Variational Approach for Joint Kidney Segmentation and Registration from DCE-MRI Using Fuzzy Clustering with Shape Priors
Abstract
1. Introduction
2. Related Work
3. Methods
3.1. Problem Statement and Notations
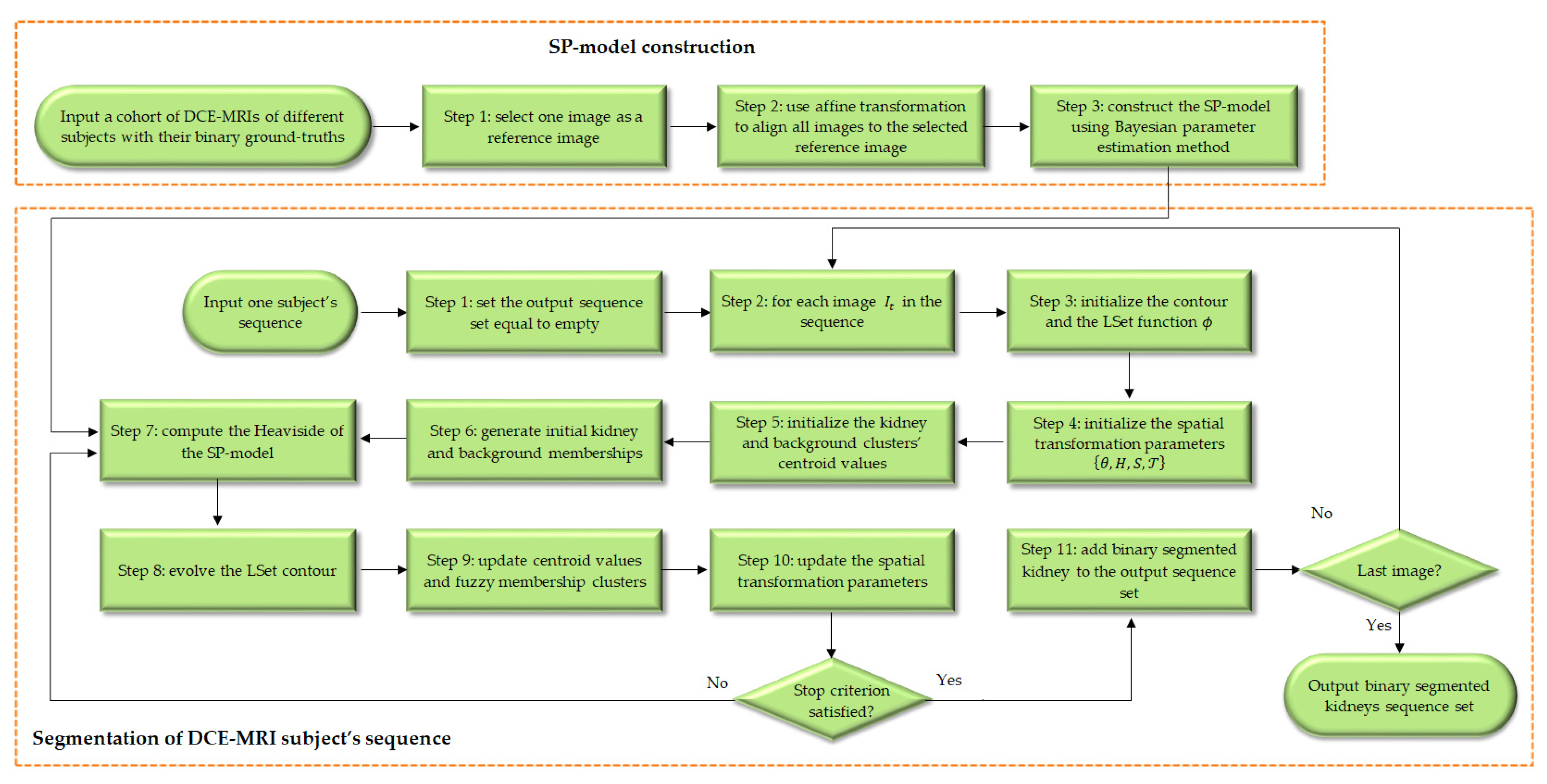
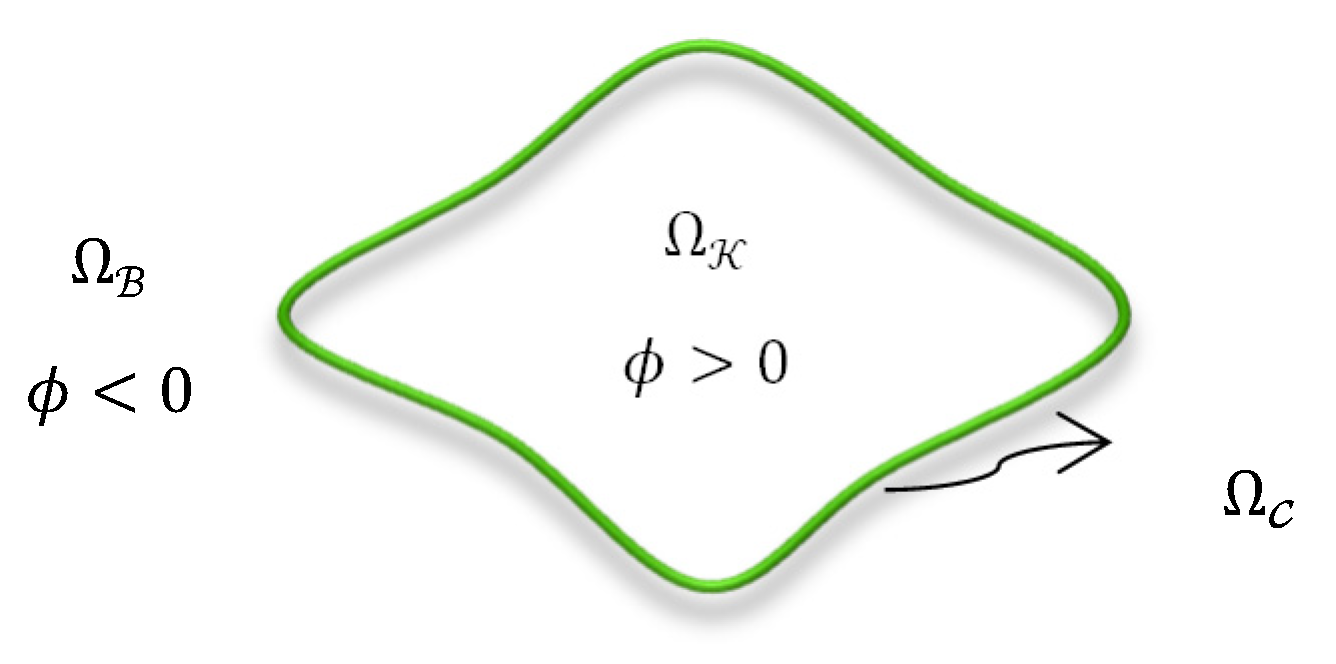
3.2. Proposed Variational Approach
3.3. Proposed Energy Functional
3.4. FCMC Membership Function
3.5. Statistical Shape Prior Model
3.6. Affine-Based Registration for the Shape Prior Model
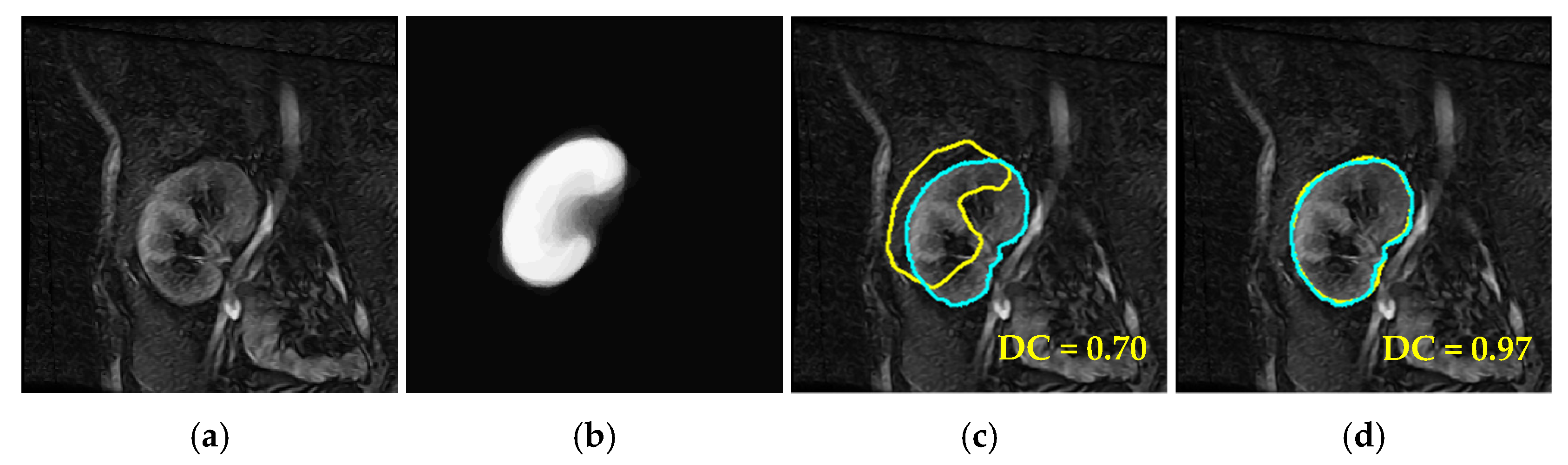
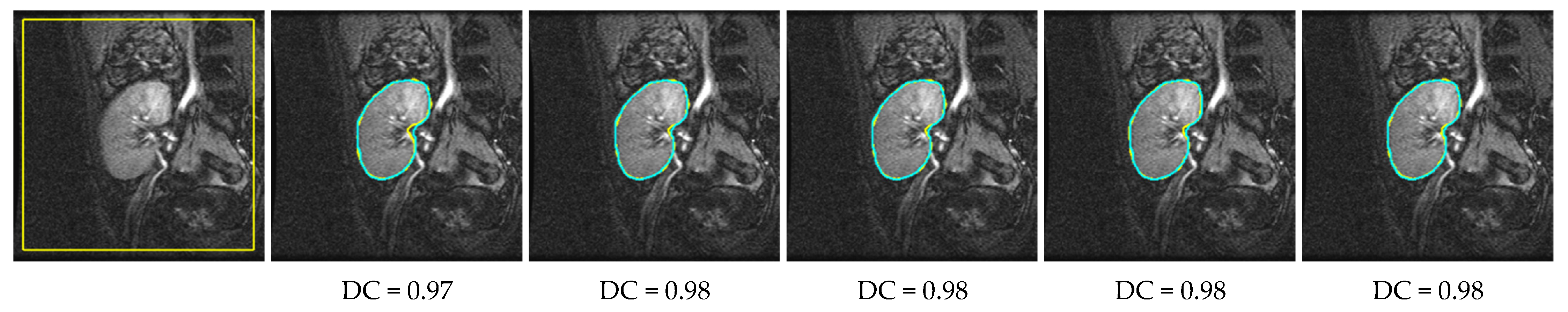
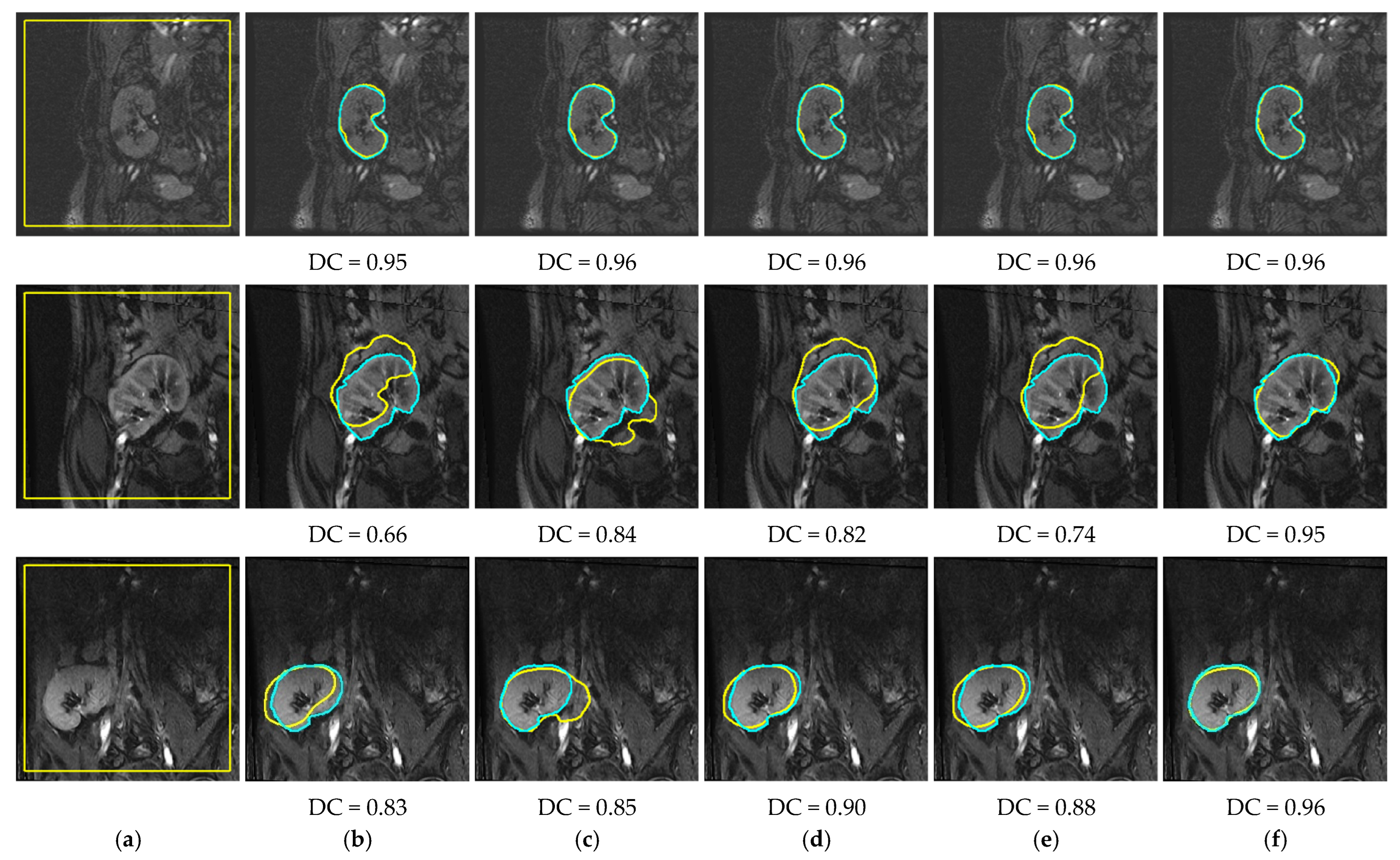
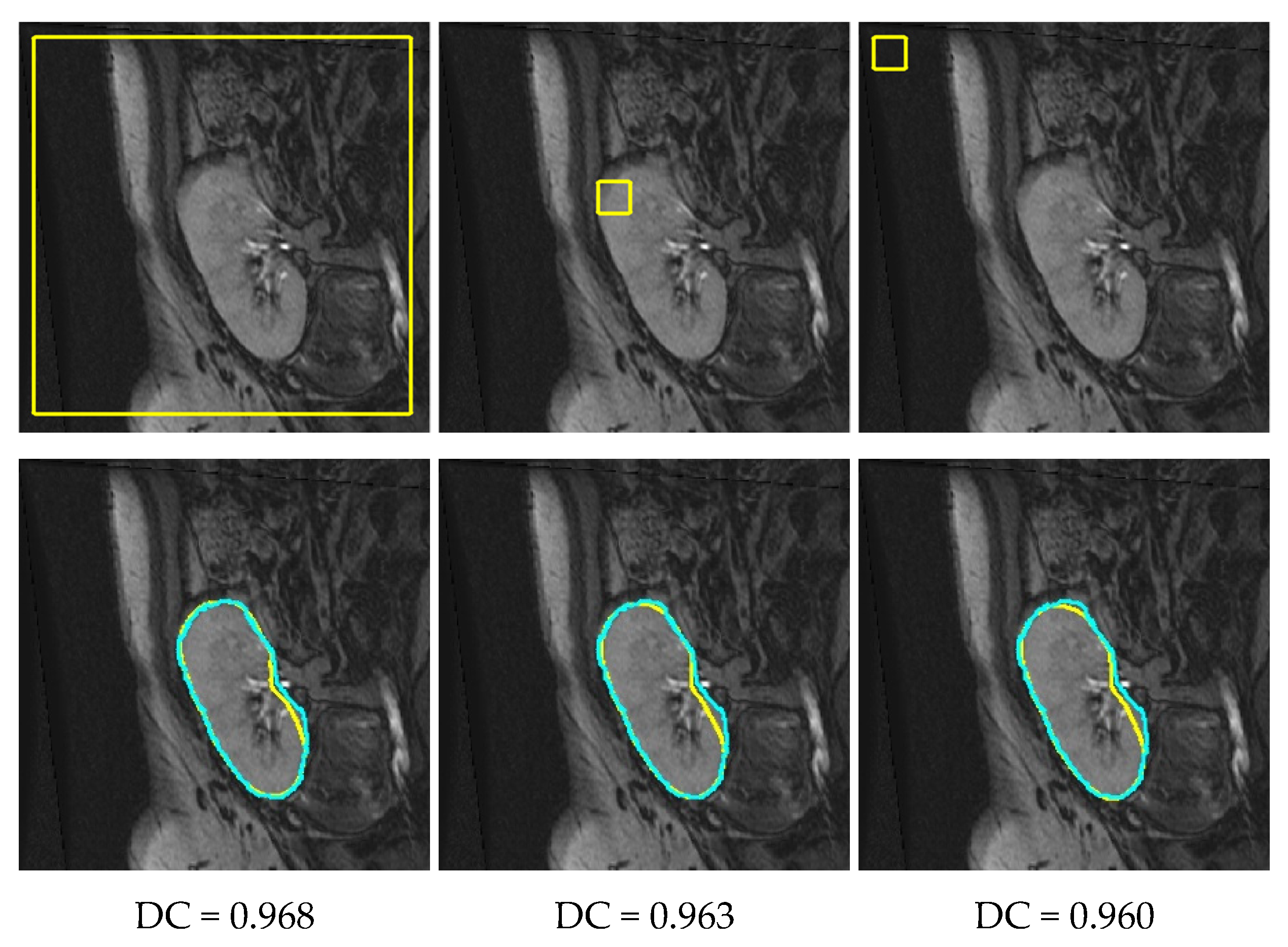
4. Results
4.1. Data
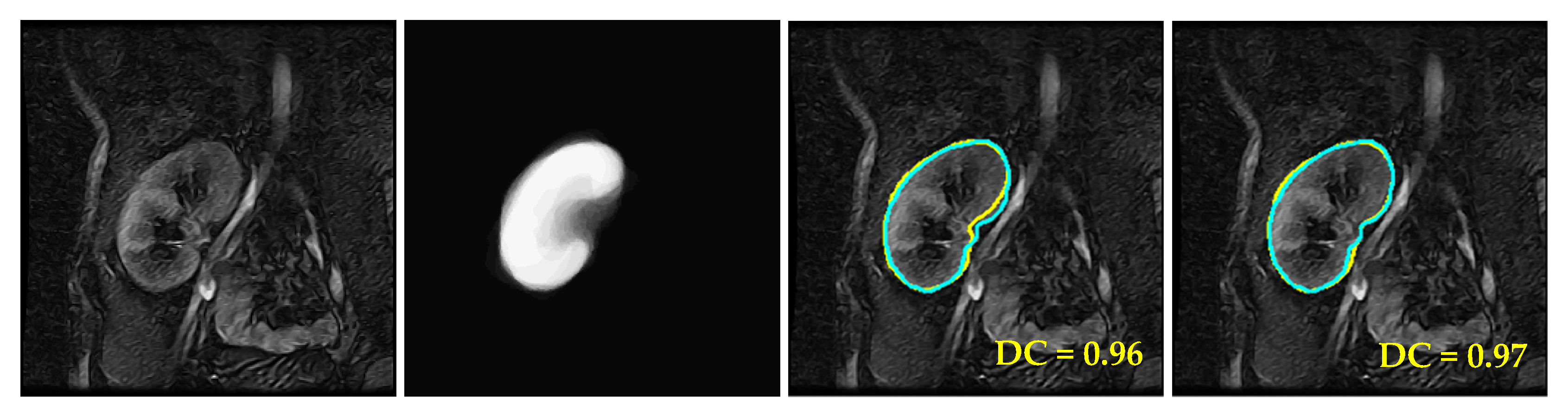
4.2. Comparison with Other LSet-Based Methods
4.3. Comparison with UNet-Based Convolutional Neural Networks
5. Conclusions
- It can be considered as the first approach in the literature to achieve accurate kidney segmentation and registration at the same time.
- It embeds FCM clustering within an LSet method in one variational approach; the membership degrees of the image pixels are updated during the LSet evolution process considering pixels’ intensities directly as well as prior shape probabilities. This promotes our approach’s performance.
- It can automatically manipulate the misalignment between the kidney in the input image and the SP-model.
- Thanks to employing smeared-out Heaviside and Dirac delta functions in the LSet method, the approach is able to accurately segment the kidney from the image regardless of where the contour has been initialized.
- It embraces an efficient statistical Bayesian parameter estimation method for SP-model construction, which can better address the cases of unobserved kidney pixels in the images while building the model.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shehata, M.; Alksas, A.; Abouelkheir, R.; Elmahdy, A.; Shaffie, A.; Soliman, A.; Ghazal, M.; Abu Khalifeh, H.; Salim, R.; Razek, A.A.; et al. A Comprehensive Computer-Assisted Diagnosis System for Early Assessment of Renal Cancer Tumors. Sensors 2021, 21, 4928. [Google Scholar] [CrossRef] [PubMed]
- Alnazer, I.; Bourdon, P.; Urruty, T.; Falou, O.; Khalil, M.; Shahin, A.; Fernandez-Maloigne, C. Recent advances in medical image processing for the evaluation of chronic kidney disease. Med. Image Anal. 2021, 69, 101960. [Google Scholar] [CrossRef] [PubMed]
- Sobrinho, A.; Queiroz, A.C.M.D.S.; Da Silva, L.D.; Costa, E.D.B.; Pinheiro, M.E.; Perkusich, A. Computer-Aided Diagnosis of Chronic Kidney Disease in Developing Countries: A Comparative Analysis of Machine Learning Techniques. IEEE Access 2020, 8, 25407–25419. [Google Scholar] [CrossRef]
- Malakar, S.; Roy, S.D.; Das, S.; Sen, S.; Velásquez, J.D.; Sarkar, R. Computer Based Diagnosis of Some Chronic Diseases: A Medical Journey of the Last Two Decades. Arch. Comput. Methods Eng. 2022, 29, 5525–5567. [Google Scholar] [CrossRef]
- Mostapha, M.; Khalifa, F.; Alansary, A.; Soliman, A.; Suri, J.; El-Baz, A.S. Computer-Aided Diagnosis Systems for Acute Renal Transplant Rejection: Challenges and Methodologies. In Abdomen and Thoracic Imaging; Springer: Boston, MA, USA, 2014; pp. 1–35. [Google Scholar] [CrossRef]
- Zollner, F.G.; Kocinski, M.; Hansen, L.; Golla, A.-K.; Trbalic, A.S.; Lundervold, A.; Materka, A.; Rogelj, P. Kidney Segmentation in Renal Magnetic Resonance Imaging—Current Status and Prospects. IEEE Access 2021, 9, 71577–71605. [Google Scholar] [CrossRef]
- Tsai, A.; Yezzi, A.; Wells, W.; Tempany, C.; Tucker, D.; Fan, A.; Grimson, W.; Willsky, A. A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans. Med Imaging 2003, 22, 137–154. [Google Scholar] [CrossRef]
- El Munim, H.E.A.; Farag, A.A. Curve/Surface Representation and Evolution Using Vector Level Sets with Application to the Shape-Based Segmentation Problem. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 945–958. [Google Scholar] [CrossRef]
- Yuksel, S.E.; El-Baz, A.; Farag, A.A.; El-Ghar, M.A.; Eldiasty, T.; Ghoneim, M.A. A Kidney Segmentation Framework for Dynamic Contrast Enhanced Magnetic Resonance Imaging. J. Vib. Control. 2007, 13, 1505–1516. [Google Scholar] [CrossRef]
- Eltanboly, A.; Ghazal, M.; Hajjdiab, H.; Shalaby, A.; Switala, A.; Mahmoud, A.; Sahoo, P.; El-Azab, M.; El-Baz, A. Level sets-based image segmentation approach using statistical shape priors. Appl. Math. Comput. 2018, 340, 164–179. [Google Scholar] [CrossRef]
- El-Melegy, M.; El-Karim, R.A.; El-Baz, A.; El-Ghar, M.A. Fuzzy Membership-Driven Level Set for Automatic Kidney Segmentation from DCE-MRI. In Proceedings of the IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–8. [Google Scholar] [CrossRef]
- El-Melegy, M.T.; El-Karim, R.M.A.; El-Baz, A.S.; El-Ghar, M.A. A Combined Fuzzy C-Means and Level Set Method for Automatic DCE-MRI Kidney Segmentation Using Both Population-Based and Patient-Specific Shape Statistics. In Proceedings of the IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar] [CrossRef]
- El-Melegy, M.; Kamel, R.; El-Ghar, M.A.; Alghamdi, N.S.; El-Baz, A. Level-Set-Based Kidney Segmentation from DCE-MRI Using Fuzzy Clustering with Population-Based and Subject-Specific Shape Statistics. Bioengineering 2022, 9, 654. [Google Scholar] [CrossRef]
- Khalifa, F.; El-Baz, A.; Gimel'Farb, G.; El-Ghar, M.A. Non-invasive Image-Based Approach for Early Detection of Acute Renal Rejection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Beijing, China, 20–24 September 2010; pp. 10–18. [Google Scholar] [CrossRef]
- Khalifa, F.; Beache, G.M.; El-Ghar, M.A.; El-Diasty, T.; Gimel'Farb, G.; Kong, M.; El-Baz, A. Dynamic Contrast-Enhanced MRI-Based Early Detection of Acute Renal Transplant Rejection. IEEE Trans. Med Imaging 2013, 32, 1910–1927. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Soliman, A.; Gimel'Farb, G.; El-Baz, A. Segmenting Kidney DCE-MRI Using 1st-Order Shape and 5th-Order Appearance Priors. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland; pp. 77–84. [Google Scholar] [CrossRef]
- El-Melegy, M.; Kamel, R.; El-Ghar, M.A.; Shehata, M.; Khalifa, F.; El-Baz, A. Kidney segmentation from DCE-MRI converging level set methods, fuzzy clustering and Markov random field modeling. Sci. Rep. 2022, 12, 18816. [Google Scholar] [CrossRef] [PubMed]
- Hodneland, E.; Hanson, E.A.; Lundervold, A.; Modersitzki, J.; Eikefjord, E.; Munthe-Kaas, A.Z. Segmentation-Driven Image Registration-Application to 4D DCE-MRI Recordings of the Moving Kidneys. IEEE Trans. Image Process. 2014, 23, 2392–2404. [Google Scholar] [CrossRef]
- Li, S.; Zöllner, F.G.; Merrem, A.D.; Peng, Y.; Roervik, J.; Lundervold, A.; Schad, L.R. Wavelet-based segmentation of renal compartments in DCE-MRI of human kidney: Initial results in patients and healthy volunteers. Comput. Med. Imaging Graph. 2012, 36, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Le Minh, H.; Cheng, K.T.T.; Sung, K.H.; Liu, W. Renal compartment segmentation in DCE-MRI images. Med. Image Anal. 2016, 32, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Yoruk, U.; Hargreaves, B.A.; Vasanawala, S.S. Automatic renal segmentation for MR urography using 3D-GrabCut and random forests. Magn. Reson. Med. 2017, 79, 1696–1707. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamasneh, A.R.; Jalab, H.A.; Palaiahnakote, S.; Obaidellah, U.H.; Ibrahim, R.W.; El-Melegy, M.T. A New Local Fractional Entropy-Based Model for Kidney MRI Image Enhancement. Entropy 2018, 20, 344. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamasneh, A.R.; Jalab, H.A.; Shivakumara, P.; Ibrahim, R.W.; Obaidellah, U.H. Kidney segmentation in MR images using active contour model driven by fractional-based energy minimization. Signal Image Video Process. 2020, 14, 1361–1368. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Rørvik, J.; Lundervold, A. Fast semi-supervised segmentation of the kidneys in DCE-MRI using convolu-tional neural networks and transfer learning. In Proceedings of the 2nd International Scientific Symposium, Functional Renal Imaging: Where Physiology, Nephrology, Radiology and Physics Meet, Berlin, Germany, 11–13 October 2017. [Google Scholar]
- Haghighi, M.; Warfield, S.K.; Kurugol, S. Automatic renal segmentation in DCE-MRI using convolutional neural networks. In Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI), Washington, DC, USA, 4–7 April 2018; pp. 1534–1537. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland; pp. 234–241. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Brunetti, A.; Cascarano, G.D.; Guerriero, A.; Pesce, F.; Moschetta, M.; Gesualdo, L. A comparison between two semantic deep learning frameworks for the autosomal dominant polycystic kidney disease segmentation based on magnetic resonance images. BMC Med Inform. Decis. Mak. 2019, 19, 244. [Google Scholar] [CrossRef]
- Brunetti, A.; Cascarano, G.D.; De Feudis, I.; Moschetta, M.; Gesualdo, L.; Bevilacqua, V. Detection and Segmentation of Kidneys from Magnetic Resonance Images in Patients with Autosomal Dominant Polycystic Kidney Disease. In Proceedings of the International Conference on Intelligent Computing, Nanchang, China, 3–6 August 2019; Springer: Cham, Switzerland, 2019; pp. 639–650. [Google Scholar] [CrossRef]
- Milecki, L.; Bodard, S.; Correas, J.-M.; Timsit, M.-O.; Vakalopoulou, M. 3d Unsupervised Kidney Graft Segmentation Based on Deep Learning and Multi-Sequence Mri. In Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI), Virtual, 13–16 April 2021; pp. 1781–1785. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2020, 18, 203–211. [Google Scholar] [CrossRef]
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.-H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS Challenge—combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2020, 69, 101950. [Google Scholar] [CrossRef] [PubMed]
- El-Melegy, M.; Mokhtar, H. Fuzzy framework for joint segmentation and registration of brain MRI with prior information. In Proceedings of the International Conference on Computer Engineering & Systems, Cairo, Egypt, 30 November–2 December 2010; pp. 9–14. [Google Scholar] [CrossRef]
- Nayak, J.; Naik, B.; Behera, H.S. Fuzzy C-Means (FCM) Clustering Algorithm: A Decade Review from 2000 to 2014. In Proceedings of the International Conference on CIDM 2015: IEEE Symposium on Computational Intelligence and Data Mining, Cape Town, South Africa, 7–12 December 2015; pp. 133–149. [Google Scholar] [CrossRef]
- Osher, S.; Fedkiw, R.; Piechor, K. Level Set Methods and Dynamic Implicit Surfaces. Appl. Mech. Rev. 2004, 57, B15. [Google Scholar] [CrossRef]
- Friedman, N.; Singer, Y. Efficient Bayesian parameter estimation in large discrete domains. In Proceedings of the 11th International Conference on Advances in Neural Information Processing Systems (NIPS'98); MIT Press: Cambridge, MA, USA, 1999; pp. 417–423. [Google Scholar]
- Nai, Y.-H.; Teo, B.W.; Tan, N.L.; O'Doherty, S.; Stephenson, M.C.; Thian, Y.L.; Chiong, E.; Reilhac, A. Comparison of metrics for the evaluation of medical segmentations using prostate MRI dataset. Comput. Biol. Med. 2021, 134, 104497. [Google Scholar] [CrossRef] [PubMed]
- Reinke, A.; Eisenmann, M.; Tizabi, M.D.; Sudre, C.H.; Rädsch, T.; Antonelli, M.; Arbel, T.; Bakas, S.; Cardoso, M.J.; Cheplygina, V.; et al. Common limitations of image processing metrics: A picture story. arXiv 2021, arXiv:2104.05642. [Google Scholar] [CrossRef]
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Bi-Directional ConvLSTM U-Net with Densley Connected Convolutions. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW), Seoul, Republic of Korea, 27–28 October 2019; pp. 1–10. [Google Scholar] [CrossRef]
- Heller, N.; Sathianathen, N.; Kalapara, A.; Walczak, E.; Moore, K.; Kaluzniak, H.; Rosenberg, J.; Blake, P.; Rengel, Z.; Oestreich, M.; et al. The kits19 challenge data: 300 kidney tumor cases with clinical context, CT semantic segmentations, and surgical outcomes. arXiv 2019, arXiv:1904.00445. [Google Scholar] [CrossRef]
- Viola, P.; Iii, W.M.W. Alignment by Maximization of Mutual Information. Int. J. Comput. Vis. 1997, 24, 137–154. [Google Scholar] [CrossRef]
- El-Melegy, M.T.; Mokhtar, H.M. Tumor segmentation in brain MRI using a fuzzy approach with class center priors. EURASIP J. Image Video Process. 2014, 2014, 21. [Google Scholar] [CrossRef]
- Heller, K.A.; Svore, K.M.; Keromytis, A.D.; Stolfo, S.J. One class support vector machines for detecting anomalous windows registry accesses. In Proceedings of the ICDM Workshop on Data Mining for Computer Security, Melbourne, FL, USA, 19 November 2003. [Google Scholar] [CrossRef]
- Shehanaz, S.; Daniel, E.; Guntur, S.R.; Satrasupalli, S. Optimum weighted multimodal medical image fusion using particle swarm optimization. Optik 2021, 231, 166413. [Google Scholar] [CrossRef]



| Method | All DCE-MRIs | Affine-Transformed DCE-MRIs | ||||
|---|---|---|---|---|---|---|
| DC | IoU | 95HD | DC | IoU | 95HD | |
| FCMLS [11] | 0.88 ± 0.10 | 0.79 ± 0.17 | 5.07 ± 7.65 | 0.83 ± 0.10 | 0.72 ± 0.14 | 8.35 ± 7.55 |
| PBPSFL [12] | 0.92 ± 0.06 | 0.87 ± 0.08 | 3.29 ± 5.65 | 0.90 ± 0.07 | 0.83 ± 0.09 | 5.4 ± 7.18 |
| PSFL [13] | 0.91 ± 0.06 | 0.84 ± 0.10 | 3.84 ± 4.56 | 0.87 ± 0.07 | 0.77 ± 0.11 | 6.57 ± 5.03 |
| FML [17] | 0.90 ± 0.08 | 0.83 ± 0.16 | 4.41 ± 6.4 | 0.87 ± 0.08 | 0.76 ± 0.12 | 7.3 ± 5.45 |
| Proposed | 0.94 ± 0.03 | 0.89 ± 0.05 | 2.2 ± 2.32 | 0.93 ± 0.05 | 0.88 ± 0.06 | 2.5 ± 2.7 |
| Method | All DCE-MRIs | Affine-Transformed DCE-MRIs | ||||
|---|---|---|---|---|---|---|
| DC | IoU | 95HD | DC | IoU | 95HD | |
| UNet [26] | 0.943 ± 0.04 | 0.90 ± 0.06 | 5.6 ± 15.7 | 0.946 ± 0.05 | 0.90 ± 0.05 | 3.5 ± 11.5 |
| BCD-UNet [38] | 0.942 ± 0.037 | 0.89 ± 0.06 | 4.4 ± 11.5 | 0.942 ± 0.035 | 0.89 ± 0.05 | 3.9 ± 10.3 |
| Proposed | 0.95 ± 0.02 | 0.92 ± 0.03 | 1.0 ± 1.2 | 0.95 ± 0.025 | 0.91 ± 0.02 | 1.9 ± 2.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Melegy, M.; Kamel, R.; Abou El-Ghar, M.; Alghamdi, N.S.; El-Baz, A. Variational Approach for Joint Kidney Segmentation and Registration from DCE-MRI Using Fuzzy Clustering with Shape Priors. Biomedicines 2023, 11, 6. https://doi.org/10.3390/biomedicines11010006
El-Melegy M, Kamel R, Abou El-Ghar M, Alghamdi NS, El-Baz A. Variational Approach for Joint Kidney Segmentation and Registration from DCE-MRI Using Fuzzy Clustering with Shape Priors. Biomedicines. 2023; 11(1):6. https://doi.org/10.3390/biomedicines11010006
Chicago/Turabian StyleEl-Melegy, Moumen, Rasha Kamel, Mohamed Abou El-Ghar, Norah S. Alghamdi, and Ayman El-Baz. 2023. "Variational Approach for Joint Kidney Segmentation and Registration from DCE-MRI Using Fuzzy Clustering with Shape Priors" Biomedicines 11, no. 1: 6. https://doi.org/10.3390/biomedicines11010006
APA StyleEl-Melegy, M., Kamel, R., Abou El-Ghar, M., Alghamdi, N. S., & El-Baz, A. (2023). Variational Approach for Joint Kidney Segmentation and Registration from DCE-MRI Using Fuzzy Clustering with Shape Priors. Biomedicines, 11(1), 6. https://doi.org/10.3390/biomedicines11010006






