Elucidating the Histone Deacetylase Gene Expression Signatures in Peripheral Blood Mononuclear Cells That Correlate Essential Cardiac Function and Aid in Classifying Coronary Artery Disease through a Logistic Regression Model
Abstract
:1. Introduction
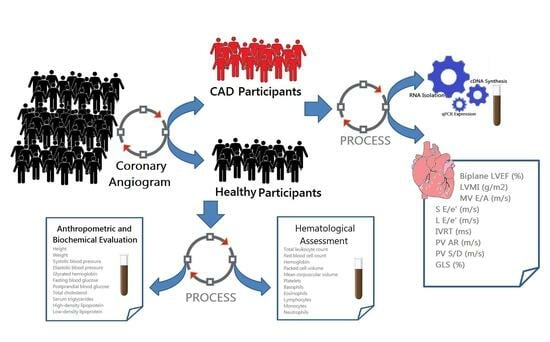
2. Materials and Methods
2.1. Clinical Specimens
2.2. Human Anthropometric and Biochemical Evaluation
2.3. Gene Expression Profiling of HDAC
2.4. Echocardiography Imaging
2.5. Diagnostic Model Using Binary Logistic Regression
2.6. Statistical Examination
3. Results
3.1. Evaluation of Anthropometric, Hematological, and Biochemical Features
3.2. Gene Expression Profiling of HDACs in PBMCs
3.3. Assessment of 2D Echocardiographic Imaging
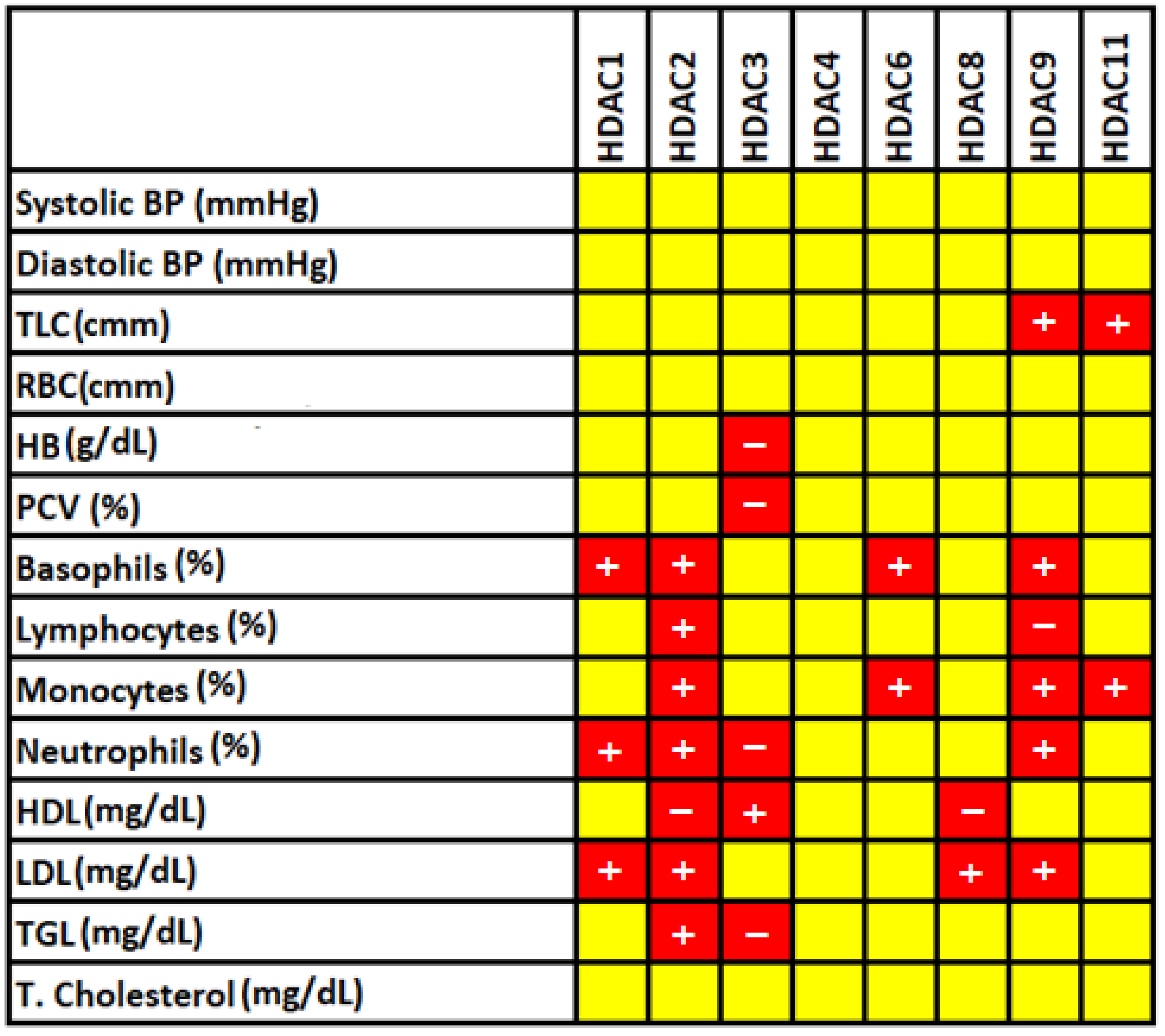
3.4. Correlation of Significantly Altered Hematological and Biochemical Features Characteristics with HDACs
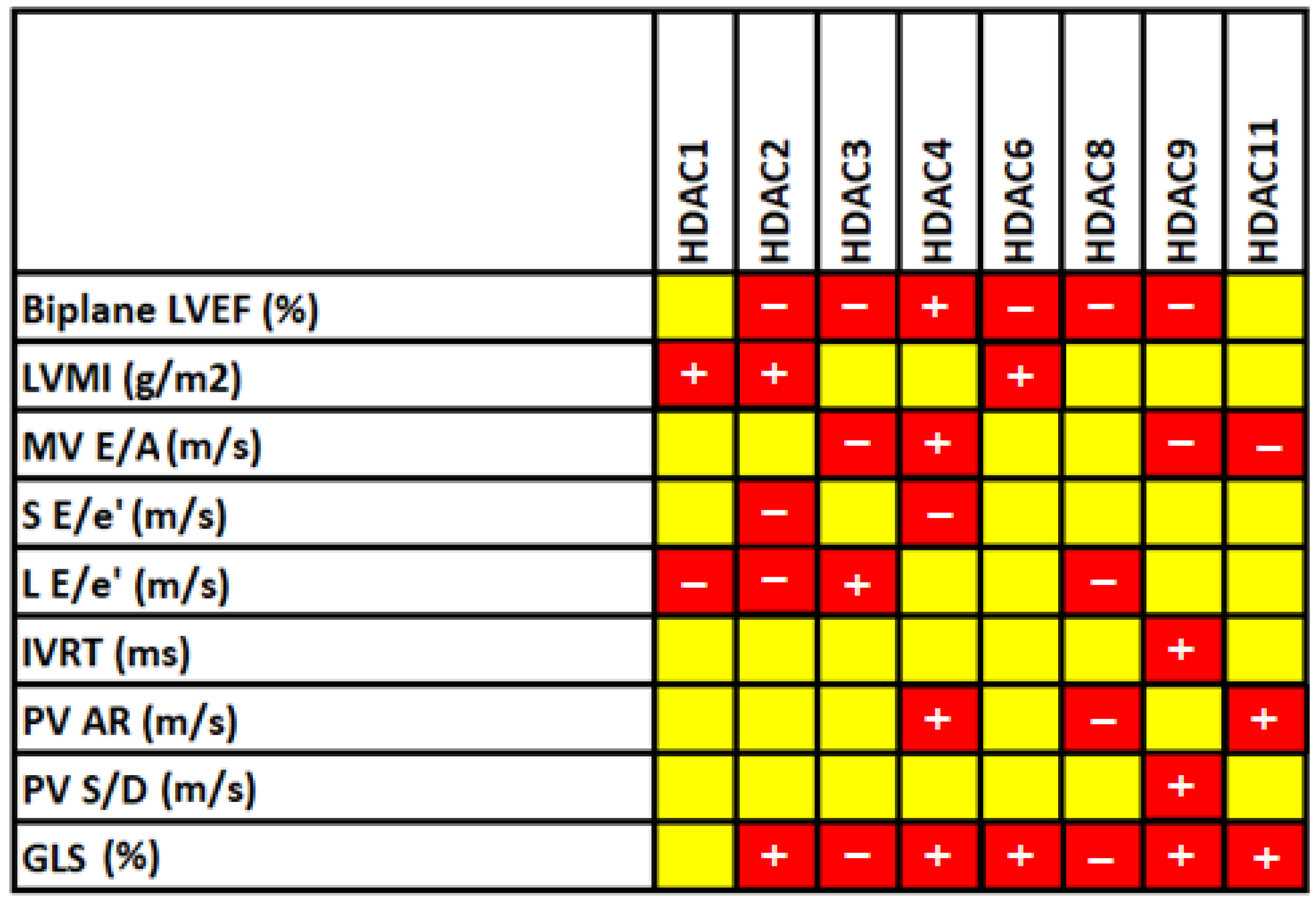
3.5. Correlation between HDAC Levels and 2D Echocardiographic Parameters
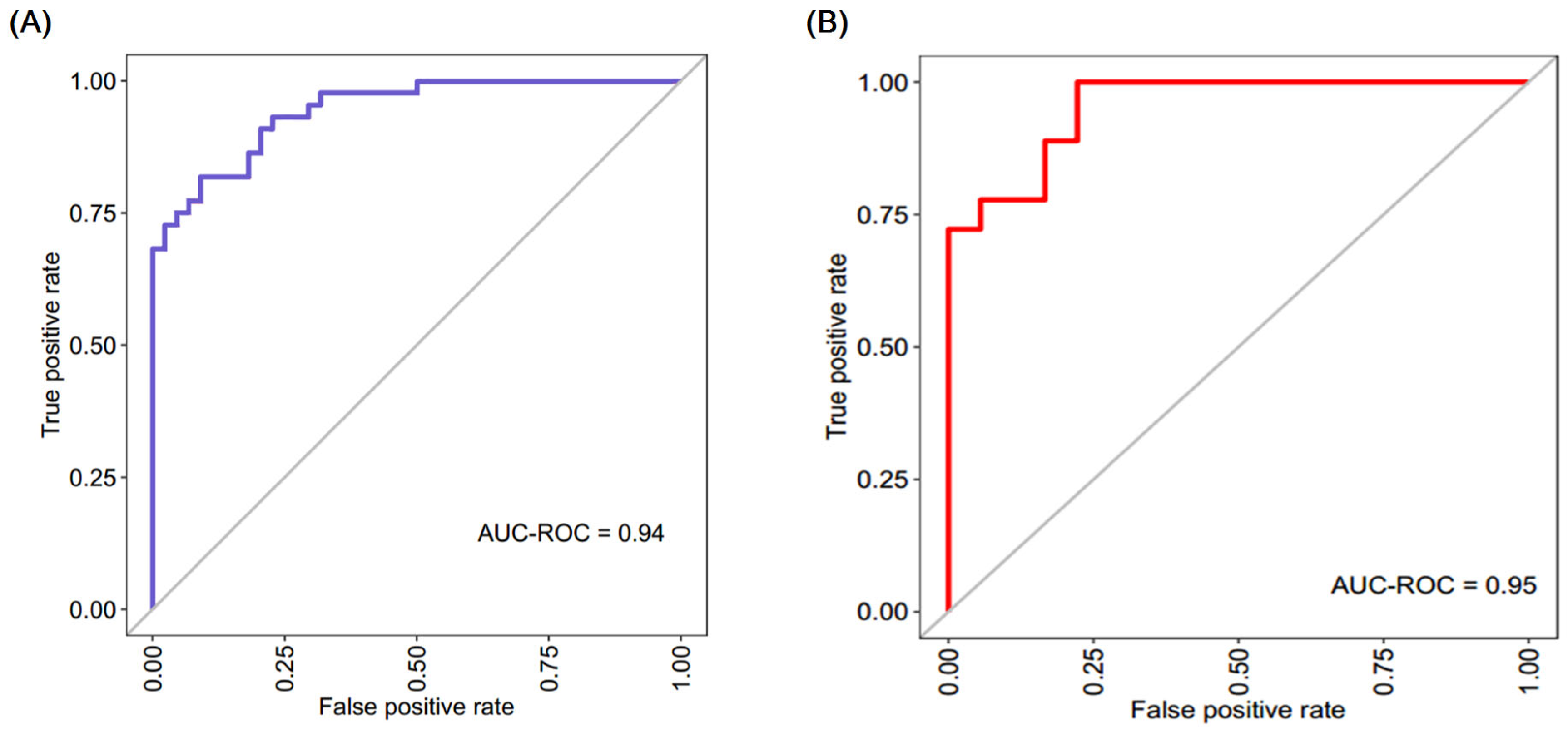
3.6. Diagnostic Model with Crucial HADCs for CAD Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, regional, and national burden of cardiovascular diseases for 10 causes. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Sumi, M.P.; Mahajan, B.; Sattar, R.S.; Nimisha Apurva Kumar, A.; Sharma, A.K.; Ahmad, E.; Ali, A.; Saluja, S.S. Elucidation of Epigenetic Landscape in Coronary Artery Disease: A Review on Basic Concept to Personalized Medicine. Epigenetics Insights 2021, 14, 2516865720988567. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Murali, S.; Lee, E.; Supramaniam, V.; Hausenloy, D.J.; Obungoloch, J.; Brecher, J.; Lin, R.; Ding, H.; Akudjedu, T.N.; et al. Sustainable low-field cardiovascular magnetic resonance in changing healthcare systems. Eur. Heart J. Cardiovasc. Imaging 2022, 23, e246–e260. [Google Scholar] [CrossRef] [PubMed]
- Monisha, K.G.; Prabu, P.; Chokkalingam, M.; Murugesan, R.; Milenkovic, D.; Ahmed, S.S.S.J. Clinical utility of brain-derived neurotrophic factor as a biomarker with left ventricular echocardiographic indices for potential diagnosis of coronary artery disease. Sci. Rep. 2020, 10, 16359. [Google Scholar] [CrossRef] [PubMed]
- Scirica, B.M. Acute coronary syndrome: Emerging tools for diagnosis and risk assessment. J. Am. Coll. Cardiol. 2010, 55, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Zarzour, A.; Kim, H.W.; Weintraub, N.L. Epigenetic regulation of vascular diseases. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Zhao, T.C.; Wang, Z.; Zhao, T.Y. The important role of histone deacetylases in modulating vascular physiology and arteriosclerosis. Atherosclerosis 2020, 303, 36–42. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Qin, W.; Yi, F. Targeting histone deacetylases: Perspectives for epigenetic-based therapy in cardio-cerebrovascular disease. J. Geriatr. Cardiol. JGC 2015, 12, 153. [Google Scholar]
- Lang, R.M.; Badano, L.P.; Mor-Avi, V.; Afilalo, J.; Armstrong, A.; Ernande, L.; Flachskampf, F.A.; Foster, E.; Goldstein, S.A.; Kuznetsova, T.; et al. Recommendations for cardiac chamber quantification by echocardiography in adults: An update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J. Am. Soc. Echocardiogr. 2015, 28, 1440–1457. [Google Scholar] [CrossRef]
- Nagueh, S.F.; Smiseth, O.A.; Appleton, C.P.; Byrd, B.F.; Dokainish, H.; Edvardsen, T. Recommendations for the evaluation of left ventricular diastolic function by echocardiography: An update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J. Am. Soc. Echocardiogr. 2016, 29, 277–314. [Google Scholar] [CrossRef] [PubMed]
- Wierda, R.J.; Geutskens, S.B.; Jukema, J.W.; Quax, P.H.; van den Elsen, P.J. Epigenetics in atherosclerosis and inflammation. J. Cell. Mol. Med. 2010, 14, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Neele, A.E.; Willemsen, L.; Chen, H.J.; Dzobo, K.E.; de Winther, M.P. Targeting epigenetics as atherosclerosis treatment: An updated view. Curr. Opin. Lipidol. 2020, 31, 324. [Google Scholar] [CrossRef] [PubMed]
- Marchio, P.; Guerra-Ojeda, S.; Vila, J.M.; Aldasoro, M.; Victor, V.M.; Mauricio, M.D. Targeting early atherosclerosis: A focus on oxidative stress and inflammation. Oxidative Med. Cell. Longev. 2019, 2019, 8563845. [Google Scholar] [CrossRef]
- Manea, S.A.; Vlad, M.L.; Fenyo, I.M.; Lazar, A.G.; Raicu, M.; Muresian, H.; Simionescu, M.; Manea, A. Pharmacological inhibition of histone deacetylase reduces NADPH oxidase expression, oxidative stress and the progression of atherosclerotic lesions in hypercholesterolemic apolipoprotein E-deficient mice; potential implications for human atherosclerosis. Redox Biol. 2020, 28, 101338. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.P.; Zhang, P.; Qi, Y.X.; Chen, S.G.; Shen, B.R.; Han, Y.; Yan, Z.Q.; Jiang, Z.L. The role of SIRT6 in the differentiation of vascular smooth muscle cells in response to cyclic strain. Int. J. Biochem. Cell Biol. 2014, 49, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Kee, H.J.; Ryu, Y.; Seok, Y.M.; Choi, S.Y.; Sun, S.; Kim, G.R.; Jeong, M.H. Selective inhibition of histone deacetylase 8 improves vascular hypertrophy, relaxation, and inflammation in angiotensin II hypertensive mice. Clin. Hypertens. 2019, 25, 13. [Google Scholar] [CrossRef]
- Usui, T.; Morita, T.; Okada, M.; Yamawaki, H. Histone deacetylase 4 controls neointimal hyperplasia via stimulating proliferation and migration of vascular smooth muscle cells. Hypertension 2014, 63, 397–403. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Qian, Z.; Lin, B.; Chen, J.; Luo, Y.; Qu, J.; Raj, J.U.; Gou, D. Phosphatidylinositol 3-Kinase-DNA Methyltransferase 1-miR-1281-Histone Deacetylase 4 Regulatory Axis Mediates Platelet-Derived Growth Factor-Induced Proliferation and Migration of Pulmonary Artery Smooth Muscle Cells. J. Am. Heart Assoc. 2018, 7, e007572. [Google Scholar] [CrossRef]
- Zhang, B.; Dong, Y.; Liu, M.; Yang, L.; Zhao, Z. miR-149-5p inhibits vascular smooth muscle cells proliferation, invasion, and migration by targeting histone deacetylase 4 (HDAC4). Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 7581. [Google Scholar] [CrossRef]
- Zheng, X.; Wu, Z.; Xu, K.; Qiu, Y.; Su, X.; Zhang, Z.; Zhou, M. Interfering histone deacetylase 4 inhibits the proliferation of vascular smooth muscle cells via regulating MEG3/miR-125a-5p/IRF1. Cell Adhes. Migr. 2019, 13, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Abend, A.; Shkedi, O.; Fertouk, M.; Caspi, L.H.; Kehat, I. Salt-inducible kinase induces cytoplasmic histone deacetylase 4 to promote vascular calcification. EMBO Rep. 2017, 18, 1166–1185. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Lin, C.C.; Luo, S.F.; Lee, H.C.; Lee, I.T.; Aird, W.C.; Yang, C.M. Tumor necrosis factor-α enhances neutrophil adhesiveness: Induction of vascular cell adhesion molecule-1 via activation of Akt and CaM kinase II and modifications of histone acetyltransferase and histone deacetylase 4 in human tracheal smooth muscle cells. Mol. Pharmacol. 2008, 73, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Leucker, T.M.; Nomura, Y.; Kim, J.H.; Bhatta, A.; Wang, V.; Wecker, A.; Jandu, S.; Santhanam, L.; Berkowitz, D.; Romer, L.; et al. Cystathionine γ-lyase protects vascular endothelium: A role for inhibition of histone deacetylase 6. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H711–H720. [Google Scholar] [CrossRef] [PubMed]
- Abouhish, H.; Thounaojam, M.C.; Jadeja, R.N.; Gutsaeva, D.R.; Powell, F.L.; Khriza, M.; Martin, P.M.; Bartoli, M. Inhibition of HDAC6 attenuates diabetes-induced retinal redox imbalance and microangiopathy. Antioxidants 2020, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Karki, P.; Ke, Y.; Tian, Y.; Ohmura, T.; Sitikov, A.; Sarich, N.; Montgomery, C.P.; Birukova, A.A. Staphylococcus aureus–induced endothelial permeability and inflammation are mediated by microtubule destabilization. J. Biol. Chem. 2019, 294, 3369–3384. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, J.; Shaik, N.F.; Yi, B.; Wei, X.; Yang, X.F.; Naik, U.P.; Summer, R.; Yan, G.; Xu, X.; et al. The histone deacetylase inhibitor tubacin mitigates endothelial dysfunction by up-regulating the expression of endothelial nitric oxide synthase. J. Biol. Chem. 2019, 294, 19565–19576. [Google Scholar] [CrossRef] [PubMed]
- Grbić, E.; Gorkič, N.; Pleskovič, A.; Zorc, M.; Ljuca, F.; Gasparini, M.; Mrđa, B.; Cilenšek, I.; MankočSanach, M.; Petrovič, D. Association between rs2107595 HDAC9 gene polymorphism and advanced carotid atherosclerosis in the Slovenian cohort. Lipids Health Dis. 2020, 19, 71. [Google Scholar] [CrossRef]
- Wang, X.B.; Han, Y.D.; Sabina, S.; Cui, N.H.; Zhang, S.; Liu, Z.J.; Li, C.; Zheng, F. HDAC9 variant Rs2107595 modifies susceptibility to coronary artery disease and the severity of coronary atherosclerosis in a Chinese Han population. PLoS ONE 2016, 11, e0160449. [Google Scholar] [CrossRef]
- Prestel, M.; Prell-Schicker, C.; Webb, T.; Malik, R.; Lindner, B.; Ziesch, N.; Rex-Haffner, M.; Röh, S.; Viturawong, T.; Lehm, M.; et al. The atherosclerosis risk variant rs2107595 mediates allele-specific transcriptional regulation of HDAC9 via E2F3 and Rb1. Stroke 2019, 50, 2651–2660. [Google Scholar] [CrossRef]
- Shroff, N.; Ander, B.P.; Zhan, X.; Stamova, B.; Liu, D.; Hull, H.; Hamade, F.R.; Dykstra-Aiello, C.; Ng, K.; Sharp, F.R.; et al. HDAC9 polymorphism alters blood gene expression in patients with large vessel atherosclerotic stroke. Transl. Stroke Res. 2019, 10, 19–25. [Google Scholar] [CrossRef]
- Asare, Y.; Campbell-James, T.A.; Bokov, Y.; Yu, L.L.; Prestel, M.; El Bounkari, O.; Roth, S.; Megens, R.T.; Straub, T.; Thomas, K.; et al. Histone deacetylase 9 activates IKK to regulate atherosclerotic plaque vulnerability. Circ. Res. 2020, 127, 811–823. [Google Scholar] [CrossRef]
- Zhang, R.; Ge, J. Proteinase-Activated Receptor-2 Modulates Ve-Cadherin Expression to Affect Human Vascular Endothelial Barrier Function. J. Cell. Biochem. 2017, 118, 4587–4593. [Google Scholar] [CrossRef] [PubMed]
- Yanginlar, C.; Logie, C. HDAC11 is a regulator of diverse immune functions. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2018, 1861, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Dorneles, G.P.; Boeira, M.C.R.; Schipper, L.L.; Silva, I.R.; Elsner, V.R.; Dal Lago, P.; Romão, P.R. Acute strenuous exercise induces an imbalance on histone H4 acetylation/histone deacetylase 2 and increases the proinflammatory profile of PBMC of obese individuals. Oxidative Med. Cell. Longev. 2017, 2017, 1530230. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Le, T.P.; Lee, S.K.; Guo, E.; Kim, D.; Lee, S.; Seo, S.Y.; Lee, S.Y.; Kim, J.H.; Lee, S.Y. Honokiol ameliorates angiotensin II-induced hypertension and endothelial dysfunction by inhibiting HDAC6-mediated cystathionine γ-lyase degradation. J. Cell. Mol. Med. 2020, 24, 10663–10676. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, Y.; Jia, W.; Sun, D.; Zhao, L.; Zhang, C.; Wang, C.; Lyu, Q.; Chen, Y.; Chen, G.; et al. Association of the Total White Blood Cell, Neutrophils, and Monocytes Count with the Presence, Severity, and Types of Carotid Atherosclerotic Plaque. Front. Med. 2020, 7, 313. [Google Scholar] [CrossRef]
- Abdi-Ali, A.; Miller, R.J.; Southern, D.; Zhang, M.; Mikami, Y.; Knudtson, M.; Heydari, B.; Howarth, A.G.; Lydell, C.P.; James, M.T.; et al. LV mass independently predicts mortality and need for future revascularization in patients undergoing diagnostic coronary angiography. JACC Cardiovasc. Imaging 2018, 11, 423–433. [Google Scholar] [CrossRef]
- Kee, H.J.; Bae, E.H.; Park, S.; Lee, K.E.; Suh, S.H.; Kim, S.W.; Jeong, M.H. HDAC inhibition suppresses cardiac hypertrophy and fibrosis in DOCA-salt hypertensive rats via regulation of HDAC6/HDAC8 enzyme activity. Kidney Blood Press. Res. 2013, 37, 229–239. [Google Scholar] [CrossRef]
- Kimbrough, D.; Wang, S.H.; Wright, L.H.; Mani, S.K.; Kasiganesan, H.; LaRue, A.C.; Cheng, Q.; Nadig, S.N.; Atkinson, C.; Menick, D.R. HDAC inhibition helps post-MI healing by modulating macrophage polarization. J. Mol. Cell. Cardiol. 2018, 119, 51–63. [Google Scholar] [CrossRef]
- Chen, Y.; Du, J.; Zhao, Y.T.; Zhang, L.; Lv, G.; Zhuang, S.; Qin, G.; Zhao, T.C. Histone deacetylase (HDAC) inhibition improves myocardial function and prevents cardiac remodeling in diabetic mice. Cardiovasc. Diabetol. 2015, 14, 99. [Google Scholar] [CrossRef]
- Jeong, M.Y.; Lin, Y.H.; Wennersten, S.A.; Demos-Davies, K.M.; Cavasin, M.A.; Mahaffey, J.H.; Monzani, V.; Saripalli, C.; Mascagni, P.; Reece, T.B.; et al. Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci. Transl. Med. 2018, 10, eaao0144. [Google Scholar] [CrossRef]
| S.No | Gene | FORWARD 5′<-----Sequence----->3′ | REVERSE 5′<-----Sequence----->3′ |
|---|---|---|---|
| 1 | HDAC1 | GAGATGACCAAGTACCACAGC | TGACAGAACTCAAACAGGCC |
| 2 | HDAC2 | TGACAAACCAGAACACTCCAG | TGACAAACCAGAACACTCCAG |
| 3 | HDAC3 | GGACTTCTACCAACCCACG | CAGCACGAGTAGAGGGATATTG |
| 4 | HDAC4 | ACAAGGAGAAGGGCAAAGAG | GCGTTTTCCCGTACCAGTAG |
| 5 | HDAC5 | TCACCGCAAAACTCCTACAG | AGTTCCCGTTGTCATAGCG |
| 6 | HDAC6 | TTCAACTCTGTGGCTGTGG | GGGACACATATAGCACACTGG |
| 7 | HDAC7 | GCAGATCATTCAACAGCCATG | TTGGTAGAAGGTTTGCTGGG |
| 8 | HDAC8 | AATTAACTGGTCTGGAGGGTG | TGCAGATCCAAATCCACGTAG |
| 9 | HDAC9 | ACACATTACCAGGAGCACAAG | CAACATTTCCATCCTTCCGC |
| 10 | HDAC10 | GTCCTTTACTTCTCCTGGCAC | GTAGTCAGCGTTTCCCATCC |
| 11 | HDAC11 | GTTTCTGTTTGAGCGTGTGG | GGTAGATGTGGCGGTTGTAG |
| 12 | GAPDH | TGTCATCAACGGAAAGGC | GCATCAGCAGAAGGAGCA |
| Parameters | Control (Mean ± Standard Deviation) | CAD (Mean ± Standard Deviation) | p-Value |
|---|---|---|---|
| Age (years) | 44.90 ± 8.96 | 48.23 ± 9.34 | 0.096 |
| Gender (male/female) | 38/24 | 33/29 | 0.360 |
| BMI (kg/m2) | 23.76 ± 2.98 | 25.02 ± 1.19 | 0.081 |
| Systolic BP (mmHg) | 103.44 ± 18.43 | 118.65 ± 18.64 | <0.001 |
| Diastolic BP (mmHg) | 69.62 ± 8.87 | 78.60 ± 14.02 | <0.001 |
| FBS (mg/dL) | 99 ± 16.59 | 107.55 ± 12.18 | <0.071 |
| PPBS (mg/dL) | 109.62 ± 38.86 | 105.34 ± 26.84 | <0.065 |
| TLC (cmm) | 7034.88 ± 1700.96 | 9783.72 ± 4165.04 | <0.001 |
| RBC (cmm) | 5.14 ± 0.65 | 4.62 ± 0.58 | <0.001 |
| HB (g/dL) | 14.57 ± 1.62 | 13.18 ± 1.60 | <0.001 |
| PCV (%) | 44.48 ± 4.96 | 40.05 ± 4.87 | <0.001 |
| MCV (fl) | 87.23 ± 7.86 | 86.57 ± 7.11 | 0.686 |
| MCH (pg) | 28.66 ± 2.86 | 28.69 ± 2.84 | 0.955 |
| MCHC (g/dL) | 32.82 ± 0.68 | 33.12 ± 0.95 | 0.096 |
| RDW (%) | 14.25 ± 1.50 | 14.41 ± 1.54 | 0.631 |
| Platelet (cmm) | 2.83 ± 0.68 | 2.81 ± 0.73 | 0.926 |
| Basophil (%) | 0.57 ± 0.44 | 0.83 ± 0.49 | 0.015 |
| Eosinophil (%) | 3.66 ± 3.24 | 3.9 ± 3.62 | 0.750 |
| Lymphocyte (%) | 29.69 ± 8.79 | 36.03 ± 12.23 | 0.007 |
| Monocyte (%) | 4.18 ± 1.80 | 7.12 ± 2.86 | <0.001 |
| Neutrophil (%) | 54.83 ± 8.77 | 66.46 ± 12.76 | <0.001 |
| HDL (mg/dL) | 38.11 ± 7.00 | 28.81 ± 7.50 | <0.001 |
| LDL (mg/dL) | 115.86 ± 27.92 | 165.83 ± 46.59 | <0.001 |
| TGL (mg/dL) | 118.90 ± 65.48 | 189.90 ± 43.70 | <0.001 |
| Total Cholesterol (mg/dL) | 182.302 ± 33.94 | 211.93 ± 52.79 | 0.003 |
| HbA1c (%) | 5.05 ± 0.43 | 5.35 ± 0.16 | 0.831 |
| HDACs | Control (Mean ± Standard Deviation) | CAD (Mean ± Standard Deviation) | p-Value |
|---|---|---|---|
| HDAC1 | 2.33 ± 1.21 | 2.91 ± 1.48 | 0.030 |
| HDAC2 | 4.35 ± 1.69 | 5.51 ± 1.52 | 0.014 |
| HDAC3 | 3.82 ± 0.73 | 2.28 ± 0.59 | 0.044 |
| HDAC4 | 3.43 ± 0.98 | 4.52 ± 1.01 | 0.032 |
| HDAC5 | 1.91 ± 0.37 | 3.48 ± 1.75 | 0.202 |
| HDAC6 | 2.26 ± 0.70 | 2.52 ± 0.71 | 0.042 |
| HDAC7 | 0.95 ± 0.17 | 1.58 ± 0.92 | 0.130 |
| HDAC8 | 3.80 ± 2.36 | 5.21 ± 3.02 | 0.004 |
| HDAC9 | 3.54 ± 1.99 | 6.25 ± 1.16 | <0.001 |
| HDAC10 | 0.84 ± 0.52 | 1.34 ± 0.55 | 0.184 |
| HDAC11 | 3.36 ± 0.21 | 4.93 ± 2.19 | <0.001 |
| Echo Parameters | Control (Mean ± Standard Deviation) | CAD (Mean ± Standard Deviation) | p-Value |
|---|---|---|---|
| Biplane LVEF (%) | 58.66 ± 1.77 | 48.51 ± 5.62 | <0.001 |
| LVMI (g/m2) | 69.95 ± 25.91 | 83.97 ± 18.74 | 0.004 |
| MV E/A (m/s) | 1.13 ± 0.30 | 0.59 ± 0.56 | <0.001 |
| S E/e′ (m/s) | 8.69 ± 0.97 | 7.08 ± 1.48 | <0.001 |
| L E/e′ (m/s) | 10.20 ± 0.98 | 7.99 ± 1.05 | <0.001 |
| IVRT (ms) | 69.54 ± 11.62 | 99.46 ± 16.33 | <0.001 |
| PV AR (m/s) | 33.37 ± 7.88 | 28.14 ± 5.79 | <0.001 |
| PV S/D (m/s) | 0.59 ± 0.19 | 0.96 ± 0.33 | <0.001 |
| GLS (%) | −15.91 ±0.20 | −14.58 ±1.01 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monisha, K.; Mahema, S.; Chokkalingam, M.; Ahmad, S.F.; Emran, T.B.; Prabu, P.; Ahmed, S.S.S.J. Elucidating the Histone Deacetylase Gene Expression Signatures in Peripheral Blood Mononuclear Cells That Correlate Essential Cardiac Function and Aid in Classifying Coronary Artery Disease through a Logistic Regression Model. Biomedicines 2023, 11, 2952. https://doi.org/10.3390/biomedicines11112952
Monisha K, Mahema S, Chokkalingam M, Ahmad SF, Emran TB, Prabu P, Ahmed SSSJ. Elucidating the Histone Deacetylase Gene Expression Signatures in Peripheral Blood Mononuclear Cells That Correlate Essential Cardiac Function and Aid in Classifying Coronary Artery Disease through a Logistic Regression Model. Biomedicines. 2023; 11(11):2952. https://doi.org/10.3390/biomedicines11112952
Chicago/Turabian StyleMonisha, K., S. Mahema, M. Chokkalingam, Sheikh F. Ahmad, Talha Bin Emran, Paramasivam Prabu, and Shiek S. S. J. Ahmed. 2023. "Elucidating the Histone Deacetylase Gene Expression Signatures in Peripheral Blood Mononuclear Cells That Correlate Essential Cardiac Function and Aid in Classifying Coronary Artery Disease through a Logistic Regression Model" Biomedicines 11, no. 11: 2952. https://doi.org/10.3390/biomedicines11112952
APA StyleMonisha, K., Mahema, S., Chokkalingam, M., Ahmad, S. F., Emran, T. B., Prabu, P., & Ahmed, S. S. S. J. (2023). Elucidating the Histone Deacetylase Gene Expression Signatures in Peripheral Blood Mononuclear Cells That Correlate Essential Cardiac Function and Aid in Classifying Coronary Artery Disease through a Logistic Regression Model. Biomedicines, 11(11), 2952. https://doi.org/10.3390/biomedicines11112952