Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. Microbiota Analysis
2.3. Real Time PCR
2.4. ELISA
2.5. Reactive Oxygen Species (ROS) and Nitric Oxide Synthase (NOS) Detection
2.6. Statistical Analysis
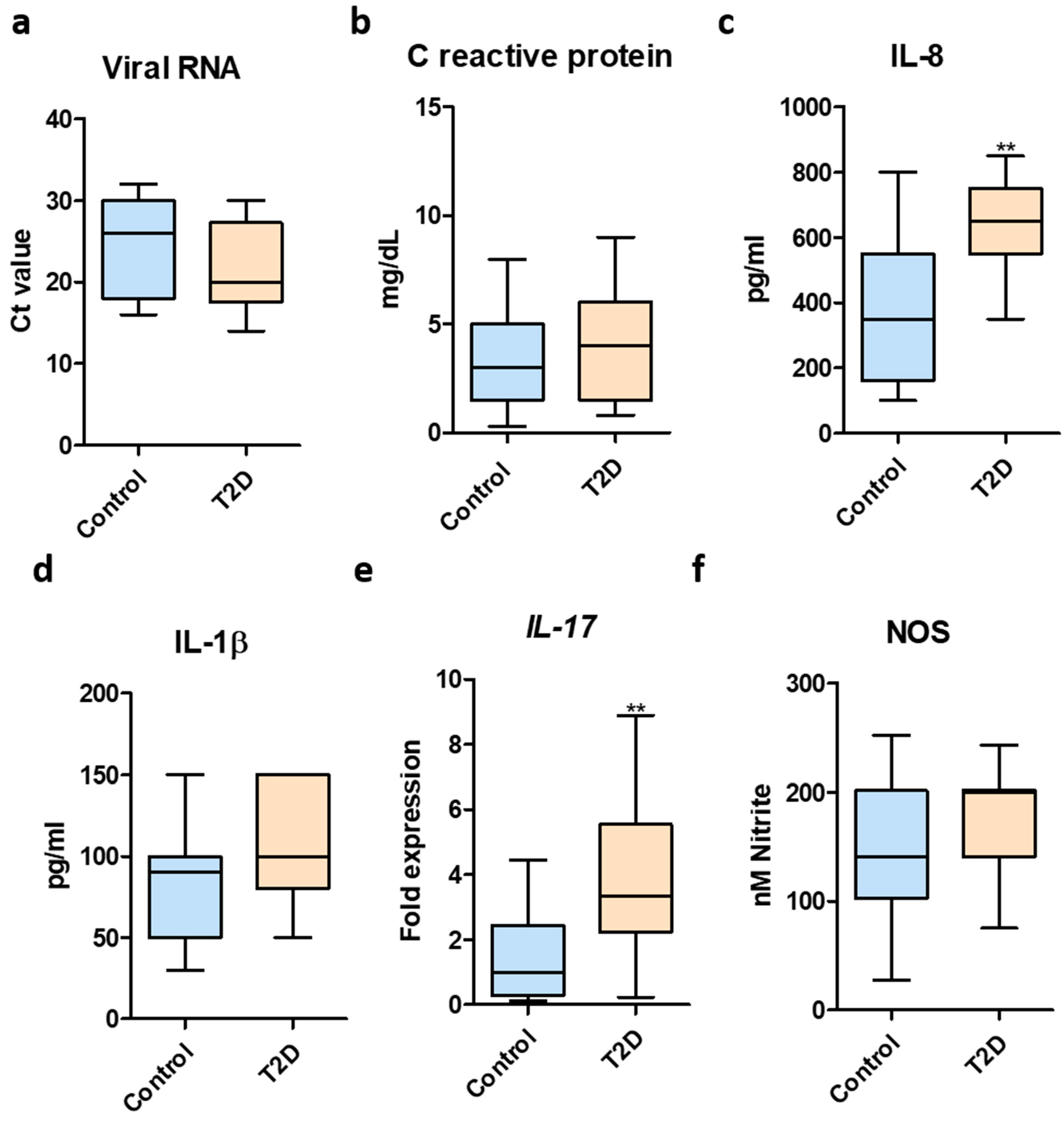
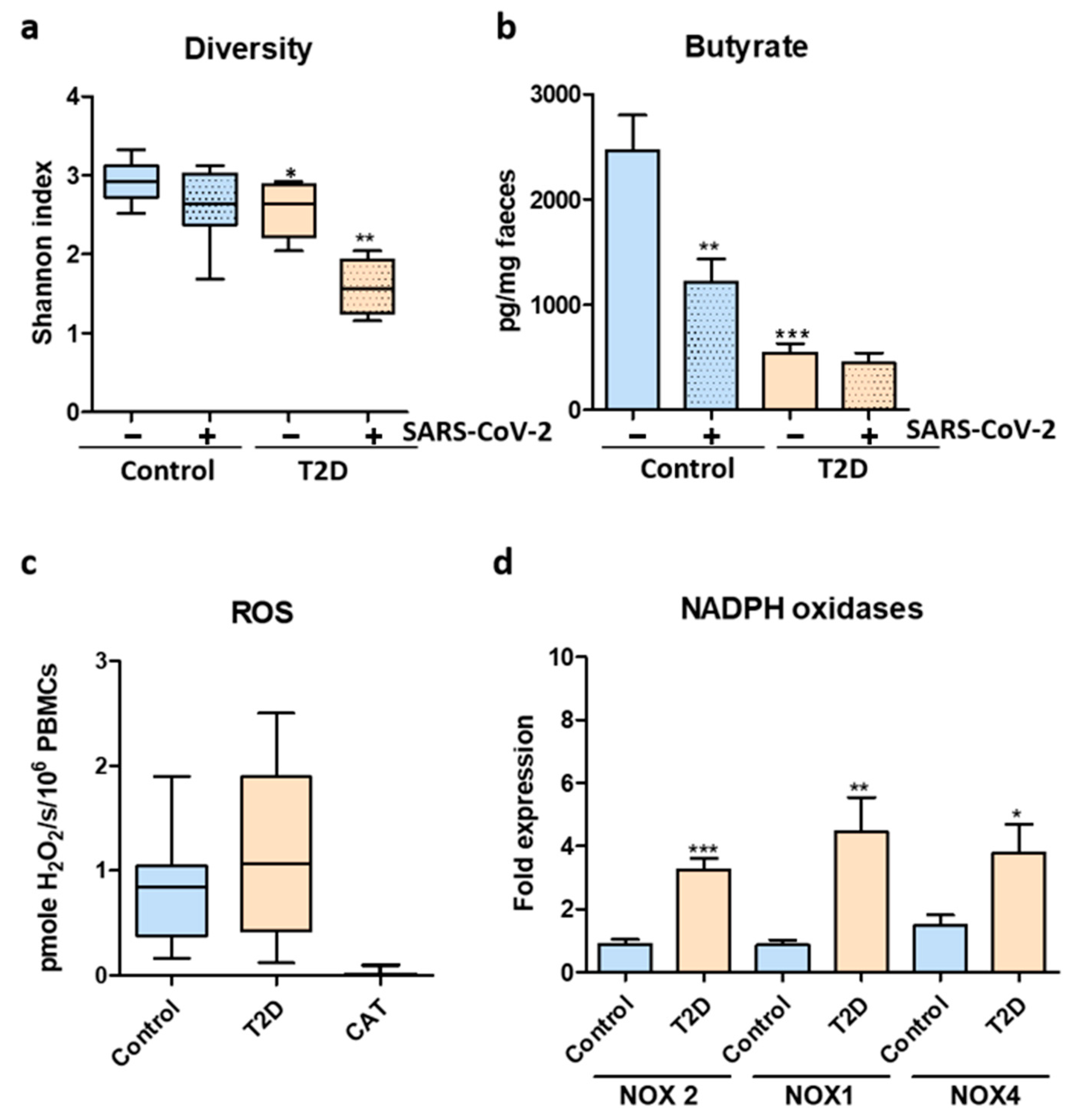
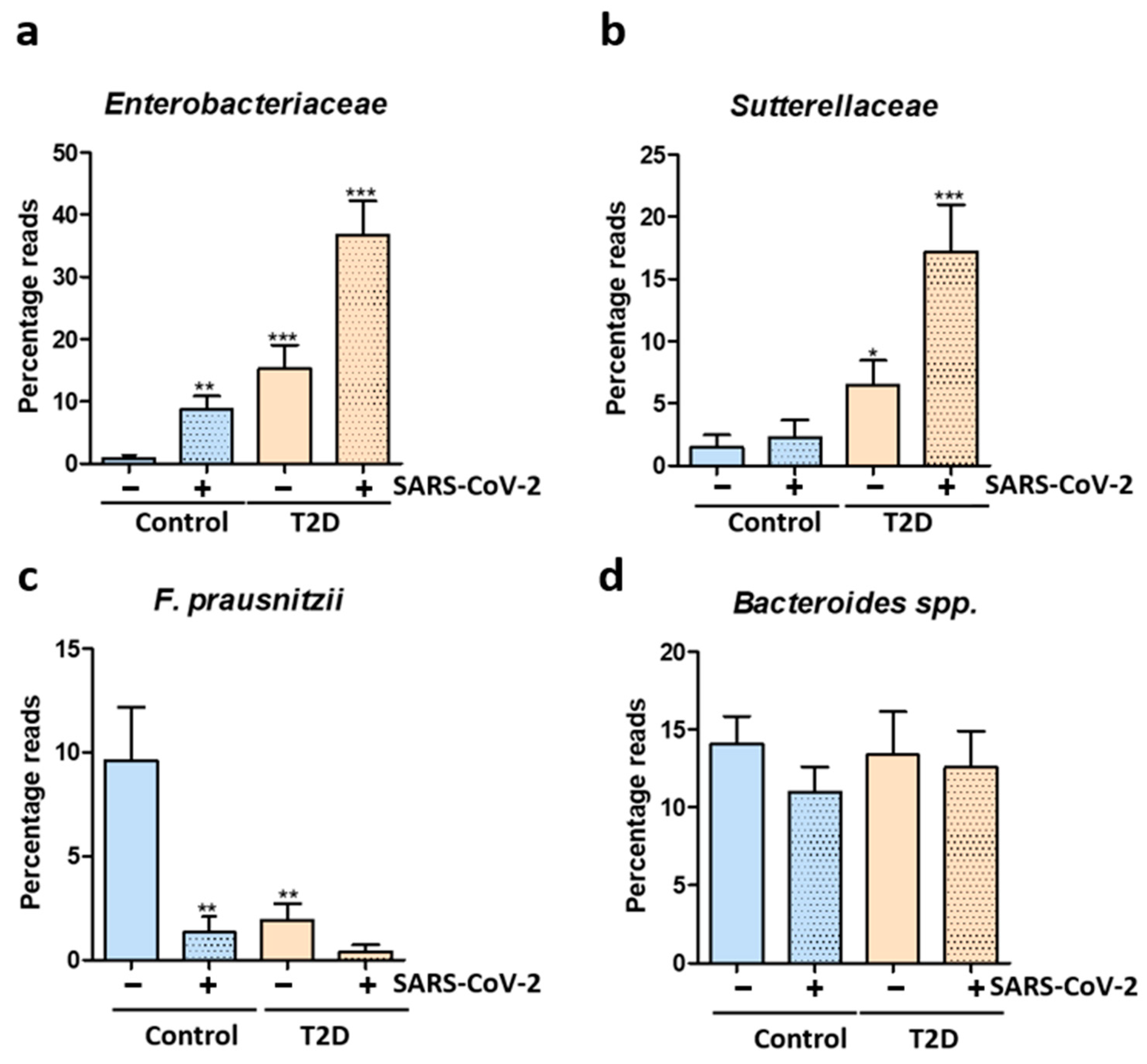
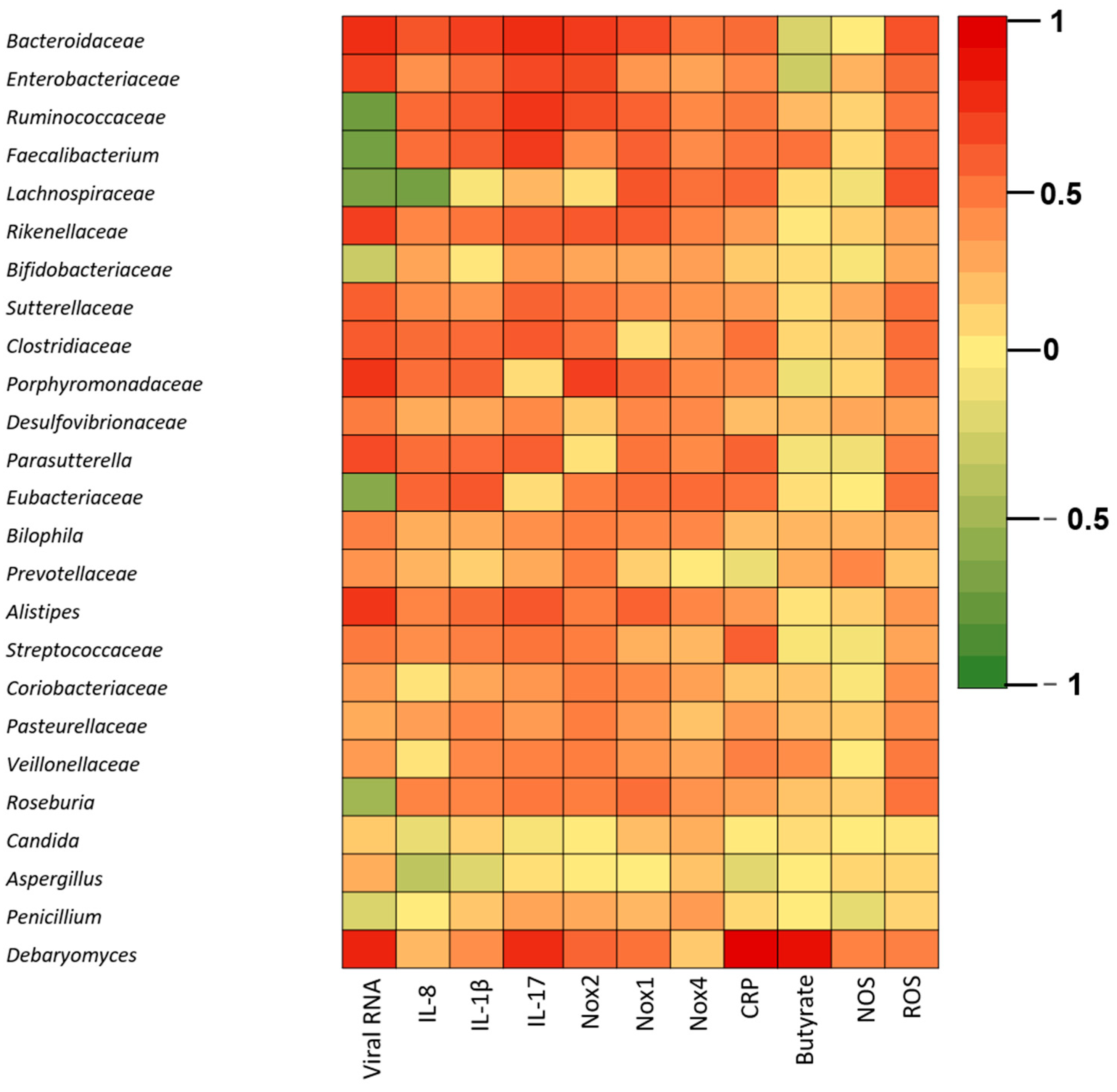
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fung, M.; Babik, J.M. COVID-19 in Immunocompromised Hosts: What We Know So Far. Clin. Infect. Dis. 2021, 72, 340–350. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 1253, 20–21. [Google Scholar] [CrossRef]
- Chan, J.F.; Yuan, S.; Kok, K.H.; To, K.K.; Chu, H.; Yang, J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Lau, R.I.; Zhang, F.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Tang, X.; Jiang, X.; Yan, X.; Liu, X.; Gong, J.; Mew, K.; Sun, H.; Chen, X.; et al. Distinct metagenomic signatures in the SARS-CoV-2 infection. Front. Cell. Infect. Microbiol. 2021, 11, 706970. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.Y.; et al. Prolonged impairment of short-chain fatty acid and L-isoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology 2022, 162, 548–561. [Google Scholar] [CrossRef]
- Vestad, B.; Ueland, T.; Lerum, T.V.; Dahl, T.B.; Holm, K.; Barratt-Due, A.; Kåsine, T.; Dyrhol-Riise, A.M.; Stiksrud, B.; Tonby, K.; et al. Respiratory dysfunction three months after severe COVID-19 is associated with gut microbiota alterations. J. Intern. Med. 2022, 291, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, Y.; He, Y.; Liu, X.; Liu, M.; Tang, Y.; Li, X.; Yang, G.; Liang, G.; Xu, S.; et al. Age-Related Risk Factors and Complications of Patients With COVID-19: A Population-Based Retrospective Study. Front. Med. 2022, 8, 757459. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chi, J.; Lv, W.; Wang, Y. Obesity and diabetes as high-risk factors for severe coronavirus disease 2019 (COVID-19). Diabetes Metab. Res. Rev. 2021, 37, e3377. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Darif, D.; Hammi, I.; Kihel, A.; El Idrissi Saik, I.; Guessous, F.; Akarid, K. The proinflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Gaulke, C.A.; Sharpton, T.J. The influence of ethnicity and geography on human gut microbiome composition. Nat. Med. 2018, 24, 1495–1496. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; D’Amico, F.; Bartoletti, M.; Lombardo, D.; Rampelli, S.; Fornaro, G.; Coladonato, S.; Siniscalchi, A.; Re, M.C.; Viale, P.; et al. The gut microbiota of critically ill patients with COVID-19. Front. Cell. Infect. Microbiol. 2021, 11, 670424. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, H.; Cui, G.; Lu, H.; Wang, L.; Luo, H.; Chen, X.; Ren, H.; Sun, R.; Liu, W.; et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 2021, 70, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Lu, R.; Zhang, T.; Wu, Q.; Cai, W.; Han, X.; Wan, Z.; Jin, X.; Zhang, Z.; Zhang, C. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun. Biol. 2021, 4, 240. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Ishizaka, A.; Koga, M.; Ikeuchi, K.; Saito, M.; Adachi, E.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Yasuhara, A.; Kiyono, H.; et al. Correlation analysis between gut microbiota alterations and the cytokine response in patients with coronavirus disease during hospitalization. Microbiol. Spectr. 2022, 10, e0168921. [Google Scholar] [CrossRef] [PubMed]
- Rafiqul Islam, S.M.; Foysal, M.J.; Hoque, M.N.; Mehedi, H.M.; Rob, M.A.; Salauddin, A.; Tanzina, A.Y.; Biswas, S.; Noyon, S.H.; Siddiki, A.M.A.M.Z.; et al. Dysbiosis of oral and gut microbiomes in SARS-CoV-2 infected patients in Bangladesh: Elucidating the role of opportunistic gut microbes. Front. Med. 2022, 9, 163. [Google Scholar] [CrossRef]
- Reinold, J.; Farahpour, F.; Fehring, C.; Dolff, S.; Konik, M.; Korth, J.; van Baal, L.; Hoffmann, D.; Buer, J.; Witzke, O.; et al. A proinflammatory gut microbiome characterizes SARS-CoV-2 infected patients and a reduction in the connectivity of an anti-inflammatory bacterial network associates with severe COVID-19. Front. Cell. Infect. Microbiol. 2021, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Gu, S.; Gong, Y.; Li, B.; Lu, H.; Li, Q.; Zhang, R.; Gao, X.; Wu, Z.; Zhang, J.; et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering 2020, 6, 1178–1184. [Google Scholar] [CrossRef]
- Tao, W.; Zhang, G.; Wang, X.; Guo, M.; Zeng, W.; Xu, Z.; Cao, D.; Pan, A.; Wang, Y.; Zhang, K.; et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020, 5, 100023. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, X.; Jiang, G.; Tang, H.; Ming, S.; Tang, L.; Lu, J.; Guo, C.; Shan, H.; Huang, X.; et al. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes 2021, 7, 61. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.; Zhang, F.; Liu, Q.; Li, A.Y.L.; Chung, A.C.K.; Cheung, C.P.; Tso, E.Y.K.; Fung, K.S.C.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Li, S.; Yang, S.; Zhou, Y.; Disoma, C.; Dong, Z.; Du, A.; Zhang, Y.; Chen, Y.; Huang, W.; Chen, J.; et al. Microbiome profiling using shotgun metagenomic sequencing identified unique microorganisms in COVID-19 patients with altered gut microbiota. Front. Microbiol. 2021, 12, 712081. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Gopinathan, S.; Sharma, A. Interplay between severities of COVID-19 and the gut microbiome: Implications of bacterial co-infections? Gut Pathog. 2021, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.R.; Daneman, N. Bacterial co-infection and secondary infection in patients with COVID-19: A living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M.; et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 2021, 27, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Rosário, A.; Marques, C.; Pinheiro, H.; Araújo, J.R.; Ribeiro, P.; Rocha, R.; Mota, I.; Pestana, D.; Ribeiro, R.; Pereira, A.; et al. Gut microbiota diversity and C-reactive protein are predictors of disease severity in COVID-19 patients. Front. Microbiol. 2021, 12, 1820. [Google Scholar] [CrossRef]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Fei, W.; Ye, Y.; Zhao, M.; Zheng, C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 2020, 62, 1–12. [Google Scholar] [CrossRef]
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.K.; Lui, G.C.Y.; Chen, N.; Li, A.; Lu, W.; Chan, F.K.L.; et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology 2020, 159, 1302–1310. [Google Scholar] [CrossRef]
- Lv, L.; Gu, S.; Jiang, H.; Yan, R.; Chen, Y.; Chen, Y.; Luo, R.; Huang, C.; Lu, H.; Zheng, B.; et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun. Biol. 2021, 4, 480. [Google Scholar] [CrossRef]
- Roudbary, M.; Kumar, S.; Kumar, A.; Černáková, L.; Nikoomanesh, F.; Rodrigues, C.F. Overview on the prevalence of fungal infections, immune response, and microbiome role in COVID-19 patients. J. Fungi. 2021, 7, 720. [Google Scholar] [CrossRef]
- Moon, S.J.; Rhee, E.J.; Jung, J.H.; Han, K.D.; Kim, S.R.; Lee, W.Y.; Yoon, K.H. Independent impact of diabetes on the severity of coronavirus disease 2019 in 5,307 patients in South Korea: A nationwide cohort study. Diabetes Metab. J. 2020, 44, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Pizano, M.D.; Campa-Mada, A.C.; Carvajal-Millan, E.; Martinez-Robinson, K.G.; Chu, A.R. The underlying mechanisms for severe COVID-19 progression in people with diabetes mellitus: A critical review. AIMS Public Health 2021, 8, 720. [Google Scholar] [CrossRef] [PubMed]
- Altonen, B.L.; Arreglado, T.M.; Leroux, O.; Murray-Ramcharan, M.; Engdahl, R. Characteristics, comorbidities and survival analysis of young adults hospitalized with COVID-19 in New York City. PLoS ONE 2020, 15, e0243343. [Google Scholar] [CrossRef] [PubMed]
- Petakh, P.; Kamyshna, I.; Nykyforuk, A.; Yao, R.; Imbery, J.F.; Oksenych, V.; Korda, M.; Kamyshnyi, A. Immunoregulatory Intestinal Microbiota and COVID-19 in Patients with Type Two Diabetes: A Double-Edged Sword. Viruses 2022, 14, 477. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Ray, A.; Sadasivam, B. Metformin in COVID-19: A possible role beyond diabetes. Diabetes Res. Clin. Pract. 2020, 164, 108183. [Google Scholar] [CrossRef]
- Sun, Z.; Song, Z.G.; Liu, C.; Tan, S.; Lin, S.; Zhu, J.; Dai, F.H.; Gao, J.; She, J.L.; Mei, Z.; et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022, 20, 24. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Delli Muti, N.; Balercia, G.; Ciavattini, A.; Giannubilo, S.R.; Marzioni, D. Preeclampsia and severe acute respiratory syndrome coronavirus 2 infection: A systematic review. J. Hypertens. 2022, 40, 1629–1638. [Google Scholar] [CrossRef]
- Delli Muti, N.; Finocchi, F.; Tossetta, G.; Salvio, G.; Cutini, M.; Marzioni, D.; Balercia, G. Could SARS-CoV-2 infection affect male fertility and sexuality? APMIS 2022, 130, 243–252. [Google Scholar] [CrossRef]
- Dale, L. Neurological Complications of COVID-19: A Review of the Literature. Cureus 2022, 14, e27633. [Google Scholar] [CrossRef]

| Characteristic | HC | T2D |
|---|---|---|
| Age | 56 ± 10.30 | 63 ± 12.25 |
| Sex (F/M) | 10/5 | 9/6 |
| BMI | 24.8 ± 2.25 | 31 ± 4.29 |
| Blood pressure (mmHg): systolic | 110 ± 2.10 | 138.5 ± 2.8 |
| Blood pressure (mmHg): diastolic | 62 ± 1.99 | 88.1 ± 1.3 |
| HbA1c (%) | 5.4 ± 0.19 | 6.5 ± 0.6 |
| HDL (mg/dL) | 65 ± 3.99 | 47 ± 6.55 |
| LDL (mg/dL) | 97 ± 15.56 | 118 ± 27.67 |
| TG (mg/dL) | 88 ± 14.27 | 132 ± 48.47 |
| Statin (number/total) | 2/15 | 3/15 |
| Metformin (number/total) | n/a | 15/15 |
| DPP4 inhibitors | n/a | 2/15 |
| Insulin | n/a | 0/15 |
| Target | Sequence |
|---|---|
| Penicillium spp. | ATTGGAGGGCAAGTCTGGTG |
| AATCCCGTCCGATCCCTAGT | |
| RNAr 18S | ATTGGAGGGCAAGTCTGGTG |
| CCGATCCCTAGTCGGCATAG | |
| Debaryomyces spp. | TAACGGGAACAATGGAGGGC |
| CAACACCCGATCCCTAGTCG | |
| Candida spp. | TTTATCAACTTGTCACACCAGA |
| ATCCCGCCTTACCACTACCG | |
| Aspergillus spp. | GTGGAGTGATTTGTCTGCTTAATTG |
| TCTAAGGGCATCACAGACCTGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gradisteanu Pircalabioru, G.; Grigore, G.A.; Czobor Barbu, I.; Chifiriuc, M.-C.; Savu, O. Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients. Biomedicines 2023, 11, 179. https://doi.org/10.3390/biomedicines11010179
Gradisteanu Pircalabioru G, Grigore GA, Czobor Barbu I, Chifiriuc M-C, Savu O. Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients. Biomedicines. 2023; 11(1):179. https://doi.org/10.3390/biomedicines11010179
Chicago/Turabian StyleGradisteanu Pircalabioru, Gratiela, Georgiana Alexandra Grigore, Ilda Czobor Barbu, Mariana-Carmen Chifiriuc, and Octavian Savu. 2023. "Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients" Biomedicines 11, no. 1: 179. https://doi.org/10.3390/biomedicines11010179
APA StyleGradisteanu Pircalabioru, G., Grigore, G. A., Czobor Barbu, I., Chifiriuc, M.-C., & Savu, O. (2023). Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients. Biomedicines, 11(1), 179. https://doi.org/10.3390/biomedicines11010179








