Somatic Copy Number Alteration in Circulating Tumor DNA for Monitoring of Pediatric Patients with Cancer
Abstract
1. Introduction
2. Materials and Methods
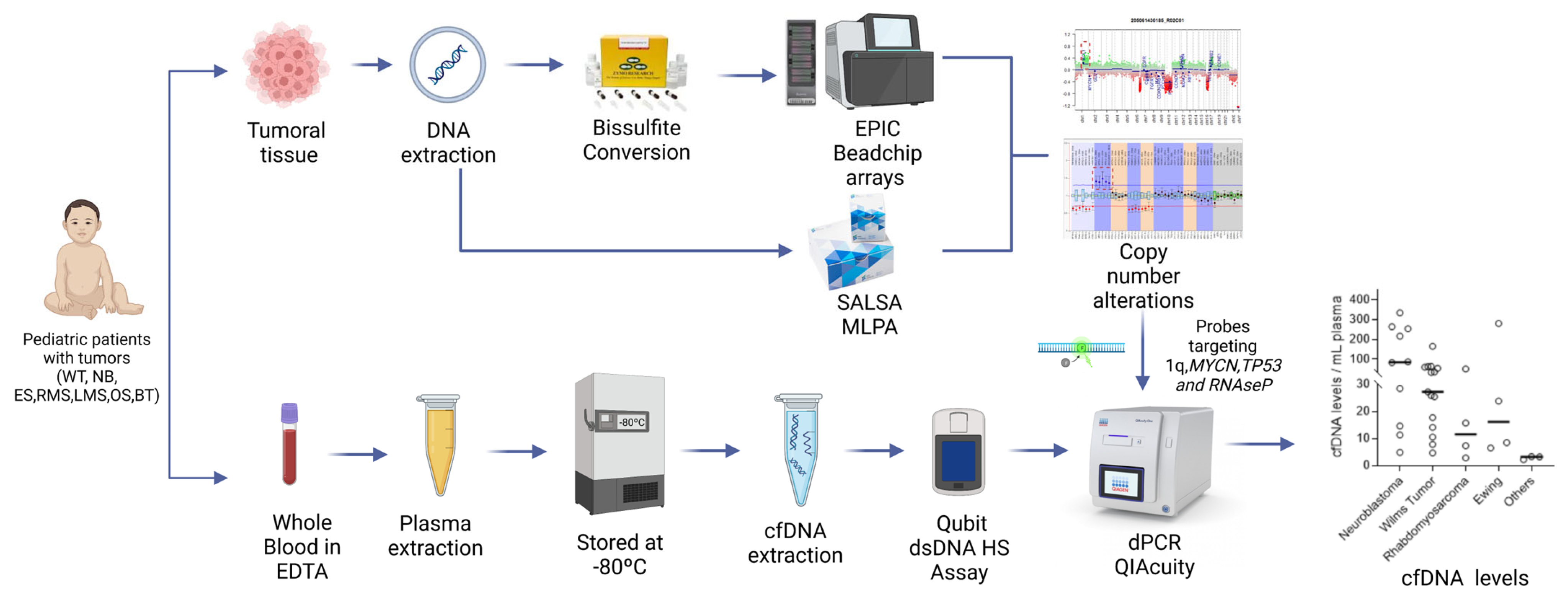
2.1. Patient Eligibility and Sample Collection
2.2. Characterization of the Copy Number Alterations (CNAs) in the Tumors
2.3. ctDNA Analysis
2.4. Data Analysis
3. Results
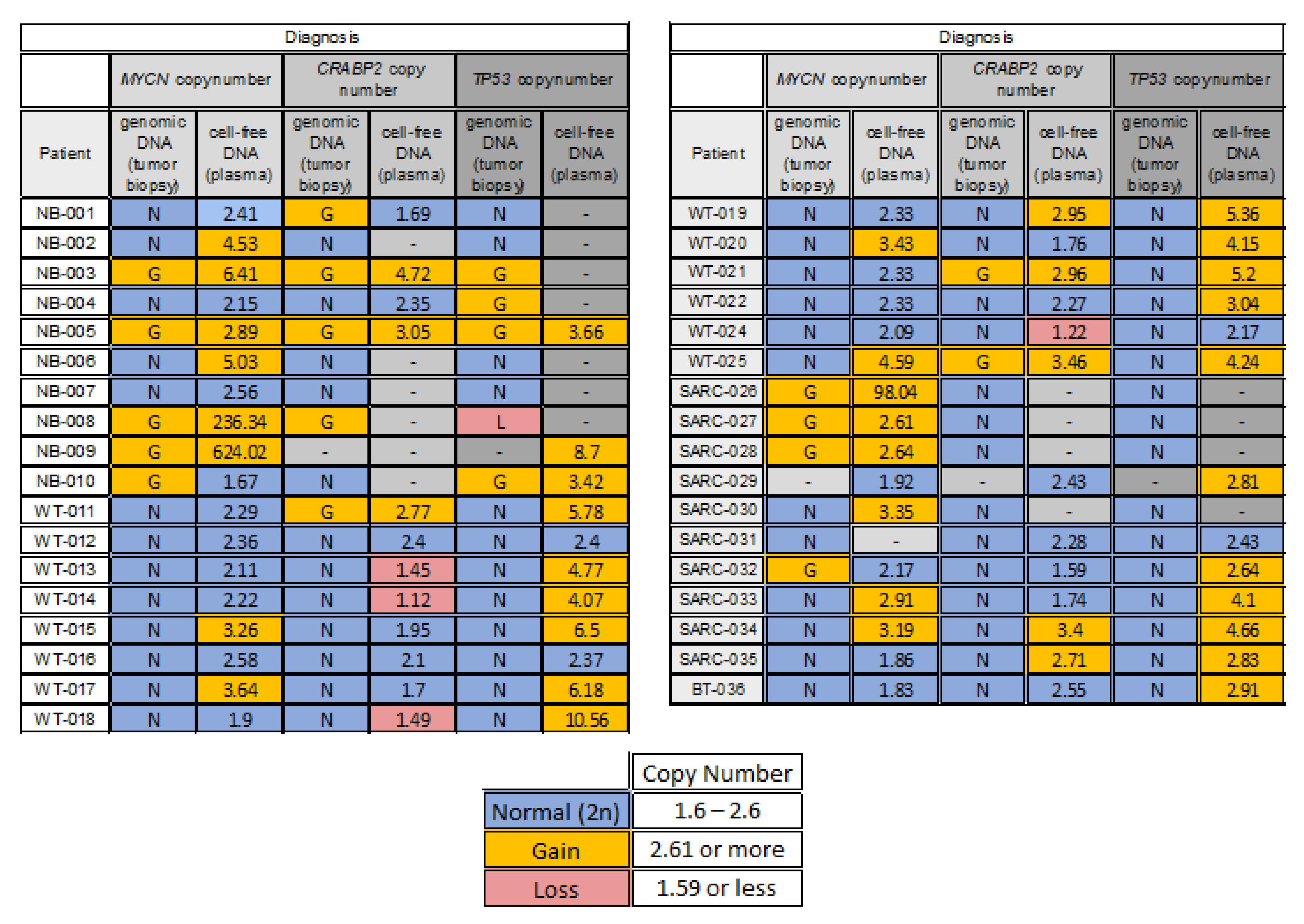
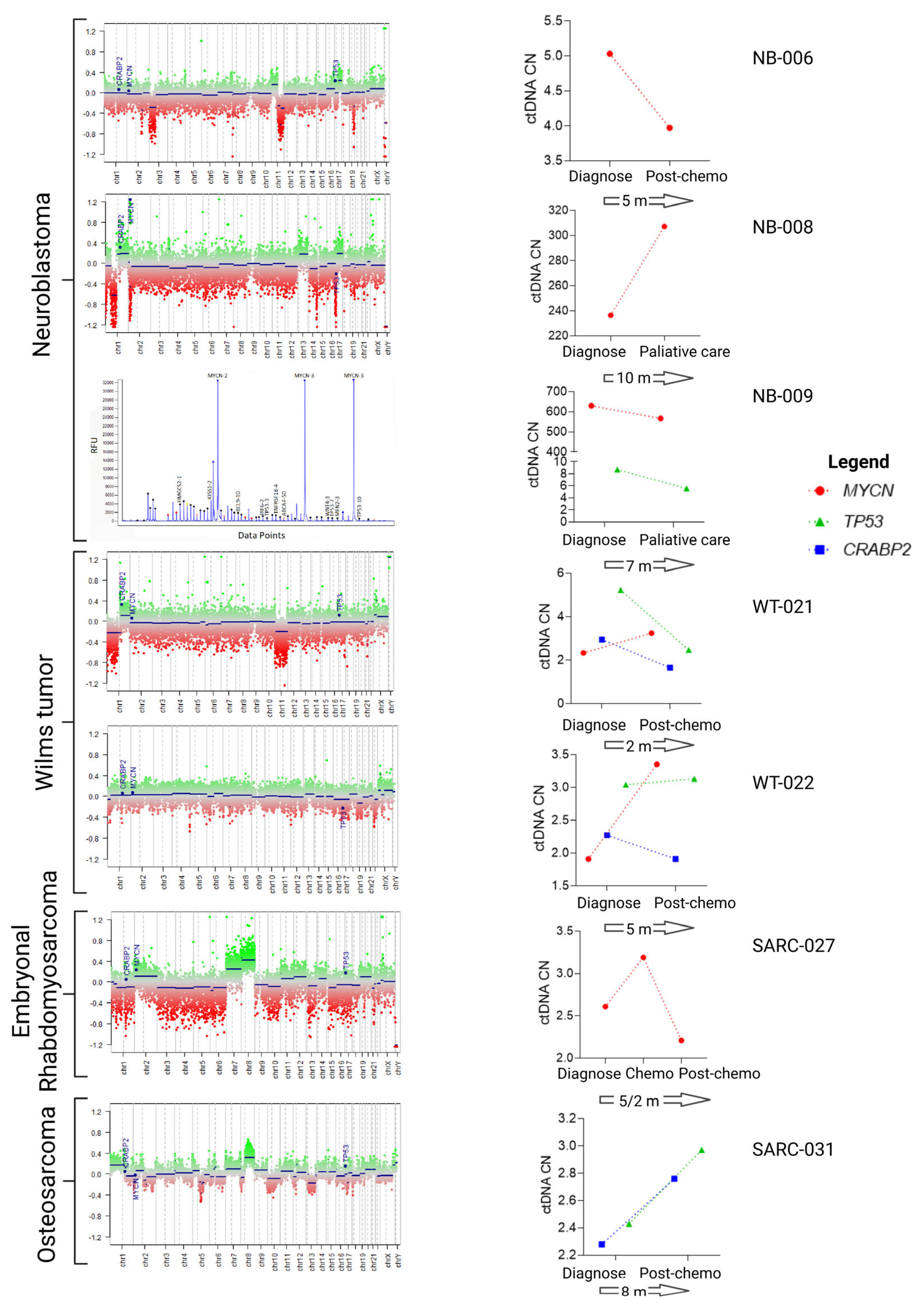
3.1. Characterization of 1q, MYCN and 17p Copy Number Status in the Tumors
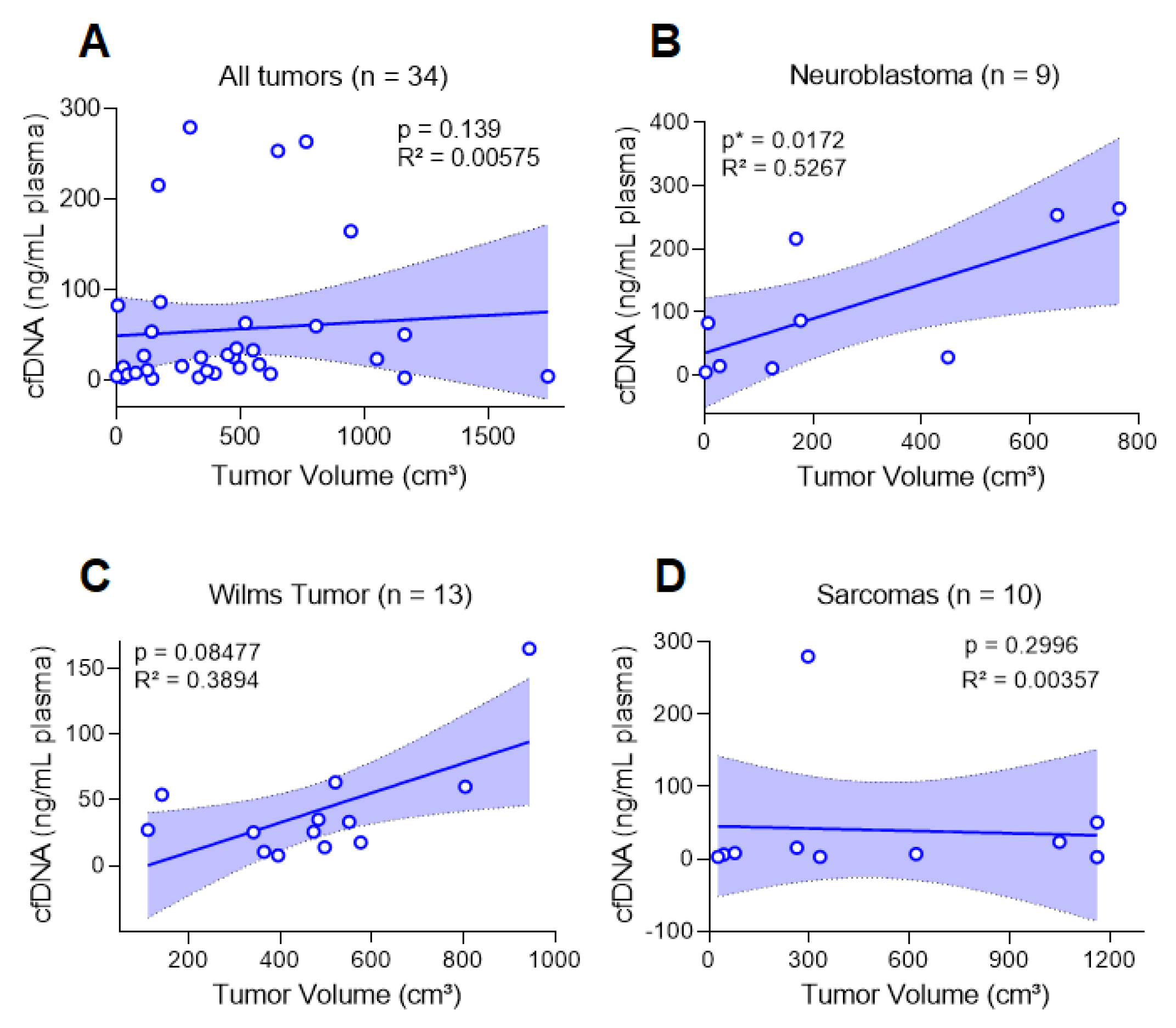
3.2. cfDNA Levels Reflect the Tumor Burden of Pediatric Patients at Diagnosis
3.3. CtDNA Plasma from Patients at Diagnosis Reflects CNA Status of the Tumor Tissues
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dehner, L.P. The Evolution of the Diagnosis and Understanding of Primitive and Embryonic Neoplasms in Children: Living through an Epoch. Mod. Pathol. 1998, 11, 669–685. Available online: https://pubmed.ncbi.nlm.nih.gov/9688189/ (accessed on 1 March 2023).
- Steliarova-Foucher, E.; Stiller, C.; Lacour, B.; Kaatsch, P. International Classification of Childhood Cancer, third edition. Cancer 2005, 103, 1457–1467. Available online: https://pubmed.ncbi.nlm.nih.gov/15712273/ (accessed on 1 March 2023). [CrossRef] [PubMed]
- Gatta, G.; Ferrari, A.; Stiller, C.A.; Pastore, G.; Bisogno, G.; Trama, A.; Capocaccia, R. Embryonal cancers in Europe. Eur. J. Cancer 2012, 48, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Tulla, M.; Berthold, F.; Graf, N.; Rutkowski, S.; von Schweinitz, D.; Spix, C.; Kaatsch, P. Incidence, Trends, and Survival of Children with Embryonal Tumors. Pediatrics 2015, 136, e623–e632. Available online: https://pubmed.ncbi.nlm.nih.gov/26304823/ (accessed on 1 March 2023). [CrossRef]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. Available online: https://pubmed.ncbi.nlm.nih.gov/29489754/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Rahal, Z.; Abdulhai, F.; Kadara, H.; Saab, R. Genomics of adult and pediatric solid tumors. Am. J. Cancer Res. 2018, 8, 1356. Available online: https://pmc/articles/PMC6129500/ (accessed on 1 March 2023). [PubMed]
- Kattner, P.; Strobel, H.; Khoshnevis, N.; Grunert, M.; Bartholomae, S.; Pruss, M.; Fitzel, R.; Halatsch, M.-E.; Schilberg, K.; Siegelin, M.D.; et al. Compare and contrast: Pediatric cancer versus adult malignancies. Cancer Metastasis Rev. 2019, 38, 673–682. Available online: https://link.springer.com/article/10.1007/s10555-019-09836-y (accessed on 1 March 2023). [CrossRef]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371–376. Available online: https://pubmed.ncbi.nlm.nih.gov/29489755/ (accessed on 25 January 2023). [CrossRef]
- Harbers, L.; Agostini, F.; Nicos, M.; Poddighe, D.; Bienko, M.; Crosetto, N. Somatic Copy Number Alterations in Human Cancers: An Analysis of Publicly Available Data from The Cancer Genome Atlas. Front. Oncol. 2021, 11, 700568. Available online: https://pubmed.ncbi.nlm.nih.gov/34395272/ (accessed on 25 January 2023). [CrossRef]
- Popov, S.D.; Vujanic, G.M.; Sebire, N.J.; Chagtai, T.; Williams, R.; Vaidya, S.; Pritchard-Jones, K. Bilateral wilms tumor with TP53-related anaplasia. Pediatr. Dev. Pathol. 2013, 16, 217–223. Available online: https://pubmed.ncbi.nlm.nih.gov/23387809/ (accessed on 29 January 2023). [CrossRef]
- Gratias, E.J.; Jennings, L.J.; Anderson, J.R.; Dome, J.S.; Grundy, P.; Perlman, E.J. Gain of 1q is associated with inferior event-free and overall survival in patients with favorable histology Wilms tumor: A report from the Children’s Oncology Group. Cancer 2013, 119, 3887–3894. Available online: https://pubmed.ncbi.nlm.nih.gov/23983061/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Williams, R.D.; Chagtai, T.; Alcaide-German, M.; Apps, J.; Wegert, J.; Popov, S.; Vujanic, G.; van Tinteren, H.; Heuvel-Eibrink, M.M.V.D.; Kool, M.; et al. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget 2015, 6, 7232–7243. Available online: https://pubmed.ncbi.nlm.nih.gov/25749049/ (accessed on 26 January 2023). [CrossRef] [PubMed]
- Maschietto, M.; Williams, R.D.; Chagtai, T.; Popov, S.D.; Sebire, N.J.; Vujanic, G.; Perlman, E.; Anderson, J.R.; Grundy, P.; Dome, J.S.; et al. TP53 mutational status is a potential marker for risk stratification in Wilms tumour with diffuse anaplasia. PLoS ONE 2014, 9, e109924. Available online: https://pubmed.ncbi.nlm.nih.gov/25313908/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Schleiermacher, G.; Michels, E.; Mosseri, V.; Ribeiro, A.; Lequin, D.; Vermeulen, J.; Couturier, J.; Peuchmaur, M.; Valent, A.; et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J. Clin. Oncol. 2009, 27, 1026–1033. Available online: https://pubmed.ncbi.nlm.nih.gov/19171713/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Lynn, M.; Wang, Y.; Slater, J.; Shah, N.; Conroy, J.; Ennis, S.; Morris, T.; Betts, D.R.; Fletcher, J.A.; O’Sullivan, M.J. High-resolution genome-wide copy-number analyses identify localized copy-number alterations in Ewing sarcoma. Diagn. Mol. Pathol. 2013, 22, 76–84. Available online: https://pubmed.ncbi.nlm.nih.gov/23628818/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Mackintosh, C.; Ordóñez, J.L.; García-Domínguez, D.J.; Sevillano, V.; Llombart-Bosch, A.; Szuhai, K.; Alberghini, M.; Sciot, R.; Sinnaeve, F.; Hogendoorn, P.C.W.; et al. 1q gain and CDT2 overexpression underlie an aggressive and highly proliferative form of Ewing sarcoma. Oncogene 2012, 31, 1287–1298. Available online: https://pubmed.ncbi.nlm.nih.gov/21822310/ (accessed on 25 January 2023). [CrossRef]
- Williamson, D.; Lu, Y.J.; Gordon, T.; Sciot, R.; Kelsey, A.; Fisher, C.; Poremba, C.; Anderson, J.; Pritchard-Jones, K.; Shipley, J. Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J. Clin. Oncol. 2005, 23, 880–888. Available online: https://pubmed.ncbi.nlm.nih.gov/15681534/ (accessed on 25 January 2023). [CrossRef]
- Lodrini, M.; Sprüssel, A.; Astrahantseff, K.; Tiburtius, D.; Konschak, R.; Lode, H.N.; Fischer, M.; Keilholz, U.; Eggert, A.; Deubzer, H.E. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget 2017, 8, 85234–85251. Available online: https://pubmed.ncbi.nlm.nih.gov/29156716/ (accessed on 25 January 2023). [CrossRef]
- Circulating MYCN DNA as a Tumor-Specific Marker in Neuroblastoma Patients1|Cancer Research|American Association for Cancer Research. Available online: https://aacrjournals.org/cancerres/article/62/13/3646/508931/Circulating-MYCN-DNA-as-a-Tumor-specific-Marker-in (accessed on 25 January 2023).
- Gotoh, T.; Hosoi, H.; Iehara, T.; Kuwahara, Y.; Osone, S.; Tsuchiya, K.; Ohira, M.; Nakagawara, A.; Kuroda, H.; Sugimoto, T. Prediction of MYCN amplification in neuroblastoma using serum DNA and real-time quantitative polymerase chain reaction. J. Clin. Oncol. 2005, 23, 5205–5210. Available online: https://pubmed.ncbi.nlm.nih.gov/16051962/ (accessed on 25 January 2023). [CrossRef]
- Ambros, P.F.; Ambros, I.M. Free DNA in the blood serum can unmask MYCN amplified tumors. Pediatr. Blood Cancer 2009, 53, 306–307. Available online: https://pubmed.ncbi.nlm.nih.gov/19489051/ (accessed on 25 January 2023). [CrossRef]
- Gallego Melcón, S.; Sánchez de Toledo Codina, J. Molecular biology of rhabdomyosarcoma. Clin. Transl. Oncol. 2007, 9, 415–419. Available online: https://pubmed.ncbi.nlm.nih.gov/17652054/ (accessed on 25 January 2023). [CrossRef]
- Wang, Q.; Diskin, S.; Rappaport, E.; Attiyeh, E.; Mosse, Y.; Shue, D.; Seiser, E.; Jagannathan, J.; Shusterman, S.; Bansal, M.; et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006, 66, 6050–6062. Available online: https://pubmed.ncbi.nlm.nih.gov/16778177/ (accessed on 25 January 2023). [CrossRef]
- Peneder, P.; Stütz, A.M.; Surdez, D.; Krumbholz, M.; Semper, S.; Chicard, M.; Sheffield, N.C.; Pierron, G.; Lapouble, E.; Tötzl, M.; et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat. Commun. 2021, 12, 3230. Available online: https://pubmed.ncbi.nlm.nih.gov/34050156/ (accessed on 29 January 2023). [CrossRef]
- Wen, X.; Pu, H.; Liu, Q.; Guo, Z.; Luo, D. Circulating Tumor DNA-A Novel Biomarker of Tumor Progression and Its Favorable Detection Techniques. Cancers 2022, 14, 6025. Available online: https://pubmed.ncbi.nlm.nih.gov/36551512/ (accessed on 1 March 2023). [CrossRef]
- Volckmar, A.L.; Sültmann, H.; Riediger, A.; Fioretos, T.; Schirmacher, P.; Endris, V.; Stenzinger, A.; Dietz, S. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosom. Cancer 2018, 57, 123–139. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/gcc.22517 (accessed on 1 March 2023). [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. Available online: https://pubmed.ncbi.nlm.nih.gov/22035192/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. Available online: https://pubmed.ncbi.nlm.nih.gov/24705333/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. Available online: https://pubmed.ncbi.nlm.nih.gov/24553385/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. Available online: https://pubmed.ncbi.nlm.nih.gov/24449238/ (accessed on 25 January 2023). [CrossRef]
- Bidard, F.C.; Madic, J.; Mariani, P.; Piperno-Neumann, S.; Rampanou, A.; Servois, V.; Cassoux, N.; Desjardins, L.; Milder, M.; Vaucher, I.; et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int. J. Cancer 2014, 134, 1207–1213. Available online: https://pubmed.ncbi.nlm.nih.gov/23934701/ (accessed on 25 January 2023). [CrossRef]
- Martignetti, J.A.; Camacho-Vanegas, O.; Priedigkeit, N.; Camacho, C.; Pereira, E.; Lin, L.; Garnar-Wortzel, L.; Miller, D.; Losic, B.; Shah, H.; et al. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia 2014, 16, 97–103. Available online: https://pubmed.ncbi.nlm.nih.gov/24563622/ (accessed on 25 January 2023). [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. Available online: https://aacrjournals.org/cancerdiscovery/article/11/4/858/665896/Liquid-Biopsy-From-Discovery-to-Clinical (accessed on 25 January 2023). [CrossRef] [PubMed]
- Shulman, D.S.; Crompton, B.D. Using Liquid Biopsy in the Treatment of Patient with OS. Adv. Exp. Med. Biol. 2020, 1257, 95–105. Available online: https://pubmed.ncbi.nlm.nih.gov/32483734/ (accessed on 25 January 2023). [PubMed]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. Available online: https://pubmed.ncbi.nlm.nih.gov/27223063/ (accessed on 25 January 2023). [CrossRef]
- Diamandis, E.P. The failure of protein cancer biomarkers to reach the clinic: Why, and what can be done to address the problem? BMC Med. 2012, 10, 87. Available online: https://pubmed.ncbi.nlm.nih.gov/22876833/ (accessed on 25 January 2023). [CrossRef]
- Kahana-Edwin, S.; Torpy, J.; Cain, L.E.; Mullins, A.; McCowage, G.; Woodfield, S.E.; Vasudevan, S.A.; Shea, D.P.T.; Minoche, A.E.; Kummerfeld, S.; et al. A quantitative universal NGS-based ctDNA assay for hepatoblastoma. medRxiv 2022. Available online: https://www.medrxiv.org/content/10.1101/2022.09.20.22279947v1 (accessed on 29 January 2023).
- Bioconductor—Conumee. Available online: https://bioconductor.org/packages/release/bioc/html/conumee.html (accessed on 25 January 2023).
- Dangoni, G.D.; Teixeira, A.C.B.; Vince, C.S.C.; Novak, E.M.; Gimenez, T.M.; Maschietto, M.; Filho, V.O.; Krepischi, A.C.V. LHX6 promoter hypermethylation in oncological pediatric patients conceived by, I.V.F. J. Dev. Orig. Health Dis. 2023, 14, 140–145. Available online: https://pubmed.ncbi.nlm.nih.gov/36154949/ (accessed on 25 January 2023). [CrossRef]
- Sánchez-Herrero, E.; Serna-Blasco, R.; Robado de Lope, L.; González-Rumayor, V.; Romero, A.; Provencio, M. Circulating Tumor DNA as a Cancer Biomarker: An Overview of Biological Features and Factors That may Impact on ctDNA Analysis. Front. Oncol. 2022, 12, 943253. Available online: https://pubmed.ncbi.nlm.nih.gov/35936733/ (accessed on 29 January 2023). [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. Available online: https://pubmed.ncbi.nlm.nih.gov/21562580/ (accessed on 29 January 2023). [CrossRef]
- Klega, K.; Imamovic-Tuco, A.; Ha, G.; Clapp, A.N.; Meyer, S.; Ward, A.; Clinton, C.; Nag, A.; Van Allen, E.; Mullen, E.; et al. Detection of Somatic Structural Variants Enables Quantification and Characterization of Circulating Tumor DNA in Children with Solid Tumors. JCO Precis Oncol. 2018, 2018, 1–13. Available online: https://pubmed.ncbi.nlm.nih.gov/30027144/ (accessed on 25 January 2023). [CrossRef]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Prim. 2016, 2, 16078. Available online: https://www.nature.com/articles/nrdp201678 (accessed on 29 January 2023). [CrossRef]
- Aguiar, T.F.M.; Rivas, M.P.; Costa, S.; Maschietto, M.; Rodrigues, T.; Sobral de Barros, J.; Barbosa, A.C.; Valieris, R.; Fernandes, G.R.; Bertola, D.R.; et al. Insights Into the Somatic Mutation Burden of Hepatoblastomas from Brazilian Patients. Front. Oncol. 2020, 10, 556. Available online: https://pubmed.ncbi.nlm.nih.gov/32432034/ (accessed on 1 March 2023). [CrossRef]
- Sumazin, P.; Chen, Y.; Treviño, L.R.; Sarabia, S.F.; Hampton, O.A.; Patel, K.; Mistretta, T.-A.; Zorman, B.; Thompson, P.; Heczey, A.; et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology 2017, 65, 104–121. Available online: https://pubmed.ncbi.nlm.nih.gov/27775819/ (accessed on 1 March 2023). [CrossRef]
- Maiti, S.; Alam, R.; Amos, C.I.; Huff, V. Frequent association of β-catenin and WT1 mutations in Wilms tumors. Cancer Res. 2000, 60, 6288–6292. Available online: https://mdanderson.elsevierpure.com/en/publications/frequent-association-of-%CE%B2-catenin-and-wt1-mutations-in-wilms-tumo (accessed on 1 March 2023).
- Maeda, S.; Ohka, F.; Okuno, Y.; Aoki, K.; Motomura, K.; Takeuchi, K.; Kusakari, H.; Yanagisawa, N.; Sato, S.; Yamaguchi, J.; et al. H3F3A mutant allele specific imbalance in an aggressive subtype of diffuse midline glioma, H3 K27M-mutant. Acta Neuropathol. Commun. 2020, 8, 8. Available online: https://pubmed.ncbi.nlm.nih.gov/32019606/ (accessed on 1 March 2023). [CrossRef] [PubMed]
- Li, D.; Bonner, E.R.; Wierzbicki, K.; Panditharatna, E.; Huang, T.; Lulla, R.; Mueller, S.; Koschmann, C.; Nazarian, J.; Saratsis, A.M. Standardization of the liquid biopsy for pediatric diffuse midline glioma using ddPCR. Sci. Rep. 2021, 11, 5098. Available online: https://pubmed.ncbi.nlm.nih.gov/33658570/ (accessed on 1 March 2023). [CrossRef] [PubMed]
- Walz, A.L.; Maschietto, M.; Crompton, B.; Evageliou, N.; Dix, D.; Tytgat, G.; Gessler, M.; Gisselsson, D.; Daw, N.C.; Wegert, J. Tumor biology, biomarkers, and liquid biopsy in pediatric renal tumors. Pediatr. Blood Cancer 2023, e30130. Available online: https://pubmed.ncbi.nlm.nih.gov/36592003/ (accessed on 25 January 2023).
- Cresswell, G.D.; Apps, J.R.; Chagtai, T.; Mifsud, B.; Bentley, C.C.; Maschietto, M.; Popov, S.D.; Weeks, M.E.; Olsen, E.; Sebire, N.J.; et al. Intra-Tumor Genetic Heterogeneity in Wilms Tumor: Clonal Evolution and Clinical Implications. EBioMedicine 2016, 9, 120–129. Available online: https://pubmed.ncbi.nlm.nih.gov/27333041/ (accessed on 26 January 2023). [CrossRef] [PubMed]
- Bellini, A.; Bernard, V.; Leroy, Q.; Frio, T.R.; Pierron, G.; Combaret, V.; Lapouble, E.; Clement, N.; Rubie, H.; Thebaud, E.; et al. Deep Sequencing Reveals Occurrence of Subclonal ALK Mutations in Neuroblastoma at Diagnosis. Clin. Cancer Res. 2015, 21, 4913–4921. Available online: https://pubmed.ncbi.nlm.nih.gov/26059187/ (accessed on 25 January 2023). [CrossRef]
- Schramm, A.; Köster, J.; Assenov, Y.; Althoff, K.; Peifer, M.; Mahlow, E.; Odersky, A.; Beisser, D.; Ernst, C.; Henssen, A.G.; et al. Mutational dynamics between primary and relapse neuroblastomas. Nat. Genet. 2015, 47, 872–877. Available online: https://pubmed.ncbi.nlm.nih.gov/26121086/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Chicard, M.; Boyault, S.; Daage, L.C.; Richer, W.; Gentien, D.; Pierron, G.; Lapouble, E.; Bellini, A.; Clement, N.; Iacono, I.; et al. Genomic Copy Number Profiling Using Circulating Free Tumor DNA Highlights Heterogeneity in Neuroblastoma. Clin. Cancer Res. 2016, 22, 5564–5573. Available online: https://pubmed.ncbi.nlm.nih.gov/27440268/ (accessed on 25 January 2023). [CrossRef]
- Madanat-Harjuoja, L.M.; Renfro, L.A.; Klega, K.; Tornwall, B.; Thorner, A.R.; Nag, A.; Dix, D.; Dome, J.S.; Diller, L.R.; Fernandez, C.V.; et al. Circulating Tumor DNA as a Biomarker in Patients with Stage III and IV Wilms Tumor: Analysis From a Children’s Oncology Group Trial, AREN0533. J. Clin. Oncol. 2022, 40, 3047–3056. Available online: https://pubmed.ncbi.nlm.nih.gov/35580298/ (accessed on 25 January 2023). [CrossRef] [PubMed]
- Lodrini, M.; Graef, J.; Thole-Kliesch, T.M.; Astrahantseff, K.; Sprussel, A.; Grimaldi, M.; Peitz, C.; Linke, R.B.; Hollander, J.F.; Lankes, E.; et al. Targeted Analysis of Cell-free Circulating Tumor DNA is Suitable for Early Relapse and Actionable Target Detection in Patients with Neuroblastoma. Clin. Cancer Res. 2022, 28, 1809–1820. Available online: https://pubmed.ncbi.nlm.nih.gov/35247920/ (accessed on 29 January 2023). [CrossRef] [PubMed]
- Krumbholz, M.; Eiblwieser, J.; Ranft, A.; Zierk, J.; Schmidkonz, C.; Stütz, A.M.; Peneder, P.; Tomazou, E.M.; Agaimy, A.; Bäuerle, T.; et al. Quantification of Translocation-Specific ctDNA Provides an Integrating Parameter for Early Assessment of Treatment Response and Risk Stratification in Ewing Sarcoma. Clin. Cancer Res. 2021, 27, 5922–5930. Available online: https://pubmed.ncbi.nlm.nih.gov/34426444/ (accessed on 25 January 2023). [CrossRef] [PubMed]

| Clinical Variable | Patients, No. (%) |
| Patients | 35 (100%) |
| Sex | |
| Male | 18 (51%) |
| Female | 17 (49%) |
| Age, years | |
| 1–10 | 29 (83%) |
| >10 | 6 (17%) |
| Diagnosis | |
| Wilms tumor | 14 (40%) |
| Neuroblastoma | 10 (29%) |
| Ewing Sarcoma | 4 (11%) |
| Rhabdomyosarcoma | 4 (11%) |
| Others | 3 (9%) |
| Tumor Volume | |
| <50 | 5 (14%) |
| 50–100 | 1 (3%) |
| 101–500 | 16 (46%) |
| 501–1000 | 8 (23%) |
| >1000 | 4 (11%) |
| Unknow | 1 (3%) |
| Clinical Stage | |
| I | 1 (3%) |
| II | 9 (26%) |
| III | 10 (29%) |
| IV | 13 (37%) |
| Unknow | 2 (6%) |
| Metastasis at diagnosis | |
| Yes | 17 (49%) |
| No | 18 (51%) |
| Metastasis during the treatment | |
| Yes | 11 (31%) |
| No | 19 (54%) |
| Unknow | 5 (14%) |
| cfDNA levels (ng/mL plasma) | |
| <10 | 10 (29%) |
| 10–50 | 13 (37%) |
| 51–100 | 6 (17%) |
| >100 | 6 (17%) |
| Clinical Variable | Patients, No. (%) | Statistical Test | ||
| Clinical Stage | WT | NB | Sarcomas | |
| I | 0 | 0 | 1 (10%) | Not applied |
| II | 6 (43%) | 2 (20%) | 1 (10%) | |
| III | 5 (36%) | 0 | 5 (50%) | |
| IV | 3 (21%) | 8 (80%) | 2 (20%) | |
| Unknow | 0 | 0 | 1 (10%) | |
| Correlation cfDNA levels vs. tumor staging | ||||
| I/II vs. III | p = 0.99 | - | p = 0.66 | Mann-Whitney test and Kruskal-Wallis test. |
| I/II vs. IV | p = 0.99 | p = 0.18 | p = 0.05 | |
| III vs. IV | p = 0.99 | - | p = 0.32 | |
| Correlation cfDNA levels vs. tumor volume | ||||
| p = 0.08 | p = 0.02 | p = 0.29 | Spearmen test. | |
| cfDNA levels and metastasis vs. no-metastasis at diagnosis | ||||
| p = 0.70 | # | p = 0.12 | Mann-Whitney test. # Statistics cannot be performed. | |
| cfDNA levels and metastasis vs. no-metastasis during treatment | ||||
| p = 0.94 | # | p = 0.50 | Mann-Whitney test. # Statistics cannot be performed. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruas, J.S.; Silva, F.L.T.; Euzébio, M.F.; Biazon, T.O.; Daiggi, C.M.M.; Nava, D.; Franco, M.T.; Cardinalli, I.A.; Cassone, A.E.; Pereira, L.H.; et al. Somatic Copy Number Alteration in Circulating Tumor DNA for Monitoring of Pediatric Patients with Cancer. Biomedicines 2023, 11, 1082. https://doi.org/10.3390/biomedicines11041082
Ruas JS, Silva FLT, Euzébio MF, Biazon TO, Daiggi CMM, Nava D, Franco MT, Cardinalli IA, Cassone AE, Pereira LH, et al. Somatic Copy Number Alteration in Circulating Tumor DNA for Monitoring of Pediatric Patients with Cancer. Biomedicines. 2023; 11(4):1082. https://doi.org/10.3390/biomedicines11041082
Chicago/Turabian StyleRuas, Juliana Silveira, Felipe Luz Torres Silva, Mayara Ferreira Euzébio, Tássia Oliveira Biazon, Camila Maia Martin Daiggi, Daniel Nava, Mayra Troiani Franco, Izilda Aparecida Cardinalli, Alejandro Enzo Cassone, Luiz Henrique Pereira, and et al. 2023. "Somatic Copy Number Alteration in Circulating Tumor DNA for Monitoring of Pediatric Patients with Cancer" Biomedicines 11, no. 4: 1082. https://doi.org/10.3390/biomedicines11041082
APA StyleRuas, J. S., Silva, F. L. T., Euzébio, M. F., Biazon, T. O., Daiggi, C. M. M., Nava, D., Franco, M. T., Cardinalli, I. A., Cassone, A. E., Pereira, L. H., Seidinger, A. L., Maschietto, M., & Jotta, P. Y. (2023). Somatic Copy Number Alteration in Circulating Tumor DNA for Monitoring of Pediatric Patients with Cancer. Biomedicines, 11(4), 1082. https://doi.org/10.3390/biomedicines11041082








