Abstract
Background: Human leukocyte antigen (HLA) genes are important in many immune processes and contribute to many adverse drug reactions. Whether genetic variations in the HLA region are associated with non-steroid anti-inflammatory drug (NSAID) hypersensitivity remains uncertain. Therefore, the aim of our study was to identify HLA genetic variations in patients with NSAID hypersensitivity in the Taiwanese population. Methods: This hospital-based, retrospective case-control study enrolled 37,156 participants with NSAID exposure from the Taiwan Precision Medicine Initiative (TPMI), who were all genotyped and imputed to fine map HLA typing. Our study assigned 1217 cases to the NSAID allergy group and 12,170 controls to a matched group. Logistic regression analyses were utilized to explore associations between HLA alleles and NSAID hypersensitivity. Results: Overall, 13,387 patients were genotyped for eight major HLA alleles. Allele frequencies were different between the two groups. In the NSAID allergy group, the genotype frequencies of HLA-A*02:01, HLA-A*34:01, and HLA-DQA1*06:01 were found to be markedly elevated compared to the control group, a significance that persisted even after applying the Bonferroni correction. Furthermore, the risk of NSAID allergy demonstrated a significant association with HLA-A*02:01 (OR = 1.29, p < 0.001) and HLA-A*34:01 (OR = 9.90, p = 0.001), in comparison to their respective counterparts. Notably, the genotype frequency of HLA-B*46:01 exhibited a significant increase in the severe allergy group when compared with the mild allergy group. Conclusions: We identified HLA genotypes linked to the onset and severity of NSAID hypersensitivity. Our findings establish a basis for precision prescription in future clinical applications.
1. Introduction
Non-steroid anti-inflammatory drugs (NSAIDs) are the most commonly prescribed medicine in the current era. They are used to treat pain, fever, and many inflammatory processes [1]. The main mechanism of action is the inhibition of the enzyme Cyclooxygenase (COX), which results in decreased synthesis of prostaglandin and other prostanoids [2]. In addition to the therapeutic usage of NSAIDs, these drugs are well known to exert multiple adverse effects, including cardiovascular, gastrointestinal, nephrotoxic, hepatotoxic, and hypersensitive reactions [3]. The clinical patterns of NSAID hypersensitivity have diverse symptoms, which can often present a diagnostic challenge [4].
NSAID hypersensitivity, an adverse drug reaction (ADR), is a significant cause of drug-related allergies, posing a potential risk of lethal outcomes [5,6]. Early detection of NSAID hypersensitivity is crucial, particularly in patients with no prior exposure to NSAIDs. Studies have shown that the prevalence of NSAID hypersensitivity in the general population ranges from 0.5 to 3.5% [7,8]. However, this prevalence is substantially higher in certain high-risk groups, with 14.89% of severe asthma patients and 9.69% of nasal polyps patients exhibiting NSAID hypersensitivity [9]. These observations suggest a potential link between NSAID hypersensitivity and human leukocyte antigen (HLA) gene expression. To delve deeper into the genetic mechanisms underlying NSAID hypersensitivity phenotypes, we employed high-throughput sequencing and characterized the genetic landscape associated with NSAID hypersensitivity phenotypes. Our results may be useful for developing new treatments and therapies for individuals with NSAID hypersensitivity.
The AC-cAMP (adenylate cyclase-cyclic adenosine monophosphate) and GC-cGMP (guanylate cyclase-cyclic guanosine monophosphate) pathways are pivotal signaling cascades within cells, intricately involved in mediating diverse physiological processes. These pathways are crucial for translating the effects of NSAIDs and are closely tied to the body’s inflammatory response. (1) The AC-cAMP pathway involves adenylate cyclase (AC), which catalyzes the conversion of ATP to cyclic AMP (cAMP). Elevated cAMP levels can modulate various cellular functions, influencing inflammation and immune responses. In the context of NSAID allergy, these pathways are disrupted, contributing to the adverse effects observed. Gaseous mediators such as nitric oxide (NO) and carbon monoxide (CO) further complicate this scenario. Some studies have implicated gaseous mediators, such as nitric oxide (NO) and carbon monoxide (CO), in the pathogenesis of NSAID allergy. NO is a signaling molecule that plays a role in various biological processes, including inflammation and vasodilation [10]. (2) The dysregulation of these pathways and the involvement of gaseous mediators contribute to the manifestation of NSAID hypersensitivity [11].
The HLA genes of the major histocompatibility complex (MHC) are important in many immune processes, and they are correlated with many autoimmune diseases and hypersensitivity reactions [12]. Studies have shown that specific HLA genes are associated with enhanced risk of drug hypersensitivity reactions. For example, in Asian populations, HLA-B*1502 was identified as a strong genetic risk factor for carbamazepine-induced Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) [13,14]. Similarly, HLA-B*5801 was associated with an increased risk of allopurinol-induced SJS and TEN in Han Chinese populations [15]. Numerous HLA genotypes have a significant association with adverse drug reactions, and several previous studies reported an association of NSAID hypersensitivity with MHC [16,17]. In addition, HLA-A*02:06, HLA-A*66:01, HLA-B*44:03, and HLA-C*12:03 were reported to be associated with NSAID hypersensitivity in the HLA –ADR web-based database [18]. Furthermore, some HLA genes were more frequent in the asthma group [19] and the chronic urticarial group [20]. However, whether genetic variations in the HLA region are associated with NSAID hypersensitivity remains largely unknown. The purpose of this study was to utilize a hospital-based database to identify HLA genetic predisposition in patients who have experienced adverse drug reactions to NSAIDs in the Taiwanese population.
2. Materials and Methods
2.1. Study Design
This case-control study included 58,091 Taiwanese and was conducted using data from the Taiwan Precision Medicine Initiative (TPMI). From June 2019 to December 2021, all participants were recruited from Taichung Veterans General Hospital (TCVGH) and their electronic health records were collected. Our study cohort comprised 13,387 patients, whose genetic profiles were connected to medical claims data in TCVGH, including demographic characteristics, procedures, examinations, diagnoses, surgeries, medication prescriptions, inpatient services, and outpatient services. This study, which involved human participants, was approved by the Ethics Committee of Taichung Veterans General Hospital Institutional Review Board (IRB no. SF19153A). Written informed consent was obtained from all participants in accordance with the principles defined in the Declaration of Helsinki.
2.2. HLA Allele Typing
Our study cohort consisted of 13,387 patients whose DNA from venous blood (2 mL) was genotyped for 8 major MHC Class I (HLA-A, -B, -C) and Class II (-DPA1, -DPB1, -DQA1, -DQB1, -DRB1) loci using an NXType™ Class I NGS HLA typing kit and an AllType™ NGS 11-Loci Amplification kit (Thermo Fisher Scientific, Waltham, MA, USA). The sequences were analyzed using Axiom HLA Analysis 1.2 (Thermo Fisher Scientific, Waltham, MA, USA), which utilizes advanced imputation methods to enable accurate HLA typing from SNP genotype data over the extended MHC region.
2.3. HLA Imputation for Genotype Data
The R library HLA Genotype Imputation with the Attribute Bagging (HIBAG) package [21] was utilized to perform HLA imputation for the HLA genes HLA-DQA1, HLA-DQB1, and HLA-DRB1. The imputation model used was specific to individuals of Asian ancestry. The resulting imputed data displayed two-field (4-digit) resolution for HLA alleles with allele frequencies (AFs) of at least 5.0%.
2.4. Participants
As illustrated in Supplementary Figure S1, the initial cohort included participants who had available genotyping information for HLA allele. We extracted participants over the age of 18 years who were exposed to NSAIDs. Data sources included 37,156 participants exposed to NSAIDs; 1217 participants who had an adverse drug reaction report (ADR) were defined as the NSAID allergy group, and 35,939 participants without ADR were defined as the NSAID non-allergy group. Both groups were matched by age and gender at a ratio of 1:10 (Supplementary Figure S1).
2.5. Covariates
In our study, clinical diagnoses were made based on the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) diagnosis codes, requiring a minimum of two outpatient diagnoses or one inpatient diagnosis between January 2009 and December 2021. We obtained comorbidity information from the electronic health records of TCVGH based on ICD-9 diagnostic codes for asthma (ICD-9-CM code 493), atopic dermatitis (ICD-9-CM code 691.8), allergic rhinitis (ICD-9-CM code 477), and urticaria (ICD-9-CM code 708). Then, we compared the allergy severity and HLA allele typing. According to the allergy severity level [22], patients with severe systemic reaction (Grade III) were defined as having severe allergy, and those with local reaction/mild-to-moderate systemic reaction (Grade I/II) were defined as having mild allergy.
2.6. Statistical Analysis
SAS version 9.4 software (SAS Institute Inc., Cary, NC, USA) was utilized to perform the data analysis. Participants’ characteristics were presented as mean values with standard deviations and percentages across the groups. The chi-square test was utilized to determine the statistical significance between categorical variables, while univariable and multivariable logistic regression analyses were conducted to estimate the associations between HLA genotypes and the risks of NSAID allergy. Odds ratios (ORs) and 95% confidence intervals (95% CIs) of HLA alleles were estimated using logistic regression models. The final model included significant covariates; the level of significance was set at a p value of less than 0.05.
3. Results
3.1. Characteristics of Participants
This study enrolled a total of 13,387 patients, consisting of 4860 males and 8527 females, who were genotyped for eight major HLA alleles. Table 1 shows the basic characteristics of the participants. The mean age of this study group was 56 ± 16 years, and compared to men, women had a greater prevalence of developing NSAID allergy. A higher IgE value was observed in the NSAID allergy group, compared to the control group. Furthermore, the patients in the NSAID allergy group had a lower creatinine level compared to the controls (p < 0.001). Compared to the control group, the NSAID allergy group had significantly higher prevalence rates of comorbidities, such as allergic rhinitis (24.7% vs. 20.6%, p < 0.001), asthma (13.3% vs. 8.0%, p < 0.001), urticaria (9.5% vs. 7.4%, p = 0.01), and atopic dermatitis (5.9% vs. 4.4%, p = 0.02).

Table 1.
Baseline characteristics of the participants.
3.2. Allele Frequencies and Association between HLA Alleles and NSAID Allergy
The allele frequencies were calculated by means of direct counting. The genotype frequencies of the HLA alleles are shown in Table 2, with only those alleles exhibiting statistical significance presented in the table. Supplementary tables have been meticulously prepared to encompass a comprehensive list of all identified alleles, offering a more thorough overview of the data. In the NSAID allergy group, the genotype frequencies of HLA-A*02:01, HLA-A*34:01, and HLA-DQA1*06:01 were found to be significantly elevated compared to the control group, even after applying the Bonferroni correction. Furthermore, the risk of NSAID allergy was significantly associated with HLA-A*02:01 (OR = 1.29, p < 0.001) and HLA-A*34:01 (OR = 9.90, p = 0.001), compared to their counterparts (Table 2).

Table 2.
The genotype frequencies of the HLA alleles in the participants.
3.3. Association of HLA Alleles and Comorbidities with the Risk of NSAID Allergy
In order to assess the association between HLA alleles and comorbidities, we utilized a multiple logistic regression model and evaluated its statistical significance. As shown in Figure 1, the risk of NSAID allergy was significantly associated with HLA-A*02:01 (OR = 1.29, 95% CI: 1.12–1.48, p < 0.001) and HLA-DRB1*16:02 (OR = 1.22, 95% CI: 1.01–1.47, p = 0.003), compared to their counterparts. Additionally, asthma (OR = 1.67, 95% CI: 1.38–2.02, p < 0.001) was an independent risk factor for the development of NSAID allergy.
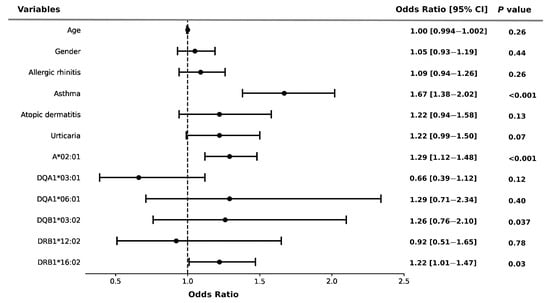
Figure 1.
Risk of NSAID allergy and HLA alleles in the participants. Error bars represent the 95% confidence intervals of the odds ratios. Multivariate logistic regression adjusted by age, gender, and potential confounders.
3.4. Distribution and Association of HLA Alleles with the Severity of Allergy
Depending on the severity of the NSAID-induced allergy, we stratified the patients into severe and mild allergy groups, respectively (Table 3). In the severe allergy group, the mean age was 57 ± 17 years, which was significantly higher than in the mild allergy group (p = 0.01). However, there were no statistically significant differences observed between the blood tests. A higher prevalence of asthma (23.4% vs. 13.2%, p = 0.01) was also observed in the severe allergy group, compared to the mild allergy group. The genotype frequencies of HLA alleles and their association with the severity of allergy are shown in Table 4. Higher frequencies of HLA-B*46:01, HLA-DPB1*02:02, and HLA-DRB*03:01 were observed in the severe allergy group compared to the mild allergy group. Specifically, the genotype frequency of HLA-B*46:01 was significantly increased in the severe allergy group after Bonferroni correction when compared with the mild allergy group. Additionally, the risk of allergy severity was linked to specific HLA allele genotypes, including HLA-B*46:01 (OR = 1.68, 95% CI: 1.04–2.71, p = 0.035), and HLA-DPB1*02:02 (OR = 1.95, 95% CI: 1.11–3.42, p = 0.020), when compared to their counterparts. Conversely, HLA-DPB1*05:01 (OR = 0.62, 95% CI: 0.39–0.99, p = 0.045) was associated with a reduced risk of severity in the severe allergy group, although these associations did not reach statistical significance after rigorous Bonferroni correction.

Table 3.
Characteristics of the study participants with allergy severity levels.

Table 4.
Genotype frequencies of allergy severity in the participants.
3.5. Association of HLA Alleles and Comorbidities with Severity of Allergy
HLA alleles and comorbidities both had significant impacts on the risk of severity of NSAID-induced allergy. As shown in Figure 2, HLA-B*46:01 (OR = 1.76, 95% CI: 1.08–2.89, p = 0.02) and HLA-DPB1*02:02 (OR = 1.86, 95% CI: 1.03–3.34, p = 0.04) were significantly associated with an increased risk of allergy severity. Similarly, the risk of severity of allergy was strongly associated with asthma (OR = 2.01, 95% CI: 1.10–3.68, p = 0.02) using the multiple logistic regression model (Figure 2).
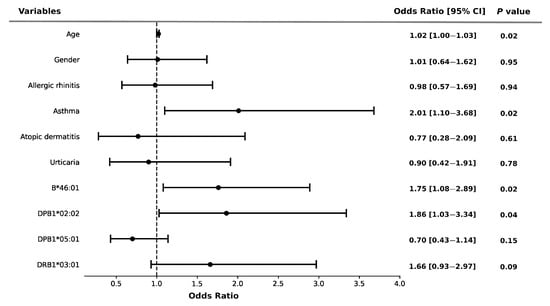
Figure 2.
Risk of NSAID allergy severity and HLA alleles in the participants. Error bars represent the 95% confidence intervals of the odds ratios. Multivariate logistic regression adjusted by age, gender, and potential confounders.
4. Discussion
NSAID hypersensitivity likely involves multiple mechanisms, including genetic predisposition, drug metabolism, pharmacokinetics, and environmental factors. Our novel finding implies that HLA allele frequency distribution differed between the NSAID allergic and control groups. Certain potential HLA alleles could potentially identify NSAID hypersensitivity in the Taiwanese population. In this study, we demonstrated an association between HLA-A*02:01, HLA-A*34:01, and NSAID-induced ADR. Patients with HLA-A*02:01 or HLA-A*34:01 had significantly increased NSAID allergic risk regardless of comorbidities of asthma. With respect to the severity of NSAID allergy, HLA-B*46:01 and HLA-DPB1*02:02 had a higher risk; in contrast, HLA-DPB1*05:01 had a decreased risk of allergy severity. In terms of the comorbidities of asthma, HLA-B*46:01 and HLA-DPB1*02:02 had a higher risk, although these associations did not reach statistical significance after rigorous Bonferroni correction.
In our study, the reason we chose HLA genes as the target for NSAID hypersensitivity is that more allergic diseases, such as allergic rhinitis, atopic dermatitis, and asthma, are found in patients with NSAID allergy [23,24], as shown in our data. In the current study, asthma had a stronger correlation (OR = 1.67, 95% CI: 1.38–2.02, p < 0.001) with the development of NSAID allergy. In previous studies, various HLA genes have been demonstrated to be associated with asthma, despite inconsistent findings [25,26,27]. Therefore, the HLA gene could be a possible anchor for genetic targeting in this group.
Previous evidence showed correlations between HLA genes and drug hypersensitivity reactions [28,29,30], and therefore, pharmacogenetic screening is a possible way to detect patients at high risk of severe drug reactions. As in our study cohort, we found that patients with certain HLA alleles tended to have a greater risk of NSAID allergy (HLA-A*02:01, HLA-A *34:01, HLA-DQA1*06:01, HLA-DRB1*12:02, HLA-DRB1*16:02). In the previous literature, HLA-DRB and HLA-DQ variability were shown to be associated with aspirin-related hypersensitivity, although the sample size was small [31,32,33,34]. For example, HLA-DRB and HLA-DPB1 showed a correlation with aspirin-induced asthma [33]. HLA-DRB1*1302, HLA-DQB1*0609, and HLA-DPB1*0201 were found to be related to aspirin-induced urticaria [32]. In the HLA–ADR web-based database, HLA-A*02:06 and HLA-B*44:03 had a higher NSAID ADR risk [18]. Our study has a much larger sample size and demonstrated a different pattern of HLA allele frequency among NSAID allergy patients; however, the alleles identified in our study were not consistent with those found in the previous studies, potentially due to variations in ethnicity and epigenetic profiles.
In addition to HLA polymorphisms, many genetic studies have been shown to have a relationship with NSAID hypersensitivity, such as the ALOX 15 gene in the arachidonic acid pathway and DAO in histamine metabolism [35,36,37]. The antigen-presenting cells are the key to activating an allergy reaction, which highlights the importance of HLA genes [38,39]. Therefore, it is rational to use them as a genetic marker for hypersensitivity reactions. HLA has an important role in antigen-presenting cells to distinguish self from non-self peptides and trigger naïve T cell immunity, and therefore, it is the initial step for many drug hypersensitivity reactions [29]. The different HLA allele variations could explain the diversity of ADR. Many studies have discussed the HLA–ADR relationship that has been identified in web-based databases. However, there are few data on NSAID hypersensitivity [18].
Genotyping HLAs can be useful for identifying individuals who are at higher risk of developing severe adverse reactions to certain drugs, allowing healthcare providers to avoid prescribing these drugs to these individuals, thus reducing the risk of adverse reactions. In the current study, we had a large cohort which contained 37,156 patients with NSAID exposure. We found different HLA allele polymorphisms between the NSAID hypersensitivity and control groups.
Nevertheless, this study had certain limitations. First, it is worth noting that our study is retrospective in nature, and as such, the diagnosis of NSAID hypersensitivity was not confirmed through provocation tests. We did not differentiate between immediate hypersensitivity, delayed hypersensitivity, or pseudo-allergic reactions. Additionally, the potential existence of cross-reactivity among different classes of NSAIDs was not explored in this study. To gain a more comprehensive understanding, future research should consider prospective study designs with a focus on confirming our findings through detailed diagnostic assessments. Second, our study did not delve into the realm of epigenetic factors. The influence of epigenetic modifications on gene expression, mediated by various environmental factors and experiences, could contribute to phenotypic variations within the Taiwanese population. Further research is needed to unravel how these epigenetic changes may impact gene expression and its role in NSAID hypersensitivity. Third, it is important to recognize that our analysis was limited to genetic variation within the HLA region. While this is a significant factor in hypersensitivity reactions, there may be other genetic pathways outside the HLA region that play a role. Future prospective studies should aim to integrate multiple genetic pathways and investigate their interplay to provide a more comprehensive understanding of NSAID hypersensitivity.
5. Conclusions
In this retrospective case-control study, our findings underscore the clinical significance of certain HLA alleles in the context of NSAID hypersensitivity. Specifically, we identified a significant association between HLA-A*02:01 and HLA-A*34:01 with NSAID hypersensitivity, shedding light on the genetic factors involved in this condition. Furthermore, our study revealed that individuals carrying HLA-B*46:01 and HLA-DPB1*02:02 alleles faced a significantly elevated risk of severe NSAID allergy. These results not only enhance our understanding of the genetic basis of NSAID hypersensitivity but also have clinical implications for risk assessment and management. The recent identification of HLA alleles associated with NSAID allergy severity highlights personalized medicine potential. Tailoring treatments based on individual HLA status holds promise for safer interventions. Future studies should integrate clinical phenotypes to grasp genetics, drug metabolism, and environmental factors’ interplay in NSAID hypersensitivity, advancing precision medicine, and understanding drug reactions in practice.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biomedicines11123273/s1. Supplementary Figure S1: Illustrative flow chart of the study design. Table S1. The genotype frequencies of the HLA-A alleles in the participants. Table S2. The genotype frequencies of the HLA-B alleles in the participants. Table S3. The genotype frequencies of the HLA-C alleles in the participants. Table S4. The genotype frequencies of the HLA-DPA1 alleles in the participants. Table S5. The genotype frequencies of the HLA-DPB1 alleles in the participants. Table S6. The genotype frequencies of the HLA-DQA1 alleles in the participants. Table S7. The genotype frequencies of the HLA-DQB1 alleles in the participants. Table S8. The genotype frequencies of the HLA-DRB1 alleles in the participants.
Author Contributions
Research idea and study design: S.-L.C. and I.-C.C.; data acquisition: S.-L.C., G.-C.L., Y.-M.C. and I.-C.C.; data analysis/interpretation: S.-L.C., C.-H.L., Y.-M.C., M.-H.L., H.-S.H. and I.-C.C.; statistical analysis: G.-C.L. and I.-C.C.; supervision or mentorship: C.-H.L., Y.-M.C., M.-H.L., H.-S.H. and I.-C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Ministry of Science and Technology, Taiwan [grant numbers, NSTC-110-2634-F-A49-005, NSTC-111-2218-E-039-001, and NSTC-111-2314-B-075A-003-MY3], Taichung Veterans General Hospital, Taiwan [grant numbers TCVGH-1127304B, TCVGH-1127301C, TCVGH-1127302D, and TCVGH-YM1120110], and Taichung Veterans General Hospital/National Health Research Institutes Joint Research Program [grant number TCVGH-YM1100103].
Institutional Review Board Statement
This research project was approved by the ethics committee of the Taichung Veterans General Hospital Institutional Review Board (IRB no. SF19153A), and all of the participants gave written informed consent.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in the study are included in the article and further inquiries can be directed to the corresponding author.
Acknowledgments
We thank all of the participants and investigators from the Taiwan Precision Medicine Initiative. The authors sincerely appreciate the assistance of the Center for Translational Medicine of Taichung Veterans General Hospital.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest. The authors have no relevant financial or non-financial interests to disclose. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
| BMI | body max index |
| WBC | white blood cell |
| HgB | hemoglobin |
| PLT | platelet |
| GOT | glutamic-oxalacetic transaminase. |
| GPT | glutamyl pyruvic transaminase |
| HLA | Human leukocyte antigen |
| NSAIDs | Non-steroid anti-inflammatory drugs |
| ADR | adverse drug reactions |
| MHC | major histocompatibility complex |
| SJS | Stevens–Johnson syndrome |
| TEN | toxic epidermal necrolysis |
| AFs | allele frequencies |
| ICD-9-CM | International Classification of Diseases, Ninth Revision, Clinical Modification |
References
- Ong, C.K.; Lirk, P.; Tan, C.H.; Seymour, R.A. An evidence-based update on nonsteroidal anti-inflammatory drugs. Clin. Med. Res. 2007, 5, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Bindu, S.; Mazumder, S.; Bandyopadhyay, U. Non-steroidal anti-inflammatory drugs (NSAIDs) and organ damage: A current perspective. Biochem. Pharmacol. 2020, 180, 114147. [Google Scholar] [CrossRef] [PubMed]
- Harirforoosh, S.; Asghar, W.; Jamali, F. Adverse effects of nonsteroidal antiinflammatory drugs: An update of gastrointestinal, cardiovascular and renal complications. J. Pharm. Pharm. Sci. 2013, 16, 821–847. [Google Scholar] [CrossRef] [PubMed]
- Yeung, W.Y.W.; Park, H.S. Update on the Management of Nonsteroidal Anti-Inflammatory Drug Hypersensitivity. Yonsei Med. J. 2020, 61, 4–14. [Google Scholar] [CrossRef]
- Hermans, M.A.W.; Otten, R.; Karim, A.F.; van Maaren, M.S. Nonsteroidal anti-inflammatory drug hypersensitivity: Not always an allergy! Neth. J. Med. 2018, 76, 52–59. [Google Scholar]
- Gomes, E.R.; Demoly, P. Epidemiology of hypersensitivity drug reactions. Curr. Opin. Allergy Clin. Immunol. 2005, 5, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Thong, B.Y. Nonsteroidal anti-inflammatory drug hypersensitivity in the Asia-Pacific. Asia Pac. Allergy 2018, 8, e38. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Dhopeshwarkar, N.; Blumenthal, K.G.; Goss, F.; Topaz, M.; Slight, S.P.; Bates, D.W. Drug allergies documented in electronic health records of a large healthcare system. Allergy 2016, 71, 1305–1313. [Google Scholar] [CrossRef]
- Rajan, J.P.; Wineinger, N.E.; Stevenson, D.D.; White, A.A. Prevalence of aspirin-exacerbated respiratory disease among asthmatic patients: A meta-analysis of the literature. J. Allergy Clin. Immunol. 2015, 135, 676–681.e1. [Google Scholar] [CrossRef]
- Denninger, J.W.; Marletta, M.A. Guanylate cyclase and the ˙NO/cGMP signaling pathway. Biochim. Biophys. Acta 1999, 1411, 334–350. [Google Scholar] [CrossRef]
- Laukeviciene, A.; Ugincius, P.; Korotkich, I.; Lazauskas, R.; Kevelaitis, E. Anaphylaxis of small arteries: Putative role of nitric oxide and prostanoids. Medicina 2010, 46, 38–44. [Google Scholar] [CrossRef]
- Choo, S.Y. The HLA system: Genetics, immunology, clinical testing, and clinical implications. Yonsei Med. J. 2007, 48, 11–23. [Google Scholar] [CrossRef]
- Chang, C.C.; Too, C.L.; Murad, S.; Hussein, S.H. Association of HLA-B*1502 allele with carbamazepine-induced toxic epidermal necrolysis and Stevens-Johnson syndrome in the multi-ethnic Malaysian population. Int. J. Dermatol. 2011, 50, 221–224. [Google Scholar] [CrossRef]
- Ferrell, P.B., Jr.; McLeod, H.L. Carbamazepine, HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics 2008, 9, 1543–1546. [Google Scholar] [CrossRef]
- Hung, S.I.; Chung, W.H.; Liou, L.B.; Chu, C.C.; Lin, M.; Huang, H.P.; Lin, Y.L.; Lan, J.L.; Yang, L.C.; Hong, H.S.; et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc. Natl. Acad. Sci. USA 2005, 102, 4134–4139. [Google Scholar] [CrossRef] [PubMed]
- Khalili, N. The Immunogenetics of Cutaneous Drug Reactions. Adv. Exp. Med. Biol. 2022, 1367, 411–431. [Google Scholar] [CrossRef] [PubMed]
- Quiralte, J.; Sánchez-García, F.; Torres, M.J.; Blanco, C.; Castillo, R.; Ortega, N.; de Castro, F.R.; Pérez-Aciego, P.; Carrillo, T. Association of HLA-DR11 with the anaphylactoid reaction caused by nonsteroidal anti-inflammatory drugs. J. Allergy Clin. Immunol. 1999, 103, 685–689. [Google Scholar] [CrossRef]
- Ghattaoraya, G.S.; Dundar, Y.; González-Galarza, F.F.; Maia, M.H.; Santos, E.J.; da Silva, A.L.; McCabe, A.; Middleton, D.; Alfirevic, A.; Dickson, R.; et al. A web resource for mining HLA associations with adverse drug reactions: HLA-ADR. Database 2016, 2016, baw069. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, D.I.; Chila, N.; Abdelouahab, F.; Bouyoucef, H.; Bougrida, M.; Rouabah, L.; Nedjar, F. Association between HLA-class II genes and asthma susceptibility in a selected Constantine population. Pan Afr. Med. J. 2020, 35, 48. [Google Scholar] [CrossRef]
- Doğan, N.; Çildağ, S.; Yenisey, Ç.; Şentürk, T. The association between chronic spontaneous urticaria and HLA class I and class II antigen. Turk. J. Med. Sci. 2020, 50, 1231–1235. [Google Scholar] [CrossRef]
- Zheng, X.; Shen, J.; Cox, C.; Wakefield, J.C.; Ehm, M.G.; Nelson, M.R.; Weir, B.S. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenom. J. 2014, 14, 192–200. [Google Scholar] [CrossRef]
- Niggemann, B.; Beyer, K. Time for a new grading system for allergic reactions? Allergy 2016, 71, 135–136. [Google Scholar] [CrossRef]
- Asero, R. Single NSAID hypersensitivity is associated with atopic status. Eur. Ann. Allergy Clin. Immunol. 2015, 47, 48–53. [Google Scholar]
- Asero, R. Intolerance to nonsteroidal anti-inflammatory drugs might precede by years the onset of chronic urticaria. J. Allergy Clin. Immunol. 2003, 111, 1095–1098. [Google Scholar] [CrossRef]
- Li, J.; Hao, Y.; Li, W.; Lv, X.; Gao, P. HLA-G in asthma and its potential as an effective therapeutic agent. Allergol. Immunopathol. 2023, 51, 22–29. [Google Scholar] [CrossRef]
- Kontakioti, E.; Domvri, K.; Papakosta, D.; Daniilidis, M. HLA and asthma phenotypes/endotypes: A review. Hum. Immunol. 2014, 75, 930–939. [Google Scholar] [CrossRef]
- Sánchez-Borges, M.; Caballero-Fonseca, F.; Capriles-Hulett, A. Cofactors and comorbidities in patients with aspirin/NSAID hypersensitivity. Allergol. Immunopathol. 2017, 45, 573–578. [Google Scholar] [CrossRef]
- Saper, V.E.; Ombrello, M.J.; Tremoulet, A.H.; Montero-Martin, G.; Prahalad, S.; Canna, S.; Shimizu, C.; Deutsch, G.; Tan, S.Y.; Remmers, E.F.; et al. Severe delayed hypersensitivity reactions to IL-1 and IL-6 inhibitors link to common HLA-DRB1*15 alleles. Ann. Rheum. Dis. 2022, 81, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Redwood, A.J.; Pavlos, R.K.; White, K.D.; Phillips, E.J. HLAs: Key regulators of T-cell-mediated drug hypersensitivity. HLA 2018, 91, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Negrini, S.; Becquemont, L. HLA-associated drug hypersensitivity and the prediction of adverse drug reactions. Pharmacogenomics 2017, 18, 1441–1457. [Google Scholar] [CrossRef] [PubMed]
- Pacor, M.L.; Di Lorenzo, G.; Mansueto, P.; Martinelli, N.; Esposito-Pellitteri, M.; Pradella, P.; Uxa, L.; Di Fede, G.; Rini, G.; Corrocher, R. Relationship between human leucocyte antigen class I and class II and chronic idiopathic urticaria associated with aspirin and/or NSAIDs hypersensitivity. Mediat. Inflamm. 2006, 2006, 62489. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Choi, J.H.; Lee, K.W.; Kim, S.H.; Shin, E.S.; Oh, H.B.; Suh, C.H.; Nahm, D.H.; Park, H.S. The human leucocyte antigen-DRB1*1302-DQB1*0609-DPB1*0201 haplotype may be a strong genetic marker for aspirin-induced urticaria. Clin. Exp. Allergy 2005, 35, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.W.; Nizankowska, E.; Schmitz-Schumann, M.; Pile, K.; Bochenek, G.; Dyczek, A.; Cookson, W.O.; Szczeklik, A. Aspirin-induced asthma and HLA-DRB1 and HLA-DPB1 genotypes. Clin. Exp. Allergy 1997, 27, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilzadeh, H.; Nabavi, M.; Amirzargar, A.A.; Aryan, Z.; Arshi, S.; Bemanian, M.H.; Fallahpour, M.; Mortazavi, N.; Rezaei, N. HLA-DRB and HLA-DQ genetic variability in patients with aspirin-exacerbated respiratory disease. Am. J. Rhinol. Allergy 2015, 29, e63–e69. [Google Scholar] [CrossRef]
- Trinh, H.K.T.; Pham, L.D.; Le, K.M.; Park, H.S. Pharmacogenomics of Hypersensitivity to Non-steroidal Anti-inflammatory Drugs. Front. Genet. 2021, 12, 647257. [Google Scholar] [CrossRef]
- Plaza-Serón, M.D.C.; García-Martín, E.; Agúndez, J.A.; Ayuso, P. Hypersensitivity reactions to nonsteroidal anti-inflammatory drugs: An update on pharmacogenetics studies. Pharmacogenomics 2018, 19, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Park, H.S. Genetic markers for differentiating aspirin-hypersensitivity. Yonsei Med. J. 2006, 47, 15–21. [Google Scholar] [CrossRef]
- Murdaca, G.; Contini, P.; Negrini, S.; Ciprandi, G.; Puppo, F. Immunoregulatory Role of HLA-G in Allergic Diseases. J. Immunol. Res. 2016, 2016, 6865758. [Google Scholar] [CrossRef]
- Mosaad, Y.M. Clinical Role of Human Leukocyte Antigen in Health and Disease. Scand. J. Immunol. 2015, 82, 283–306. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).