76 | −2.35 | −4.29 | 0.025 | 0.520 |
| rno-miR-873-3p | −0.71 | 6.71 | −2.35 | −4.29 | 0.026 | 0.520 |
| rno-miR-9a-5p | 0.95 | 14.95 | 2.32 | −4.22 | 0.027 | 0.520 |
| rno-miR-384-5p | 0.73 | 6.72 | 2.28 | −4.31 | 0.030 | 0.520 |
| rno-miR-872-5p | 1.11 | 4.52 | 2.13 | −4.41 | 0.041 | 0.520 |
| rno-miR-487b-5p | −0.65 | 2.92 | −2.09 | −4.47 | 0.045 | 0.520 |
| rno-miR-338-3p* | 0.80 | 3.54 | 2.08 | −4.44 | 0.046 | 0.520 |
| rno-miR-21-5p | 0.89 | 6.17 | 2.06 | −4.38 | 0.048 | 0.520 |
| rno-miR-500-3p | −0.63 | 0.84 | −2.05 | −4.52 | 0.049 | 0.520 |
| Nucleus Accumbens miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
| rno-miR-338-3p * | −0.84 | 3.54 | −2.17 | −4.43 | 0.038 | 0.847 |
| rno-miR-382-3p ** | −1.08 | 2.54 | −2.05 | −4.48 | 0.049 | 0.847 |
| Infralimbic Prefrontal Cortex miRNAs | Log2FC | AveExpr | t | B | p. Value | adj.p.Val |
|---|---|---|---|---|---|---|
| rno-miR-873-5p | −0.68 | 5.75 | −3.81 | −0.46 | 0.001 | 0.231 |
| rno-miR-101a-3p | 1.44 | 4.99 | 3.47 | −1.23 | 0.002 | 0.231 |
| rno-miR-935 * | 0.81 | 6.30 | 3.41 | −1.31 | 0.002 | 0.231 |
| rno-miR-666-3p | 1.07 | 0.82 | 3.08 | −2.47 | 0.004 | 0.358 |
| rno-miR-29b-3p | 0.65 | 5.37 | 2.99 | −2.17 | 0.005 | 0.358 |
| rno-miR-27b-5p | −0.76 | 4.14 | −2.87 | −2.44 | 0.007 | 0.358 |
| rno-miR-379-5p | −0.99 | 11.08 | −2.86 | −2.41 | 0.008 | 0.358 |
| rno-miR-325-5p | −0.70 | 8.24 | −2.84 | −2.44 | 0.008 | 0.358 |
| rno-miR-25-5p | 0.95 | 2.19 | 2.80 | −2.70 | 0.009 | 0.358 |
| rno-miR-187-3p | −0.78 | 7.84 | −2.65 | −2.82 | 0.013 | 0.358 |
| rno-miR-132-5p | −0.68 | 10.79 | −2.60 | −2.92 | 0.014 | 0.358 |
| rno-miR-218a-5p | −0.88 | 11.47 | −2.52 | −3.06 | 0.017 | 0.368 |
| rno-miR-770-5p ** | 0.83 | 6.89 | 2.51 | −3.06 | 0.017 | 0.368 |
| rno-miR-146b-5p | −0.83 | 10.78 | −2.51 | −3.08 | 0.018 | 0.368 |
| rno-miR-92b-3p | 0.71 | 12.03 | 2.48 | −3.14 | 0.019 | 0.370 |
| rno-miR-702-3p | 0.67 | 7.21 | 2.42 | −3.23 | 0.021 | 0.401 |
| rno-miR-3594-3p | 0.75 | 3.26 | 2.40 | −3.27 | 0.023 | 0.405 |
| rno-miR-1843-5p | −0.80 | 9.52 | −2.36 | −3.34 | 0.025 | 0.418 |
| rno-miR-136-3p | 1.31 | 1.64 | 2.30 | −3.47 | 0.028 | 0.440 |
| rno-miR-434-5p | −0.80 | 9.85 | −2.21 | −3.60 | 0.034 | 0.462 |
| rno-miR-22-5p | −0.68 | 6.18 | −2.14 | −3.70 | 0.040 | 0.462 |
| rno-miR-192-5p | −1.07 | 5.14 | −2.14 | −3.68 | 0.041 | 0.462 |
| rno-miR-1249 | 0.60 | 8.07 | 2.08 | −3.81 | 0.046 | 0.487 |
| Prelimbic Prefrontal Cortex miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
| rno-miR-128-1-5p | −0.98 | 4.04 | −3.26 | −2.27 | 0.003 | 0.513 |
| rno-miR-873-3p | −0.76 | 6.71 | −2.45 | −3.26 | 0.020 | 0.823 |
| rno-miR-410-5p | −0.60 | 1.67 | −2.19 | −3.88 | 0.036 | 0.823 |
| rno-miR-664-2-5p | −0.62 | 6.21 | −2.18 | −3.63 | 0.037 | 0.823 |
| rno-miR-199a-5p | 0.84 | 2.79 | 2.15 | −3.80 | 0.039 | 0.823 |
| rno-miR-501-3p | −0.60 | 4.44 | −2.14 | −3.71 | 0.040 | 0.823 |
| rno-miR-300-5p | −0.83 | 0.95 | −2.13 | −3.97 | 0.041 | 0.823 |
| Nucleus Accumbens miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
| rno-miR-935 * | 0.70 | 6.30 | 3.05 | −4.09 | 0.005 | 0.853 |
| rno-miR-3068-3p | −0.97 | 3.13 | −2.70 | −4.37 | 0.011 | 0.853 |
| rno-miR-30b-5p | −0.73 | 3.43 | −2.36 | −4.40 | 0.025 | 0.853 |
| rno-miR-770-5p ** | 0.66 | 6.89 | 2.07 | −4.36 | 0.047 | 0.853 |
| rno-miR-582-3p | −0.71 | 2.98 | −2.04 | −4.47 | 0.050 | 0.853 |
| Infralimbic Prefrontal Cortex miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
|---|---|---|---|---|---|---|
| rno-miR-187-3p | −1.21 | 7.84 | −4.24 | 0.40 | 0.0002 | 0.071 |
| rno-miR-448-3p | −1.08 | 1.90 | −3.15 | −2.24 | 0.004 | 0.302 |
| rno-miR-382-3p | 1.66 | 2.54 | 3.13 | −2.23 | 0.004 | 0.302 |
| rno-miR-541-5p | −0.61 | 11.65 | −3.11 | −1.92 | 0.004 | 0.302 |
| rno-miR-101a-3p | 1.17 | 4.99 | 2.94 | −2.32 | 0.006 | 0.333 |
| rno-miR-425-3p | 0.67 | 2.66 | 2.77 | −2.77 | 0.009 | 0.439 |
| rno-miR-137-3p | 1.15 | 2.76 | 2.59 | −3.05 | 0.015 | 0.540 |
| rno-miR-499-5p | 1.03 | 3.45 | 2.55 | −3.05 | 0.016 | 0.540 |
| rno-miR-551b-3p | 1.04 | 1.07 | 2.46 | −3.36 | 0.020 | 0.564 |
| rno-miR-764-5p | −1.05 | 2.51 | −2.43 | −3.28 | 0.021 | 0.565 |
| rno-miR-1298 | −0.89 | 8.11 | −2.30 | −3.43 | 0.028 | 0.710 |
| rno-miR-496-3p | 0.78 | 3.46 | 2.24 | −3.52 | 0.032 | 0.714 |
| rno-miR-106b-5p | 0.70 | 2.09 | 2.24 | −3.57 | 0.032 | 0.714 |
| rno-miR-30b-5p | 0.67 | 3.43 | 2.14 | −3.66 | 0.040 | 0.730 |
| rno-miR-292-5p | 2.07 | 1.20 | 2.13 | −3.71 | 0.041 | 0.730 |
| rno-miR-299a-5p | 0.80 | 2.16 | 2.09 | −3.76 | 0.045 | 0.763 |
| Prelimbic Prefrontal Cortex miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
| rno-miR-1188-3p | 1.51 | 0.97 | 2.76 | −4.45 | 0.010 | 0.998 |
| rno-miR-300-5p | −0.87 | 0.95 | −2.19 | −4.51 | 0.037 | 0.998 |
| Nucleus Accumbens miRNAs | Log2FC | AveExpr | t | B | p.Value | adj.p.Val |
| rno-miR-29c-3p | 0.61 | 3.55 | 2.64 | −4.34 | 0.013 | 0.998 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumaresan, V.; Lim, Y.; Juneja, P.; Tipton, A.E.; de Guglielmo, G.; Carrette, L.L.G.; Kallupi, M.; Maturin, L.; Liu, Y.; George, O.; et al. Abstinence from Escalation of Cocaine Intake Changes the microRNA Landscape in the Cortico-Accumbal Pathway. Biomedicines 2023, 11, 1368. https://doi.org/10.3390/biomedicines11051368
Kumaresan V, Lim Y, Juneja P, Tipton AE, de Guglielmo G, Carrette LLG, Kallupi M, Maturin L, Liu Y, George O, et al. Abstinence from Escalation of Cocaine Intake Changes the microRNA Landscape in the Cortico-Accumbal Pathway. Biomedicines. 2023; 11(5):1368. https://doi.org/10.3390/biomedicines11051368
Chicago/Turabian StyleKumaresan, Vidhya, Yolpanhchana Lim, Poorva Juneja, Allison E. Tipton, Giordano de Guglielmo, Lieselot L. G. Carrette, Marsida Kallupi, Lisa Maturin, Ying Liu, Olivier George, and et al. 2023. "Abstinence from Escalation of Cocaine Intake Changes the microRNA Landscape in the Cortico-Accumbal Pathway" Biomedicines 11, no. 5: 1368. https://doi.org/10.3390/biomedicines11051368
APA StyleKumaresan, V., Lim, Y., Juneja, P., Tipton, A. E., de Guglielmo, G., Carrette, L. L. G., Kallupi, M., Maturin, L., Liu, Y., George, O., & Zhang, H. (2023). Abstinence from Escalation of Cocaine Intake Changes the microRNA Landscape in the Cortico-Accumbal Pathway. Biomedicines, 11(5), 1368. https://doi.org/10.3390/biomedicines11051368







