Identification of New Key Genes and Their Association with Breast Cancer Occurrence and Poor Survival Using In Silico and In Vitro Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of Datsets and Extraction of Differentially Expressed Genes (DEGs)
2.2. Construction of Protein-Protein Interaction (PPI) Network
2.3. Characterization of Networks Topological Properties
2.4. Probability of Degree Distribution
2.5. Betweenness Centrality
2.6. Closeness Centrality
2.7. Community Detection: Leading Eigen Vector Approach
2.8. Genes Tracing across the Networks
2.9. Gene Ontology and Pathway Analysis of Key Genes
2.10. GEPIA Analysis
2.11. bc-GenExMiner Analysis
2.12. UALCAN Analysis
2.13. OncoLnc Analysis
2.13.1. Survival Analysis
2.13.2. Cell Lines, Culture and Validation of Key Regulatory Genes by qRT-PCR
3. Results
3.1. Characteristics of Datasets Used to Extract Common DEGs
3.2. Identification of Common DEGs
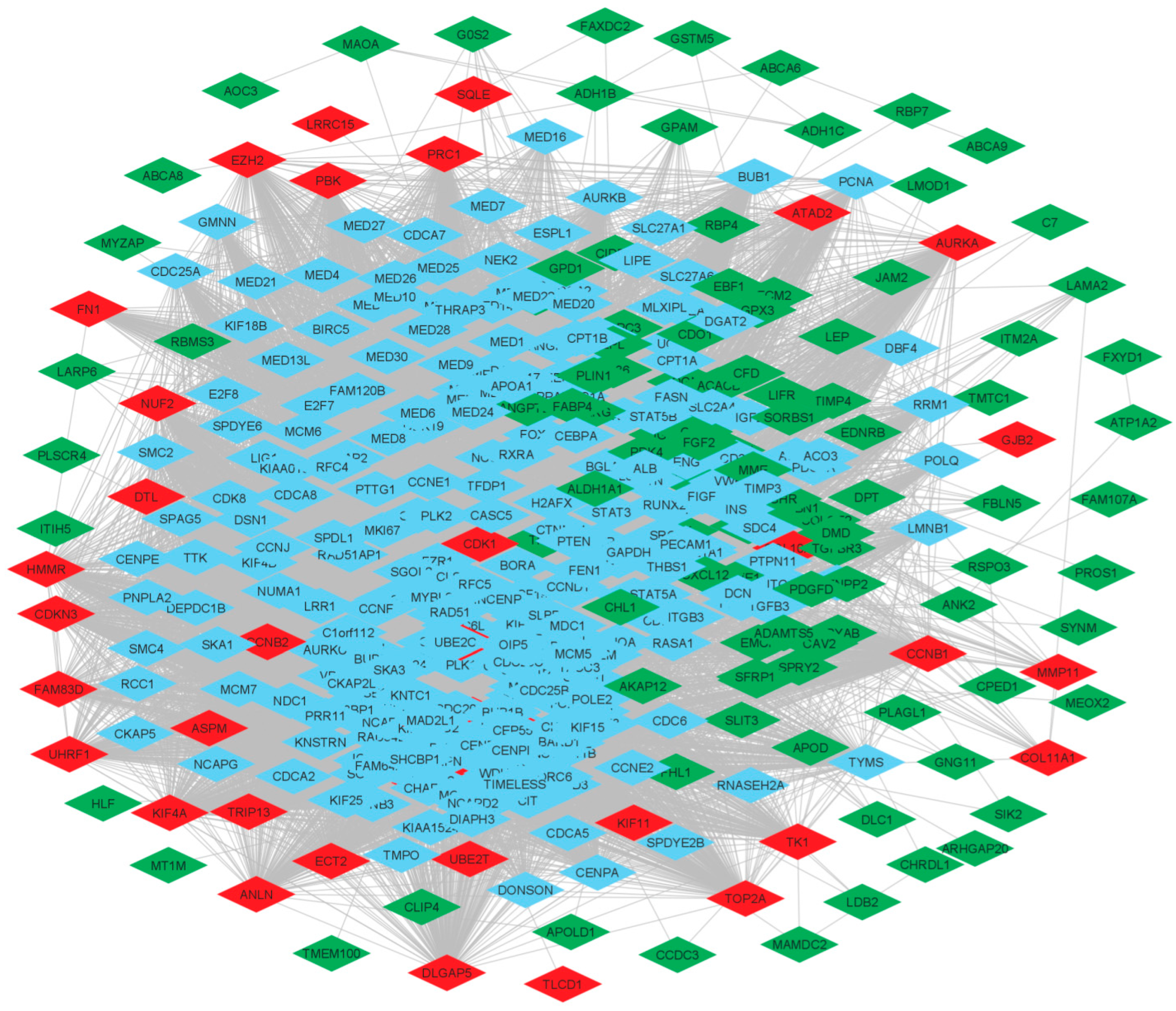
3.3. Protein-Protein Interaction (PPI) Network of DEGs
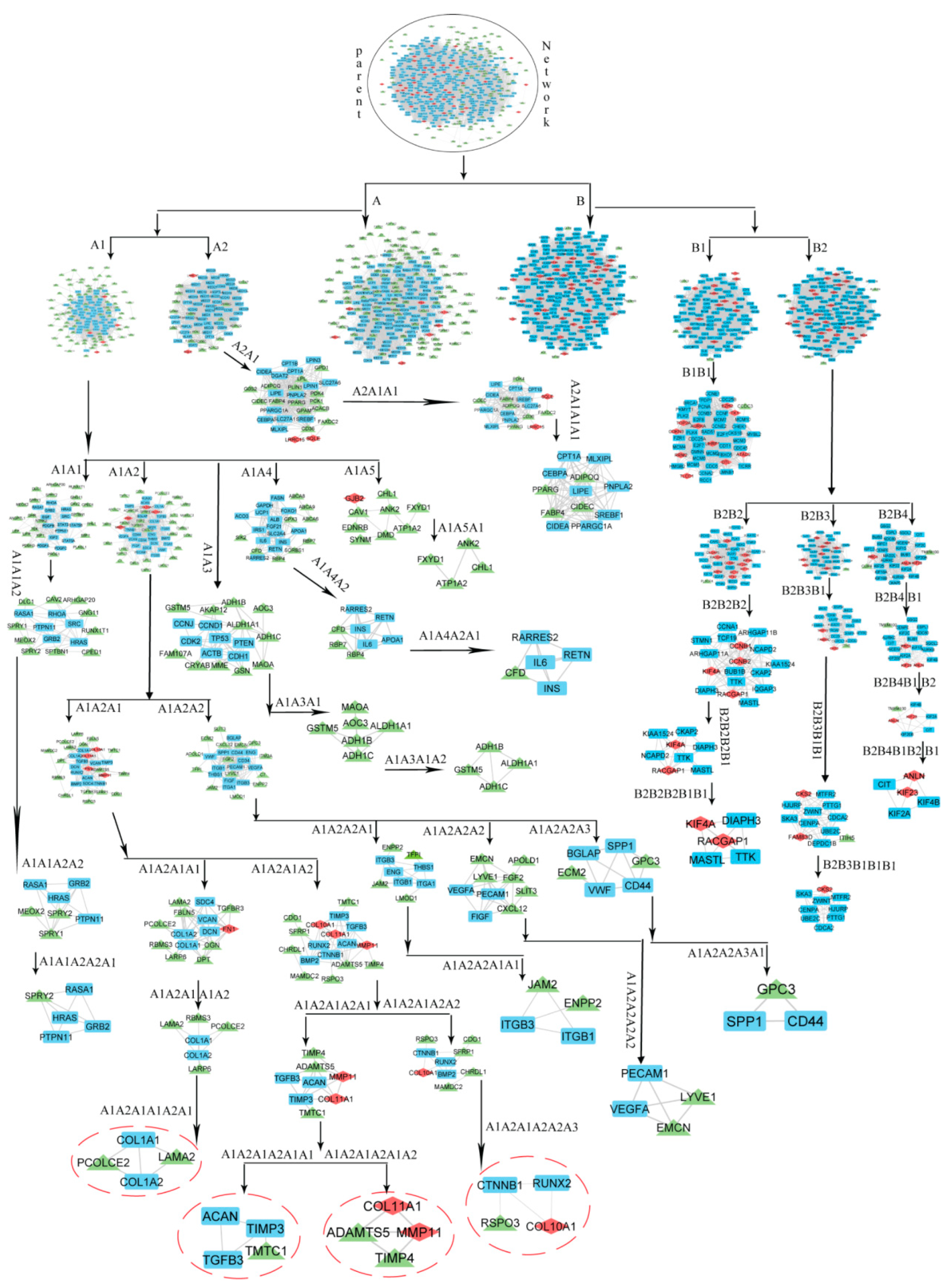
3.4. Community Detection by Leading Eigen Vector Method
3.5. Identification of Key Regulators and Properties of Breast Cancer Network
3.6. Gene Ontology and KEGG Pathway Analysis of Key DEGs
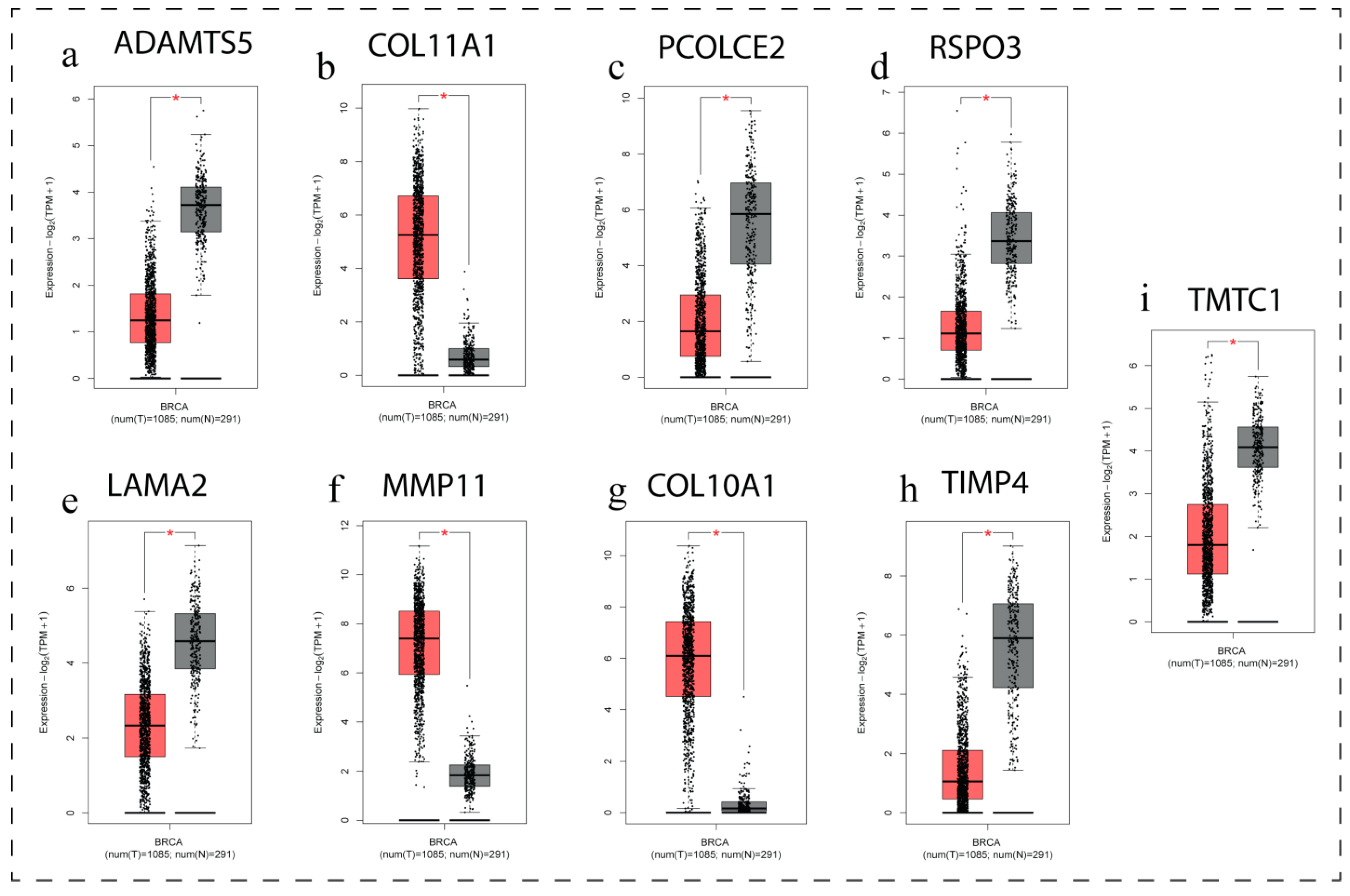
3.7. Gene Expression Profiling of Key DEGs
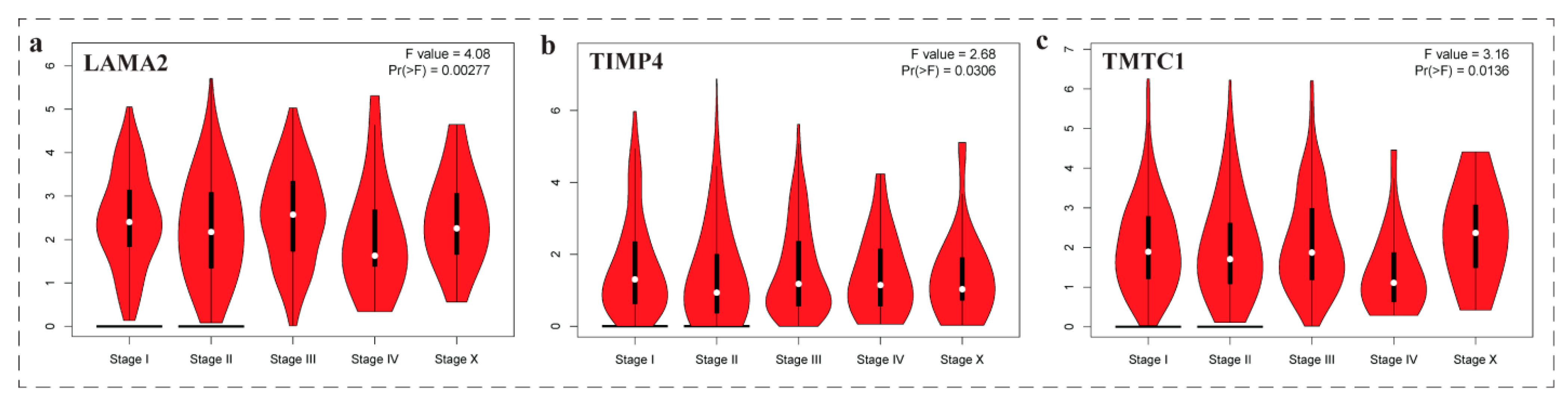
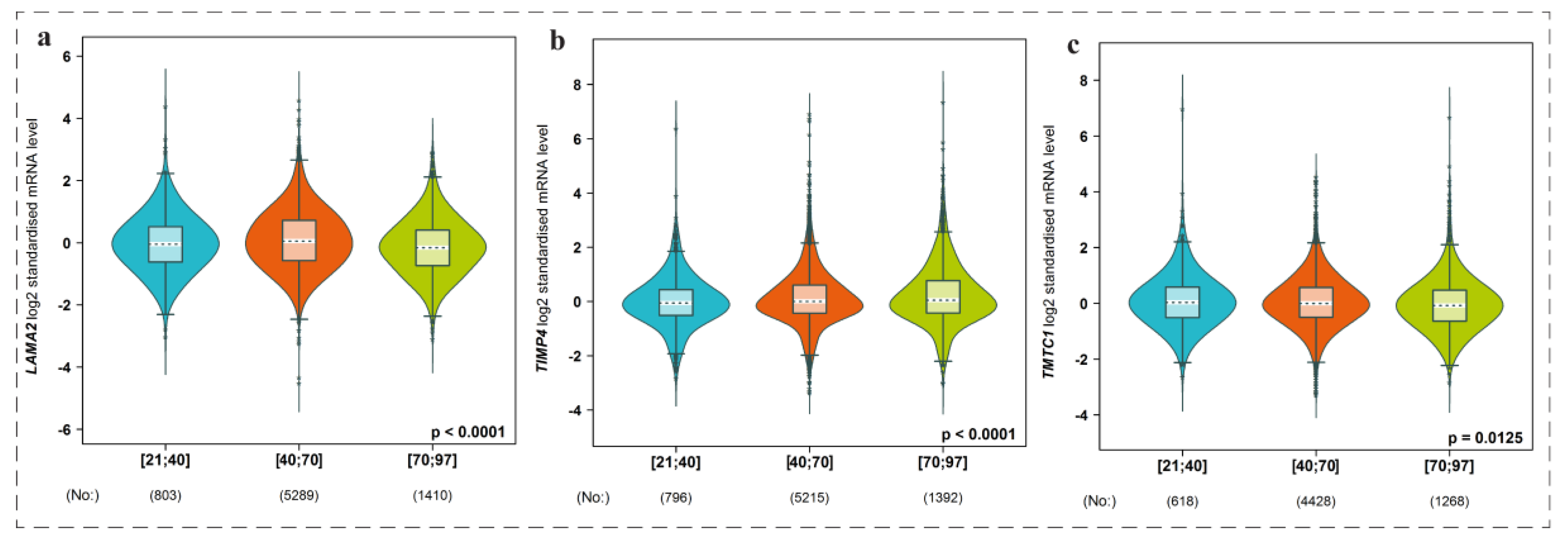
3.8. Expression Level of Genes among Groups of Patients with Respect to Age Factor
3.9. Pan-Cancer View of LAMA2, TIMP4, and TMTC1 Expression Level Using UALCAN Analysis
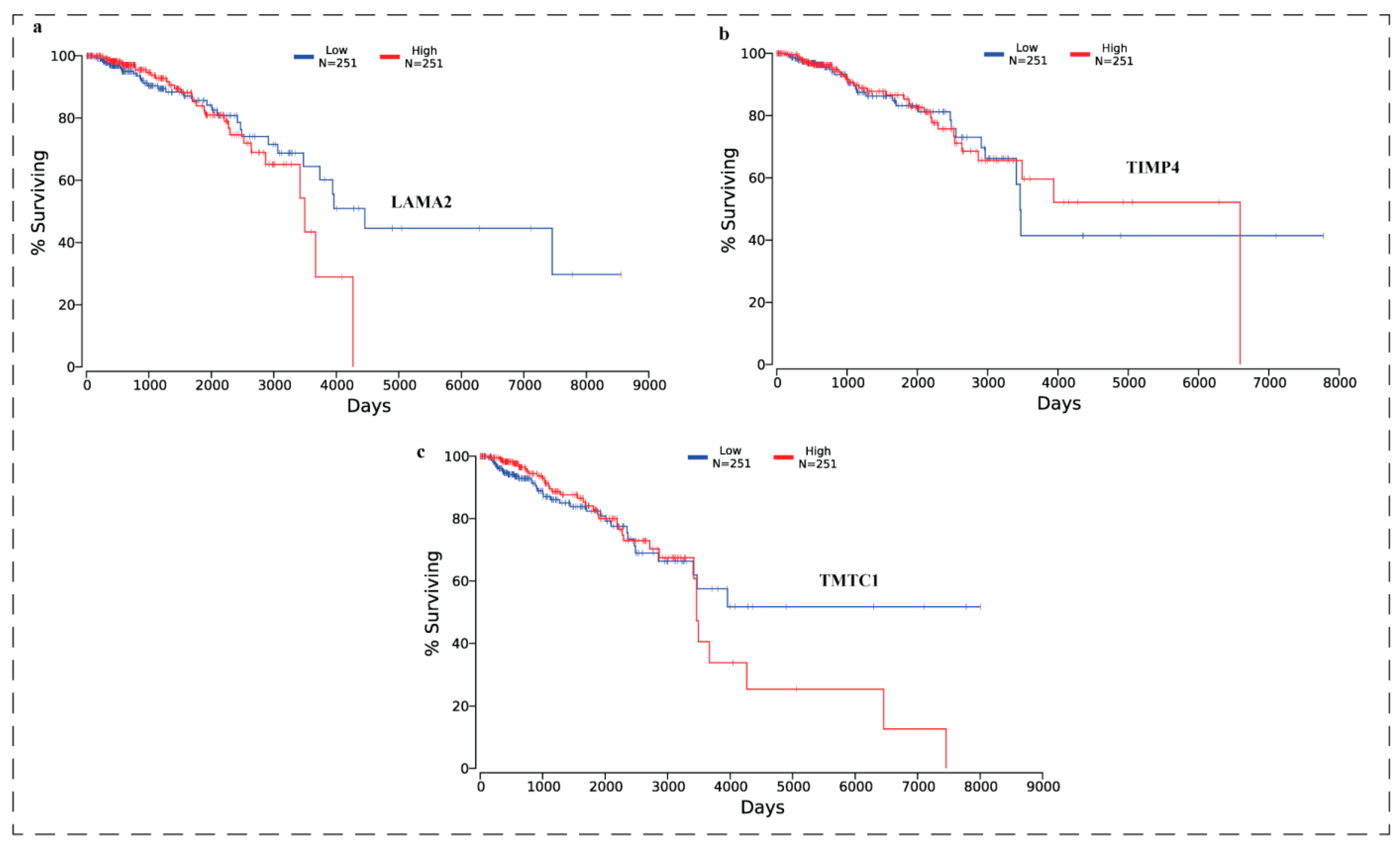
3.10. Effect of Expression Levels of LAMA2, TIMP4, and TMTC1 on the Survival of Breast Cancer Patient Oncolnc Analysis
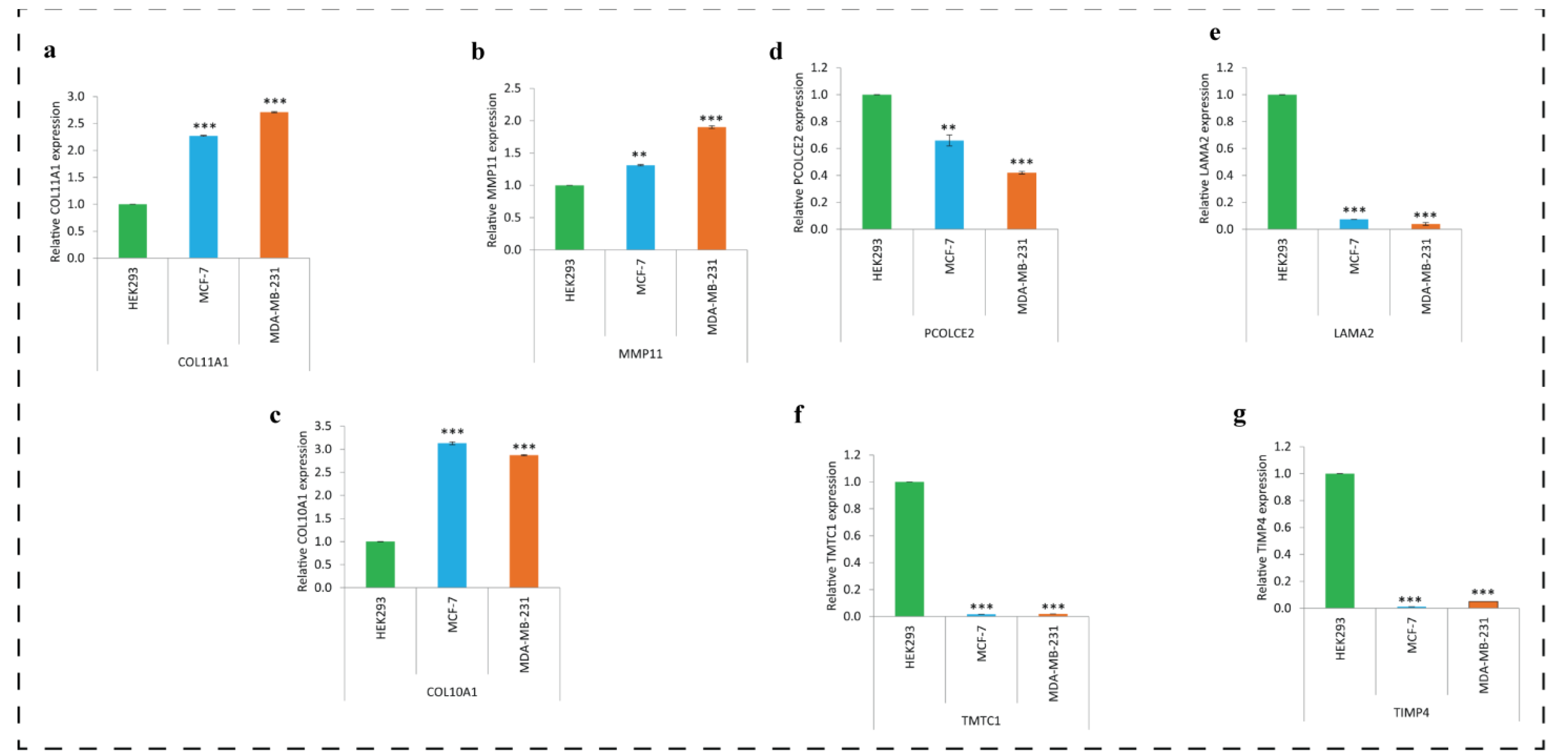
3.11. Expression of Key DEGs in Human Breast Cancer Cell Lines Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and Future Burden of Breast Cancer: Global Statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Soerjomataram, I.; Bray, F. Planning for Tomorrow: Global Cancer Incidence and the Role of Prevention 2020–2070. Nat. Rev. Clin. Oncol. 2021, 18, 663–672. [Google Scholar] [CrossRef]
- Lei, S.; Zheng, R.; Zhang, S.; Wang, S.; Chen, R.; Sun, K.; Zeng, H.; Zhou, J.; Wei, W. Global Patterns of Breast Cancer Incidence and Mortality: A Population-based Cancer Registry Data Analysis from 2000 to 2020. Cancer Commun. 2021, 41, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Li, C.; Tan, X. An Age Stratified Analysis of the Biomarkers in Patients with Colorectal Cancer. Sci. Rep. 2021, 11, 22464. [Google Scholar] [CrossRef]
- Francies, F.Z.; Hull, R.; Khanyile, R.; Dlamini, Z. Breast Cancer in Low-Middle Income Countries: Abnormality in Splicing and Lack of Targeted Treatment Options. Am. J. Cancer Res. 2020, 10, 1568–1591. [Google Scholar]
- Wilkinson, L.; Gathani, T. Understanding Breast Cancer as a Global Health Concern. BJR 2022, 95, 20211033. [Google Scholar] [CrossRef]
- Britt, K.L.; Cuzick, J.; Phillips, K.-A. Key Steps for Effective Breast Cancer Prevention. Nat. Rev. Cancer 2020, 20, 417–436. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.; Mao, F.; Yao, R.; Sun, Q. Ki-67 Index, Progesterone Receptor Expression, Histologic Grade and Tumor Size in Predicting Breast Cancer Recurrence Risk: A Consecutive Cohort Study. Cancer Commun. 2020, 40, 181–193. [Google Scholar] [CrossRef]
- Momenimovahed, Z.; Salehiniya, H. Epidemiological Characteristics of and Risk Factors for Breast Cancer in the World. BCTT 2019, 11, 151–164. [Google Scholar] [CrossRef]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. A Review of Computational Drug Repositioning: Strategies, Approaches, Opportunities, Challenges, and Directions. J. Cheminform. 2020, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Abubaker Bagabir, H.; Sultan, A.; Siddiqui, M.F.; Imam, N.; Alkhanani, M.F.; Alsulimani, A.; Haque, S.; Ishrat, R. An Integrative Network Approach to Identify Common Genes for the Therapeutics in Tuberculosis and Its Overlapping Non-Communicable Diseases. Front. Pharmacol. 2022, 12, 770762. [Google Scholar] [CrossRef] [PubMed]
- Kamel, H.F.M.; Al-Amodi, H.S.A.B. Exploitation of Gene Expression and Cancer Biomarkers in Paving the Path to Era of Personalized Medicine. Genom. Proteom. Bioinform. 2017, 15, 220–235. [Google Scholar] [CrossRef]
- Mishra, A.; Verma, M. Cancer Biomarkers: Are We Ready for the Prime Time? Cancers 2010, 2, 190–208. [Google Scholar] [CrossRef]
- Zarei Ghobadi, M.; Emamzadeh, R. Integration of Gene Co-Expression Analysis and Multi-Class SVM Specifies the Functional Players Involved in Determining the Fate of HTLV-1 Infection toward the Development of Cancer (ATLL) or Neurological Disorder (HAM/TSP). PLoS ONE 2022, 17, e0262739. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Shi, G.; He, Q.; Zhu, P. Screening and Predicted Value of Potential Biomarkers for Breast Cancer Using Bioinformatics Analysis. Sci. Rep. 2021, 11, 20799. [Google Scholar] [CrossRef]
- You, G.; Zu, B.; Wang, B.; Fu, Q.; Li, F. Identification of MiRNA–MRNA–TFs Regulatory Network and Crucial Pathways Involved in Tetralogy of Fallot. Front. Genet. 2020, 11, 552. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Assenov, Y.; Domingues, F.S.; Albrecht, M. Topological Analysis and Interactive Visualization of Biological Networks and Protein Structures. Nat. Protoc. 2012, 7, 670–685. [Google Scholar] [CrossRef]
- Albert, R.; Barabási, A.-L. Statistical Mechanics of Complex Networks. Rev. Mod. Phys. 2002, 74, 47–97. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Albert, R. Emergence of Scaling in Random Networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef]
- Brandes, U. A Faster Algorithm for Betweenness Centrality*. J. Math. Sociol. 2001, 25, 163–177. [Google Scholar] [CrossRef]
- Mason, O.; Verwoerd, M. Graph Theory and Networks in Biology. IET Syst. Biol. 2007, 1, 89–119. [Google Scholar] [CrossRef] [PubMed]
- Canright, G.; Engø-Monsen, K. Roles in Networks. Sci. Comput. Program. 2004, 53, 195–214. [Google Scholar] [CrossRef]
- Farooqui, A.; Tazyeen, S.; Ahmed, M.M.; Alam, A.; Ali, S.; Malik, M.Z.; Ali, S.; Ishrat, R. Assessment of the Key Regulatory Genes and Their Interologs for Turner Syndrome Employing Network Approach. Sci. Rep. 2018, 8, 10091. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. Finding Community Structure in Networks Using the Eigenvectors of Matrices. Phys. Rev. E 2006, 74, 036104. [Google Scholar] [CrossRef]
- Newman, M.E.J.; Girvan, M. Finding and Evaluating Community Structure in Networks. Phys. Rev. E 2004, 69, 026113. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. InterJournal Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Ud-Dean, S.M.M.; Heise, S.; Klamt, S.; Gunawan, R. TRaCE+: Ensemble Inference of Gene Regulatory Networks from Transcriptional Expression Profiles of Gene Knock-out Experiments. BMC Bioinform. 2016, 17, 252. [Google Scholar] [CrossRef]
- Fan, S.; Liang, Z.; Gao, Z.; Pan, Z.; Han, S.; Liu, X.; Zhao, C.; Yang, W.; Pan, Z.; Feng, W. Identification of the Key Genes and Pathways in Prostate Cancer. Oncol. Lett. 2018, 16, 6663–6669. [Google Scholar] [CrossRef]
- Liu, M.; Xu, Z.; Du, Z.; Wu, B.; Jin, T.; Xu, K.; Xu, L.; Li, E.; Xu, H. The Identification of Key Genes and Pathways in Glioma by Bioinformatics Analysis. J. Immunol. Res. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Ning, P.; Wu, Z.; Hu, A.; Li, X.; He, J.; Gong, X.; Xia, Y.; Shang, Y.; Bian, H. Integrated Genomic Analyses of Lung Squamous Cell Carcinoma for Identification of a Possible Competitive Endogenous RNA Network by Means of TCGA Datasets. PeerJ 2018, 6, e4254. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Salavaty, A.; Rezvani, Z.; Najafi, A. Survival Analysis and Functional Annotation of Long Non-coding RNAs in Lung Adenocarcinoma. J. Cell. Mol. Med. 2019, 23, 5600–5617. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Thumkeo, D.; Watanabe, S.; Narumiya, S. Physiological Roles of Rho and Rho Effectors in Mammals. Eur. J. Cell Biol. 2013, 92, 303–315. [Google Scholar] [CrossRef]
- Boguslawska, J.; Kedzierska, H.; Poplawski, P.; Rybicka, B.; Tanski, Z.; Piekielko-Witkowska, A. Expression of Genes Involved in Cellular Adhesion and Extracellular Matrix Remodeling Correlates with Poor Survival of Patients with Renal Cancer. J. Urol. 2016, 195, 1892–1902. [Google Scholar] [CrossRef]
- Bonacich, P. Power and Centrality: A Family of Measures. Am. J. Sociol. 1987, 92, 1170–1182. [Google Scholar] [CrossRef]
- Raglow, Z.; Thomas, S.M. Tumor Matrix Protein Collagen XIα1 in Cancer. Cancer Lett. 2015, 357, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Villa, F.; García-Ocaña, M.; Galván, J.A.; García-Martínez, J.; García-Pravia, C.; Menéndez-Rodríguez, P.; Rey, C.G.; Barneo-Serra, L.; de los Toyos, J.R. COL11A1/(pro)Collagen 11A1 Expression Is a Remarkable Biomarker of Human Invasive Carcinoma-Associated Stromal Cells and Carcinoma Progression. Tumor Biol. 2015, 36, 2213–2222. [Google Scholar] [CrossRef]
- Farmer, P.; Bonnefoi, H.; Anderle, P.; Cameron, D.; Wirapati, P.; Becette, V.; André, S.; Piccart, M.; Campone, M.; Brain, E.; et al. A Stroma-Related Gene Signature Predicts Resistance to Neoadjuvant Chemotherapy in Breast Cancer. Nat. Med. 2009, 15, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-Y.; Kandel, J.J.; Yamashiro, D.J.; Canoll, P.; Anastassiou, D. A Multi-Cancer Mesenchymal Transition Gene Expression Signature Is Associated with Prolonged Time to Recurrence in Glioblastoma. PLoS ONE 2012, 7, e34705. [Google Scholar] [CrossRef] [PubMed]
- Cheon, D.-J.; Tong, Y.; Sim, M.-S.; Dering, J.; Berel, D.; Cui, X.; Lester, J.; Beach, J.A.; Tighiouart, M.; Walts, A.E.; et al. A Collagen-Remodeling Gene Signature Regulated by TGF-β Signaling Is Associated with Metastasis and Poor Survival in Serous Ovarian Cancer. Clin. Cancer Res. 2014, 20, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-H.; Chang, T.-H.; Huang, Y.-F.; Huang, H.-D.; Chou, C.-Y. COL11A1 Promotes Tumor Progression and Predicts Poor Clinical Outcome in Ovarian Cancer. Oncogene 2014, 33, 3432–3440. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, S.; Guo, J.; Zhou, L.; You, L.; Zhang, T.; Zhao, Y. Insights into the Distinct Roles of MMP-11 in Tumor Biology and Future Therapeutics (Review). Int. J. Oncol. 2016, 48, 1783–1793. [Google Scholar] [CrossRef]
- Peruzzi, D.; Mori, F.; Conforti, A.; Lazzaro, D.; De Rinaldis, E.; Ciliberto, G.; La Monica, N.; Aurisicchio, L. MMP11: A Novel Target Antigen for Cancer Immunotherapy. Clin. Cancer Res. 2009, 15, 4104–4113. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, M.G.; Min, K.-W.; Jung, U.S.; Kim, D.-H. High MMP-11 Expression Associated with Low CD8+ T Cells Decreases the Survival Rate in Patients with Breast Cancer. PLoS ONE 2021, 16, e0252052. [Google Scholar] [CrossRef]
- Chapman, K.B.; Prendes, M.J.; Sternberg, H.; Kidd, J.L.; Funk, W.D.; Wagner, J.; West, M.D. COL10A1 Expression Is Elevated in Diverse Solid Tumor Types and Is Associated with Tumor Vasculature. Future Oncol. 2012, 8, 1031–1040. [Google Scholar] [CrossRef]
- Huang, H.; Li, T.; Ye, G.; Zhao, L.; Zhang, Z.; Mo, D.; Wang, Y.; Zhang, C.; Deng, H.; Li, G.; et al. High Expression of COL10A1 Is Associated with Poor Prognosis in Colorectal Cancer. OTT 2018, 11, 1571–1581. [Google Scholar] [CrossRef]
- Li, T.; Huang, H.; Shi, G.; Zhao, L.; Li, T.; Zhang, Z.; Liu, R.; Hu, Y.; Liu, H.; Yu, J.; et al. TGF-Β1-SOX9 Axis-Inducible COL10A1 Promotes Invasion and Metastasis in Gastric Cancer via Epithelial-to-Mesenchymal Transition. Cell Death Dis 2018, 9, 849. [Google Scholar] [CrossRef]
- Giussani, M.; Landoni, E.; Merlino, G.; Turdo, F.; Veneroni, S.; Paolini, B.; Cappelletti, V.; Miceli, R.; Orlandi, R.; Triulzi, T.; et al. Extracellular Matrix Proteins as Diagnostic Markers of Breast Carcinoma. J. Cell Physiol. 2018, 233, 6280–6290. [Google Scholar] [CrossRef] [PubMed]
- Xiang, A.; Lin, X.; Xu, L.; Chen, H.; Guo, J.; Zhou, F. PCOLCE Is Potent Prognostic Biomarker and Associates With Immune Infiltration in Gastric Cancer. Front. Mol. Biosci. 2020, 7, 544895. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, A.; Griffith, O.L.; Soroceanu, L.; Leonoudakis, D.; Luciani-Torres, M.G.; Daemen, A.; Gray, J.W.; Muschler, J.L. Loss of Cell-Surface Laminin Anchoring Promotes Tumor Growth and Is Associated with Poor Clinical Outcomes. Cancer Res. 2012, 72, 2578–2588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zeng, L.; Peng, Y.; Fan, B.; Chen, P.; Liu, J. Immune-Related Genes LAMA2 and IL1R1 Correlate with Tumor Sites and Predict Poor Survival in Pancreatic Adenocarcinoma. Future Oncol. 2021, 17, 3061–3076. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Q.; Chekouo, T. Filtering High-Dimensional Methylation Marks With Extremely Small Sample Size: An Application to Gastric Cancer Data. Front. Genet. 2021, 12, 705708. [Google Scholar] [CrossRef]
- Porter, S.; Scott, S.D.; Sassoon, E.M.; Williams, M.R.; Jones, J.L.; Girling, A.C.; Ball, R.Y.; Edwards, D.R. Dysregulated Expression of Adamalysin-Thrombospondin Genes in Human Breast Carcinoma. Clin. Cancer Res. 2004, 10, 2429–2440. [Google Scholar] [CrossRef]
- Li, C.; Xiong, Y.; Yang, X.; Wang, L.; Zhang, S.; Dai, N.; Li, M.; Ren, T.; Yang, Y.; Gan, L.; et al. Lost Expression of ADAMTS5 Protein Associates with Progression and Poor Prognosis of Hepatocellular Carcinoma. DDDT 2015, 9, 1773. [Google Scholar] [CrossRef]
- Lizarraga, F.; Espinosa, M.; Ceballos-Cancino, G.; Vazquez-Santillan, K.; Bahena-Ocampo, I.; Schwarz-Cruz y Celis, A.; Vega-Gordillo, M.; Garcia Lopez, P.; Maldonado, V.; Melendez-Zajgla, J. Tissue Inhibitor of Metalloproteinases-4 (TIMP-4) Regulates Stemness in Cervical Cancer Cells: TIMP-4 MODULATES CERVICAL CANCER CELLS STEMNESS. Mol. Carcinog. 2016, 55, 1952–1961. [Google Scholar] [CrossRef]
- Gu, H.; Tu, H.; Liu, L.; Liu, T.; Liu, Z.; Zhang, W.; Liu, J. RSPO3 Is a Marker Candidate for Predicting Tumor Aggressiveness in Ovarian Cancer. Ann. Transl. Med. 2020, 8, 1351. [Google Scholar] [CrossRef]
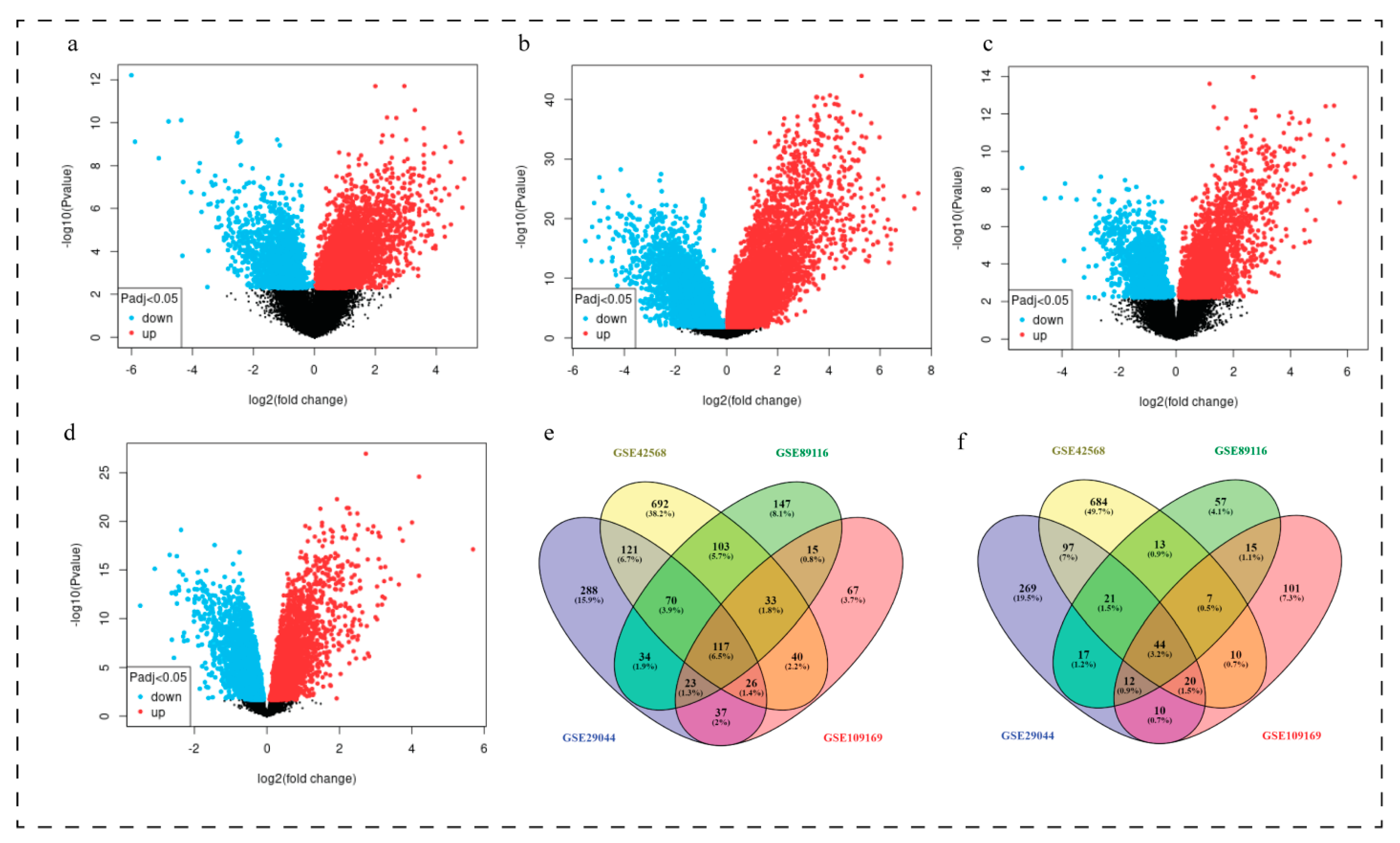


| Datasets | Total Samples | Normal (No. of Samples and Age) | Disease (No. of Samples and Age) | Up Regulated | Down Regulated | Country | Platform | Author | ||
|---|---|---|---|---|---|---|---|---|---|---|
| GSE29044 | 43 | 5 (Age 20–35 years) | 7 (Age > 55 years) | 6 (Age 20–35 years) | 25 (Age > 55 years) | 490 | 716 | Saudi Arabia | GPL570 | Colak D |
| GSE89116 | 33 | 4 (Age upto 35 years) | 5 (Age upto 80 years) | 11 (Age upto 38 years) | 13 (Age upto 80 years) | 186 | 542 | India | GPL6947 | Malvia S |
| GSE109169 | 50 | 5 (Age upto 40 years) | 20 (Age > 40 years) | 5 (Age upto 40 years) | 20 (Age > 40 years) | 219 | 358 | Taiwan | GPL570 | Chang JW |
| GSE42568 | 121 | 17 (Age not mentioned) | 104 (Age 31–89 years) | 896 | 1202 | Ireland | GPL570 | Clarke C | ||
| IGF1, LIFR, SFRP1, TSHZ2, SPTBN1, TMEM47, MME, NTRK2, PLAGL1, DMD, MAMDC2, LPL, HLF, PROS1, FABP4, ABCA9, ADAMTS5, RBMS3, APOLD1, SPRY2, ANGPTL1, EBF1, ITM2A, MEOX2, CAV1, TGFBR3, OGN, ANKRD29, CRYAB, CLIP4, CD36, EDNRB, CAV2, FAM13A, SYNM, ADH1B, FHL1, CDO1, TFPI, GPAM, CHRDL1, ABCA8, LYVE1, CACHD1, ITIH5, PLSCR4, SORBS1, LARP6, RUNX1T1, GPC3, PCDH18, AKAP12, PDK4, ACACB, ECM2, SPRY1, PAMR1, CXCL12, ADIPOQ, EMCN, FGF2, ATP1A2, ADH1C, PCOLCE2, ABCA6, SIK2, PCDH9, TMEM100, ARHGAP20, CHL1, MAOA, MTURN, LEP, SLIT3, IGSF10, GSTM5, VIT, RBP7, GPD1, CIDEC, MYZAP, RSPO3, FAXDC2, CFD, FBLN5, LDB2, GNG11, TEK, RBP4, FAM107A, ENPP2, PLIN1, CPED1, TIMP4, GSN, LMOD1, ALDH1A1, LAMA2, GPX3, AOC3, MT1M, JAM2, FXYD1, PPARG, APOD, DPT, G0S2, SH3BGRL2, VGLL3, DLC1, PCK1, C7, PDGFD, ANK2, GHR, TMTC1, CCDC3, PRC1, RRM2, TPX2, CDKN3, DTL, KIF4A, FAM83D, UBE2T, NUF2, MMP11, COL10A1, CCNB1, CDK1, DEPDC1, AURKA, CENPF, KIF23, KIF11, ANLN, FN1, EZH2, TRIP13, PBK, DLGAP5, UHRF1, TK1, CCNB2, MELK, CENPU, ATAD2, HMMR, ECT2, NUSAP1, ASPM, FANCI, SQLE, TOP2A, KIF20A, RACGAP1, GJB2, TLCD1, COL11A1, LRRC15, CKS2 |
| Genes | LogFC | p Value | Adj. p Value |
|---|---|---|---|
| PCOLCE2 | −4.45 | 2.44 × 10−11 | 1.67 × 10−7 |
| LAMA2 | −1.89 | 3.21 × 10−4 | 1.26 × 10−2 |
| TMTC1 | −2.49 | 2.97 × 10−3 | 4.98 × 10−2 |
| ADAMTS5 | −2.31 | 1.80 × 10−4 | 9.13 × 10−3 |
| TIMP4 | −2.54 | 1.40 × 10−4 | 7.60 × 10−3 |
| RSPO3 | −2.58 | 5.07 × 10−5 | 4.03 × 10−3 |
| COL11A1 | 5.29 | 9.91 × 10−14 | 3.60 × 10−12 |
| MMP11 | 4.62 | 4.13 × 10−6 | 8.01 × 10−4 |
| COL10A1 | 3.49 | 6.29 × 10−13 | 2.01 × 10−11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, R.; Sultan, A.; Ishrat, R.; Haque, S.; Khan, N.J.; Prieto, M.A. Identification of New Key Genes and Their Association with Breast Cancer Occurrence and Poor Survival Using In Silico and In Vitro Methods. Biomedicines 2023, 11, 1271. https://doi.org/10.3390/biomedicines11051271
Ali R, Sultan A, Ishrat R, Haque S, Khan NJ, Prieto MA. Identification of New Key Genes and Their Association with Breast Cancer Occurrence and Poor Survival Using In Silico and In Vitro Methods. Biomedicines. 2023; 11(5):1271. https://doi.org/10.3390/biomedicines11051271
Chicago/Turabian StyleAli, Rafat, Armiya Sultan, Romana Ishrat, Shafiul Haque, Nida Jamil Khan, and Miguel Angel Prieto. 2023. "Identification of New Key Genes and Their Association with Breast Cancer Occurrence and Poor Survival Using In Silico and In Vitro Methods" Biomedicines 11, no. 5: 1271. https://doi.org/10.3390/biomedicines11051271
APA StyleAli, R., Sultan, A., Ishrat, R., Haque, S., Khan, N. J., & Prieto, M. A. (2023). Identification of New Key Genes and Their Association with Breast Cancer Occurrence and Poor Survival Using In Silico and In Vitro Methods. Biomedicines, 11(5), 1271. https://doi.org/10.3390/biomedicines11051271









